Genotype X Environment Response of ‘Matooke’ Hybrids (Naritas) to Pseudocercospora fijiensis, the Cause of Black Sigatoka in Banana
Abstract
1. Introduction
2. Materials and Methods
2.1. Planting Materials
2.2. Field Trials
2.3. Black Sigatoka Confirmation
2.4. Disease Evaluations
2.5. Data Analysis
- YLS—rank of the youngest leaf counting from bottom bearing at least 10 necrotic lesions with a dry center,
- NSL—the number of leaves starting from the youngest,
- INSL—the proportion of standing leaves without the typical black Sigatoka late-stage necrotic lesions.
- n = number of leaves in each disease severity group, b = % severity group; N = number of severity groups used in the scale (7); T = total number of leaves scored.
3. Results
3.1. Test Locations Characteristics
3.2. Black Sigatoka Confirmation
3.3. Black Sigatoka Severity Evaluation Parameters
3.4. Genotype Response at Each Crop Cycle
3.5. Genotype Response to Black Sigatoka at Each Environment
3.6. Genotype Response between Countries
3.7. Multi-Location Analysis of NARITA Hybrids
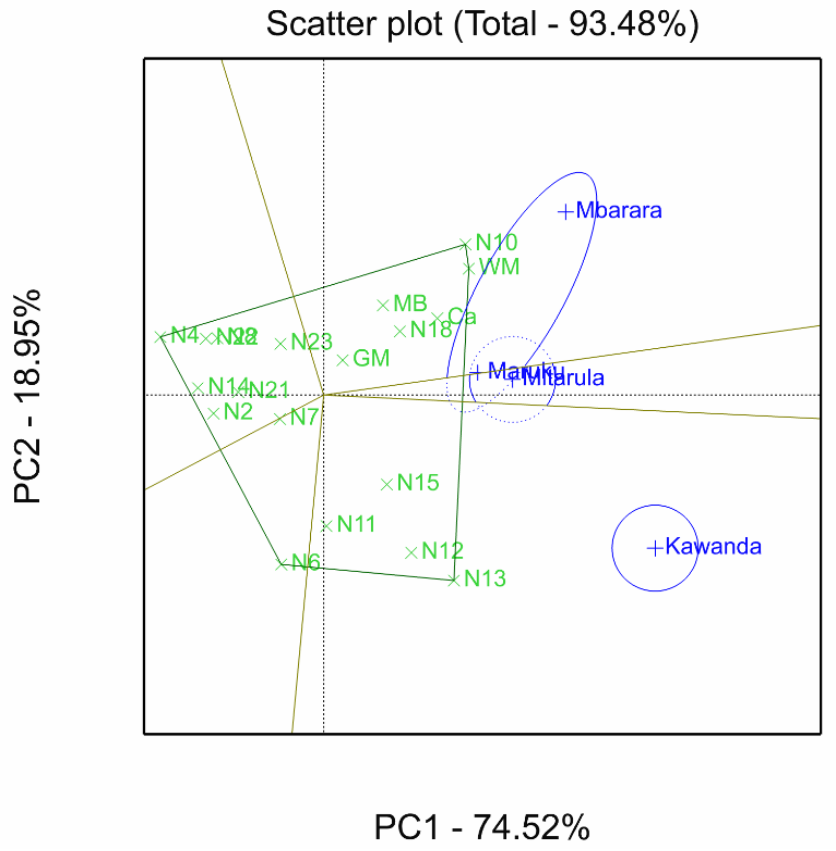
3.8. Discrimination Power and Representativeness of Test Environments
3.9. Influence of Weather Variables on Disease Severity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faostat, F. Agriculture Organization of the United Nations Statistics Division. In Economic and Social Development Department; FAOSTAT: Rome, Italy, 2016; Available online: http://faostat3.fao.org/home/E (accessed on 31 December 2016).
- Gambart, C.; Swennen, R.; Blomme, G.; Groot, J.C.; Remans, R.; Ocimati, W. Impact and opportunities of agroecological intensification strategies on farm performance: A case study of banana-based systems in central and south-western Uganda. Front. Sustain. Food Syst. 2020, 4. [Google Scholar] [CrossRef]
- Karamura, D.; Karamura, E.; Tinzaara, W. Banana Cultivar Names, Synonyms and Their Usage in Eastern Africa; Bioversity International: Kampala, Uganda, 2012. [Google Scholar]
- Karamura, E.; Frison, E.; Karamura, D.; Sharrock, S. Banana production systems in eastern and southern Africa. In Bananas and Food Security; INIBAP: Montpellier, France, 1998; pp. 401–412. [Google Scholar]
- Christelová, P.; De Langhe, E.; Hřibová, E.; Čížková, J.; Sardos, J.; Hušáková, M.; Sutanto, A.; Kepler, A.K.; Swennen, R.; Roux, N.; et al. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 2017, 26, 801–824. [Google Scholar] [CrossRef]
- Kitavi, M.; Cashell, R.; Ferguson, M.; Lorenzen, J.; Nyine, M.; McKeown, P.; Spillane, C. Heritable epigenetic diversity for conservation and utilization of epigenetic germplasm resources of clonal East African Highland banana (EAHB) accessions. Theor. Appl. Genet. 2020, 133, 2605–2625. [Google Scholar] [CrossRef] [PubMed]
- Kitavi, M.; Downing, T.; Lorenzen, J.; Karamura, D.; Onyango, M.; Nyine, M.; Ferguson, M.; Spillane, C. The triploid East African Highland Banana (EAHB) genepool is genetically uniform arising from a single ancestral clone that underwent population expansion by vegetative propagation. Theor. Appl. Genet. 2016, 129, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jenny, C.; Bakry, F.; Karamura, D.; Kitavi, M.; Dubois, C.; Hervouet, C.; Philippson, G.; De Langhe, E. East African diploid and triploid bananas: A genetic complex transported from South-East Asia. Ann. Bot. 2019, 123, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Němečková, A.; Christelová, P.; Čížková, J.; Nyine, M.; Svačina, R.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Molecular and cytogenetic study of east African highland banana. Front. Plant Sci. 2018, 9, 1371. [Google Scholar] [CrossRef] [PubMed]
- Tugume, A.; Lubega, G.; Rubaihayo, P. Genetic diversity of East African Highland bananas using AFLP. Infomusa 2002, 11, 28–32. [Google Scholar]
- Brown, A.; Tumuhimbise, R.; Amah, D.; Uwimana, B.; Nyine, M.; Mduma, H.; Talengera, D.; Karamura, D.; Kuriba, J.; Swennen, R. Bananas and plantains (Musa spp.). In Genetic Improvement of Tropical Crops; Springer: Berlin/Heidelberg, Germany, 2017; pp. 219–240. [Google Scholar]
- Alakonya, A.; Kimunye, J.; Mahuku, G.; Amah, D.; Uwimana, B.; Brown, A.; Swennen, R. Progress in understanding Pseudocercospora banana pathogens and the development of resistant Musa germplasm. Plant Pathol. 2018, 67, 759–770. [Google Scholar] [CrossRef]
- Churchill, A.C. Mycosphaerella fijiensis, the black leaf streak pathogen of banana: Progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Mol. Plant Pathol. 2011, 12, 307–328. [Google Scholar] [CrossRef]
- Marin, D.H.; Romero, R.A.; Guzmán, M.; Sutton, T.B. Black Sigatoka: An increasing threat to banana cultivation. Plant Dis. 2003, 87, 208–222. [Google Scholar] [CrossRef]
- Mobambo, K.; Gauhl, F.; Vuylsteke, D.; Ortiz, R.; Pasberg-Gauhl, C.; Swennen, R. Yield loss in plantain from black sigatoka leaf spot and field performance of resistant hybrids. Field Crop. Res. 1993, 35, 35–42. [Google Scholar] [CrossRef]
- Bellaire, L.d.L.; Fouré, E.; Abadie, C.; Carlier, J. Black Leaf Streak Disease is challenging the banana industry. Fruits 2010, 65, 327–342. [Google Scholar] [CrossRef][Green Version]
- Swennen, R.; Vuylsteke, D. Breeding black sigatoka resistant plantains with a wild banana. Trop. Agric. 1993, 70, 74–77. [Google Scholar]
- Tenkouano, A.; Swennen, R. Plantains and banana: Progress in breeding and delivering improved plantain and banana to African farmers. Chron. Hortic. 2004, 44, 9–15. [Google Scholar]
- Tenkouano, A.; Lamien, N.y.; Agogbua, J.; Amah, D.; Swennen, R.; Traoré, S.; Thiemele, D.; Aby, N.; Kobenan, K.; Gnonhouri, G. Promising high-yielding tetraploid plantain-bred hybrids in West Africa. Int. J. Agron. 2019. [Google Scholar] [CrossRef]
- Batte, M.; Swennen, R.; Uwimana, B.; Akech, V.; Brown, A.; Tumuhimbise, R.; Hovmalm, H.P.; Geleta, M.; Ortiz, R. Crossbreeding East African highland bananas: Lessons learnt relevant to the botany of the crop after 21 years of genetic enhancement. Front. Plant Sci. 2019, 10, 81. [Google Scholar] [CrossRef]
- Pillay, M.; Ssebuliba, R.; Hartman, J.; Vuylsteke, D.; Talengera, D.; Tushemereirwe, W. Conventional breeding strategies to enhance the sustainability of Musa biodiversity conservation for endemic cultivars. Afr. Crop Sci. J. 2004, 12, 58–65. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.; Dai, X.; Gong, Q.; Huang, X.; Xiao, W.; Zhao, J.; Huang, X. Non-conventional breeding of banana (Musa spp.). Acta Hortic. 2011, 39–46. [Google Scholar] [CrossRef]
- Swennen, R.; Sharrock, S.; Frison, E. Biotechnology in support of smallholders cultivating bananas in tropics. Proc. Sustain. Agric. New Millenn. Impact Biotechnol. Dev. Ctries. Bruss. Belg. 2000, 28–31. [Google Scholar]
- Kosky, R.G.; Chong-Pérez, B.; López-Torres, J.; Reyes, M.; Bermúdez-Caraballoso, I.; Martín, N.M.; Machado-Rodriguez, J.M.; Portal, O.; Ocaña, B.; Alvarado-Capó, Y.; et al. Plantain (Musa spp. cv.‘Navolean’ AAB) transgenic plants from Agrobacterium tumefaciens-mediated transformation of embryogenic cell suspensions. Biotecnol. Veg. 2010, 10, 209–218. [Google Scholar]
- Kovács, G.; Sági, L.; Jacon, G.; Arinaitwe, G.; Busogoro, J.-P.; Thiry, E.; Strosse, H.; Swennen, R.; Remy, S. Expression of a rice chitinase gene in transgenic banana (‘Gros Michel’, AAA genome group) confers resistance to black leaf streak disease. Transgenic Res. 2013, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Vishnevetsky, J.; White, T.L.; Palmateer, A.J.; Flaishman, M.; Cohen, Y.; Elad, Y.; Velcheva, M.; Hanania, U.; Sahar, N.; Dgani, O. Improved tolerance toward fungal diseases in transgenic Cavendish banana (Musa spp. AAA group) cv. Grand Nain. Transgenic Res. 2011, 20, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N.; Kumar, L.P. Application of CRISPR/Cas for Diagnosis and Management of Viral Diseases of Banana. Front. Microbiol. 2021, 11, 3622. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. CRISPR/Cas9 based genome editing of banana for disease resistance. Curr. Opin. Plant Biol. 2020, 56, 118–126. [Google Scholar] [CrossRef]
- Swennen, R.; Vuylsteke, D. Bananas in Africa: Diversity, uses and prospects for improvement. Crop Genet. Resour. Afr. 1991, 2, 151–160. [Google Scholar]
- Tushemereirwe, W.; Batte, M.; Nyine, M.; Tumuhimbise, R.; Barekye, A.; Tendo, S.; Kubiriba, J.; Lorenzen, J.; Swennen, R. Performance of Narita Banana Hybrids in the Preliminary Yield Trial, Uganda; Report NARO; IITA-CGIAR: Kampala, Uganda, 2015. [Google Scholar]
- Batte, M.; Nyine, M.; Uwimana, B.; Swennen, R.; Akech, V.; Brown, A.; Hovmalm, H.P.; Geleta, M.; Ortiz, R. Significant progressive heterobeltiosis in banana crossbreeding. BMC Plant Biol. 2020, 20, 489. [Google Scholar] [CrossRef]
- Jones, D.R. Diseases of Banana, Abaca and Enset; CABI Publishing: Wallingford, UK, 2000. [Google Scholar]
- Craenen, K.; Ortiz, R. Effect of the bs1 gene in plantain-banana hybrids on response to black sigatoka. Theor. Appl. Genet. 1997, 95, 497–505. [Google Scholar] [CrossRef]
- Fullerton, R.; Olsen, T. Pathogenic variability in Mycosphaerella fijiensis Morelet, cause of black Sigatoka in banana and plantain. N. Z. J. Crop Hortic. Sci. 1995, 23, 39–48. [Google Scholar] [CrossRef][Green Version]
- Ortiz, R. Plant Breeding in the Omics Era; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Kimunye, J.; Were, E.; Mussa, F.; Tazuba, A.; Jomanga, K.; Viljoen, A.; Swennen, R.; Muthoni, F.; Mahuku, G. Distribution of Pseudocercospora species causing Sigatoka leaf diseases of banana in Uganda and Tanzania. Plant Pathol. 2019, 69, 50–59. [Google Scholar] [CrossRef]
- Gauhl, F. Epidemiology and Ecology of Black Sigatoka (Mycosphaerella Fijiensis Morelet) on Plantain and Banana (Musa spp.) in Costa Rica, Central America. Ph.D Thesis, Göttingen University, Göttingen, Germany, 1994. [Google Scholar]
- Ortiz, R.; Vuylsteke, D.; Ferris, R.; Okoro, J.; N’Guessan, A.; Hemeng, O.; Yeboah, D.; Afreh-Nuamah, K.; Ahiekpor, E.; Foure, E. Developing new plantain varieties for Africa. Plant Var. Seeds 1997, 10, 39–57. [Google Scholar]
- Orjeda, G. Evaluation of Musa Germplasm for Resistance to Sigatoka Diseases and Fusarium Wilt. INIBAP Technical Guidelines 3; International Plant Genetic Resources Institute: Rome, Italy; International Network for the Improvement of Banana and Plantain: Montpellier, France, 1998; p. 62. [Google Scholar]
- Shaner, G.; Finney, R. The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology 1977, 67, 1051–1056. [Google Scholar] [CrossRef]
- Yan, W.; Falk, D.E. Biplot analysis of host-by-pathogen data. Plant Dis. 2002, 86, 1396–1401. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Yan, W.; Hunt, L.; Sheng, Q.; Szlavnics, Z. Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Parihar, A.; Basandrai, A.K.; Saxena, D.; Kushwaha, K.; Chandra, S.; Sharma, K.; Singha, K.; Singh, D.; Lal, H.; Gupta, S. Biplot evaluation of test environments and identification of lentil genotypes with durable resistance to fusarium wilt in India. Crop Pasture Sci. 2017, 68, 1024–1030. [Google Scholar] [CrossRef]
- Akankwasa, K.; Marimo, P.; Tumuhimbise, R.; Asasira, M.; Khakasa, E.; Mpirirwe, I.; Kleih, U.; Forsythe, L.; Fliedel, G.; Dufour, D. The East African highland cooking bananas ‘Matooke’ preferences of farmers and traders: Implications for variety development. Int. J. Food Sci. Technol. 2020. [Google Scholar] [CrossRef]
- Nowankunda, K.; Barekye, A.; Ssali, R.T.; Namaganda, J.; Tushemereirwe, W.K.; Nabulya, G.; Erima, R.; Akankwasa, K.; Hilman, E.; Batte, M. ‘Kiwangaazi’ (syn ‘KABANA 6H’) Black Sigatoka Nematode and Banana Weevil Tolerant ‘Matooke’ Hybrid Banana Released in Uganda. HortScience 2015, 50, 621–623. [Google Scholar] [CrossRef]
- Escobar-Tovar, L.; Guzmán-Quesada, M.; Sandoval-Fernández, J.A.; Gómez-Lim, M.A. Comparative analysis of the in vitro and in planta secretomes from Mycosphaerella fijiensis isolates. Fungal Biol. 2015, 119, 447–470. [Google Scholar] [CrossRef]
- Tumuhimbise, R.; Buregyeya, H.; Barekye, A.; Ssali, R.T.; Talengera, D.; Kubiriba, J.; Muhangi, S.; Namagembe, B.; Namanya, P.; Arinaitwe, G.; et al. Selection of cooking banana genotypes for yield and black Sigatoka resistance in different locations in Uganda. J. Plant Breed. Crop Sci. 2016, 8, 60–71. [Google Scholar]
- Jacome, L.; Schuh, W.; Stevenson, R. Effect of temperature and relative humidity on germination and germ tube development of Mycosphaerella fijiensis var. difformis. Phytopathology 1991, 81, 1480–1485. [Google Scholar] [CrossRef]
- Khan, M.; Hossain, I.; Ahmad, M. Impact of Weather on Sigatoka Leaf Spot of Banana (Musa spp. L.) and its Ecofriendly Management. Agronomy 2015, 13, 44–53. [Google Scholar] [CrossRef]
- Jacome, L.H.; Schuh, W. Effects of leaf wetness duration and temperature on development of Black Sigatoka disease on banana infected by Mycosphaerella fijiensis var. difformis. Phytopathology 1992, 82, 515–520. [Google Scholar] [CrossRef]
- Bananuka, J.; Rubaihayo, P. Backyard banana cultivation in Uganda. Infomusa 1994, 3, 17. [Google Scholar]
- Kablan, L.; Lagauche, A.; Delvaux, B.; Legrve, A. Silicon reduces black sigatoka development in banana. Plant Dis. 2012, 96, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Oluma, H.; Onyezili, F. Effect of cultural practices on the severity of black sigatoka leaf spot diseases of plantain banana in Nigerian savanna. MusAfrica 1995, 8, 10–11. [Google Scholar]
- Milgroom, M.G. Recombination and the multilocus structure of fungal populations. Annu. Rev. Phytopathol. 1996, 34, 457–477. [Google Scholar] [CrossRef] [PubMed]
- Kimunye, J.N.; Muzhinji, N.; Mostert, D.; Viljoen, A.; van der Merwe, A.E.; Mahuku, G. Genetic Diversity and Mating Type Distribution of Pseudocercospora fijiensis on Banana in Uganda and Tanzania. Phytopathology 2020. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef]
- Barekye, A.; Tongoona, P.; Derera, J.; Laing, M.; Tushemereirwe, W. Appraisal of methods for assessing black Sigatoka resistance in diploid banana populations. Afr. J. Plant Sci. 2011, 5, 900–908. [Google Scholar] [CrossRef]
- Carlier, J.; De Waele, D.; Escalant, J.-V. Global Evaluation of Musa Germplasm for Resistance to Fusarium Wilt, Mycosphaerella Leaf Spot Diseases and Nematodes: In-dept Evaluation; Vezina, A., Picq, C., Eds.; INIBAP Technical Guidelines 7; The International Network for the Improvement of Banana and Plantain: Montpellier, France, 2003; p. 62. [Google Scholar]
- Craenen, K.; Ortiz, R. Influence of black Sigatoka disease on the growth and yield of diploid and tetraploid hybrid plantains. Crop Prot. 1998, 17, 13–18. [Google Scholar] [CrossRef]




| Country | Location | Grid reference (Decimal Degrees) Longitude Latitude | Altitude (masl) | Rainfall (mm) | Average Temperatures °C | pH | OM (%) | P (ppm) | Ca (ppm) | Mg (ppm) | K (ppm) | Soil Type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uganda | Kawanda | 0.414833 | 32.53238 | 1196 | 900–1500 | >22 | 5.1 | 2.7 | 9.5 | 2516.9 | 611.0 | 341.4 | Sandy clay |
| Mbarara | −0.60032 | 30.59843 | 1412 | 900–1500 | >22 | 4.7 | 1.8 | 11.0 | 1757.7 | 668.4 | 456.8 | Sandy loam | |
| Tanzania | Maruku | −1.42446 | 31.77358 | 1364 | 1500–2500 | >22 | 5.3 | 3.0 | 18.0 | 1262.9 | 291.2 | 964.8 | Clay loam |
| Mitarula | −9.39769 | 33.62753 | 1055 | >900 | >22 | 4.6 | 7.1 | 4.2 | 361.3 | 47.1 | 176.8 | Sandy loam | |
| Disease Parameter | DSI a | AUDPC b | INSL c | YLS d | R2 (Coefficient of Determination) |
|---|---|---|---|---|---|
| DSI | 1 | 0.86 | |||
| AUDPC | 0.83 * | 1 | 0.87 | ||
| INSL | −0.85 * | −0.66 * | 1 | 0.84 | |
| YLS | −0.57 * | −0.49 | 0.60 * | 1 | 0.78 |
| Area Under Disease Progress Curve (AUDPC) * | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kawanda | Mbarara | Maruku | Mitarula | |||||||||
| Genotype | Cycle 1 | Cycle 2 | Cycle 3 | Cycle 1 | Cycle 2 | Cycle 3 | Cycle 1 | Cycle 2 | Cycle 3 | Cycle 1 | Cycle 2 | Cycle 3 |
| NARITA 2 | 175.8 a | 197.6 a | 175.8 a | 23.4 a | 140.4 b | 91.7 b | 94.1 b | 93.4 b | 34.9 a | 80.4 a | 62.3 a | 66.1 a |
| NARITA 4 | 101.3 a | 126.7 a | 133.1 a | 33.7 a | 136.1 b | 96.7 b | 78.8 a | 131.9 b | 51.4 a | 59.2 a | 65.0 a | 71.3 a |
| NARITA 6 | 219.8 a | 279.0 b | 201.6 a | 62.4 a | 93.7 a | 78.0 a | 149.7 b | 154.9 b | 87.2 a | 72.3 a | 130.1 b | 78.6 a |
| NARITA 7 | 142.2 a | 240.8 b | 205.8 b | 41.2 a | 166.2 b | 152.0 b | 80.7 a | 165.8 b | 49.9 a | 84.8 a | 115.8 ab | 102.5 a |
| NARITA 8 | 172.6 a | 164.9 a | 146.4 a | 18.4 a | 161.4 c | 84.4 b | 78.8 ab | 104.15 b | 45.7 a | 71.4 a | 95.4 a | 77.1 a |
| NARITA 10 | 229.1 a | 313.2 b | 258.8 a | 58.8 a | 330.5 c | 208.8 b | 74.7 a | 129.5 b | 39.4 a | 79.4 a | 202.3 c | 152.1 b |
| NARITA 11 | 187.6 a | 311.2 c | 249.2 b | 23.9 a | 147.8 c | 90.6 b | 107.1 b | 133.0 b | 57.7 a | 96.4 a | 122.6 ab | 133.7 b |
| NARITA 12 | 220.1 a | 384.8 c | 293.4 b | 35.3 a | 185.5 b | 175.2 b | 34.5 ab | 70.48 b | 27.0 a | 67.5 a | 147.9 b | 64.3 a |
| NARITA 13 | 152.0 a | 419.0 c | 298.1 b | 44.4 a | 190.7 c | 96.5 b | 68.8 a | 144.6 b | 53.5 a | 113.1 a | 190.9 c | 126.8 b |
| NARITA 14 | 139.6 a | 170.3 a | 162.1 a | 24.6 a | 133.9 b | 101.6 b | 106.4 b | 165.7 c | 65.2 a | 65.2 a | 76.3 a | 79.7 a |
| NARITA 15 | 312.1 b | 333.3 b | 206.9 a | 43.8 a | 187.0 b | 176.1 b | 35.7 a | 68.8 a | 35.0 a | 112.4 a | 163.3 b | 107.7 a |
| NARITA 18 | 259.5 b | 285.7 b | 188.4 a | 63.2 a | 248.9 b | 105.5 a | 33.1 a | 79.5 b | 38.4 a | 79.8 a | 179 b | 161.5 b |
| NARITA 21 | 167.1 a | 199.3 a | 187.7 a | 30.2 a | 152.1 b | 137.7 b | 102.2 ab | 70.9 a | 42.4 a | 38.7 a | 100.4 a | 94.1 a |
| NARITA 22 | 142.9 a | 157.3 a | 178.6 a | 27.2 a | 155.9 b | 113.3 b | 59.4 a | 100.2 b | 39.3 a | 42.4 a | 89.1 ab | 116.2 b |
| NARITA 23 | 197.7 a | 210.6 a | 165.8 a | 21.3 a | 190.2 b | 148.8 b | 50.5 a | 50.6 a | 26.3 a | 39.1 a | 125.2 b | 61.4 a |
| Mbwazirume | 189.6 a | 266.7 b | 259.4 b | 79.4 a | 253.1 c | 150.8 b | 43.0 a | 77.3 a | 46.9 a | 86.8 a | 175.8 b | 150.6 b |
| Williams | 250.1 a | 320.6 b | 303.8 b | 73.0 a | 317.8 c | 157.8 b | 138.3 b | 184.9 c | 93.4 a | 82.7 a | 200.6 b | 180.7 b |
| Cachaco | 244.7 a | 349.2 b | 286.7 a | 59.8 a | 324.1 c | 222.6 b | 59.5 a | 105.8 b | 36.9 a | 72.5 a | 132 b | 121.5 b |
| Gros Michel | 174.7 a | 244.8 b | 253.5 b | 52.4 a | 197.7 b | 151.0 b | 54.4 ab | 84.2 b | 34.2 a | 81.4 a | 195.7 b | 156.7 b |
| Average | 193.6 a | 261.8 c | 218.7 b | 43.0 a | 195.4 c | 133.6 b | 75.8 b | 111.7 c | 49.6 a | 81.1 a | 135.2 c | 110.7 b |
| Genotype | Kawanda * | Mbarara * | Maruku * | Mitarula * | Uganda * | Tanzania * |
|---|---|---|---|---|---|---|
| NARITA 2 | 197.6 b–e (6) | 140.4 a–d (6) | 68.8 a–c (3) | 62.3 a (1) | 169.0 a–d (5) | 65.4 ab (2) |
| NARITA 4 | 126.8 a (1) | 136.1 a–c (4) | 50.6 a (1) | 65.0 ab (2) | 132.0 a (1) | 58.1 a (1) |
| NARITA 6 | 279.0 g–k (13) | 93.7 a (1) | 105.8 d–h (13) | 130.1 e–I (13) | 242.0 c–f (10) | 117.4 e–I (15) |
| NARITA 7 | 240.8 b–h (9) | 166.2 b–h (11) | 84.1 b–f (9) | 115.8 c–f (10) | 203.5 c–e (9) | 100 d–h (10) |
| NARITA 8 | 164.9 a–c (3) | 161.4 b–h (10) | 77.2 a–e (7) | 95.4 a–e (8) | 163.5 a–d (4) | 86.3 b–f (7) |
| NARITA 10 | 314.0 j–m (18) | 330.5 k (22) | 165.8 j–m (23) | 202.3 kl (24) | 315.1 f–m (21) | 190.1 m–o (23) |
| NARITA 11 | 311.4 i–m (17) | 147.8 a–e (7) | 104.2 c–h (12) | 122.6 d–g (11) | 275.0 f–l (16) | 116.1 e–I (12) |
| NARITA 12 | 384.9 no (22) | 185.5 e–h (12) | 129.5 g–j (16) | 147.9 f–j (17) | 285.2 f–l (17) | 137.9 i–k (17) |
| NARITA 13 | 419.0 o (23) | 190.7 e–h (13) | 133.0 h–k (18) | 190.9 jk (21) | 293.2 f–m (18) | 163.1 j–m (19) |
| NARITA 14 | 170.4 a–c (4) | 133.9 a–c (3) | 70.5 a–c (4) | 76.3 ab (3) | 152.0 ab (2) | 73.3 a–c (4) |
| NARITA 15 | 333.3 j–n (20) | 187.0 e–h (14) | 144.6 i–l (19) | 163.3 g–j (18) | 268.7 f–k (15) | 153.1 j–l (18) |
| NARITA 16 | 296.2 h–m (15) | 137.9 a–d (5) | _ | 124.7 d–h (12) | 296.2 f–m (19) | 125.4 c–j (16) |
| NARITA 17 | 245.1 b–j (11) | _ | _ | _ | 245.1 c–h (12) | _ |
| NARITA 18 | 284.4 h–l (14) | 248.9 gi (17) | 165.7 j–m (22) | 179.0 h–k (20) | 262.2 f–I (13) | 172.4 j–o (21) |
| NARITA 19 | _ | _ | 153.6 i–l (20) | 232.1 l (25) | _ | 198.1 o (25) |
| NARITA 20 | _ | _ | 109.6 d–h (14) | 127.2 e–I (14) | _ | 117.2 e–I (14) |
| NARITA 21 | 197.2 b–f (7) | 152.1 a–f (8) | 79.4 a–e (8) | 100.4 b–e (9) | 169.9 b–d (6) | 92 c–g (8) |
| NARITA 22 | 157.3 ab (2) | 155.9 b–g (9) | 71 a–d (5) | 89.1 a–e (7) | 156.9 a–c (3) | 83.1 a–e (6) |
| NARITA 23 | 210.7 b–g (8) | 190.2 e–h (15) | 100.2 c–g (11) | 125.2 e–I (15) | 200.4 c–e (8) | 112.7 e–I (11) |
| NARITA 24 | 297.7 i–m (16) | 295.3 jk (19) | 297.7 f–m (20) | _ | ||
| NARITA 25 | _ | _ | 73.8 a–d (6) | 78.4 a–c (4) | _ | 74.8 a–d (5) |
| NARITA 26 | _ | _ | 60.5 ab (2) | 84.2 a–d (5) | _ | 71.5 a–c (30 |
| NARITA 27 | _ | _ | 120.5 g–I (15) | 86.7 a–d (6) | _ | 97.5 c–h (9) |
| Mbwazirume | 266.7 g–j (12) | 253.1 g–j (18) | 154.9 i–m (21) | 175.8 h–k (19) | 266.7 f–j (14) | 166.5 j–n (20) |
| Williams | 320.6 j–m (19) | 317.8 k (20) | 184.9 m (24) | 200.6 j–l (23) | 319.2 h–m (22) | 192.1 no (24) |
| Pisang Ceylan | 192.0 b–d (5) | 128.3 ab (2) | 192.0 b–e (7) | _ | ||
| Cachaco | 349.2 k–o (21) | 324.1 k (21) | 93.4 c–g (10) | 132.0 e–I (16) | 349.2 m (23) | 116.6 e–I (13) |
| Gros Michel | 244.8 b–j (10) | 197.7 e–h (16) | 131.9 g–j (17) | 195.7 j–l (22) | 244.8 e–g (11) | 176.9 j–o (22) |
| Source of Variation | Degrees of Freedom | Sum of Squares | Mean Sum of SQUARES | % Variance Explained |
|---|---|---|---|---|
| Treatments | 75 | 2,277,733 | 30,370 * | - |
| Genotypes | 18 | 854,825 | 47,490 * | 37.5 |
| Environments | 3 | 890,810 | 296,937 * | 39.1 |
| Block | 12 | 61,113 | 5093 ns | - |
| Interactions | 54 | 532,098 | 9854 * | 23.4 |
| IPCA 1 | 20 | 263,413 | 13,171 * | 49.5 |
| IPCA 2 | 18 | 226,642 | 12,591 * | 42.6 |
| Residuals | 16 | 42,044 | 2628 ns | - |
| Error | 216 | 819,161 | 3792 | - |
| Total | 303 | 3,158,007 | 10,422 | - |
| Monthly Rainfall (mm) * | Relative Humidity (RH %) * | Minimum Temperature (Tmin °C) * | Maximum Temperature (Tmax °C) * | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cycle 1 | Cycle 2 | Cycle 3 | Combined | Cycle 1 | Cycle 2 | Cycle 3 | Combined | Cycle 1 | Cycle 2 | Cycle 3 | Combined | Cycle 1 | Cycle 2 | Cycle 3 | Combined | |
| Kawanda | 43.1 a | 56.6 ab | 88.0 b | 60.6 ab | 68.4 ab | 68.8 ab | 68.7 ab | 68.2 ab | 17.6 cd | 18.4 d | 18.6 de | 18.1 d | 26.2 cd | 26.3 cd | 26.6 cd | 26.4 cd |
| Mbarara | 65.3 ab | 77.0 ab | 97.2 b | 78.4 ab | 68.3 ab | 67.2 ab | 68.9 ab | 67.7 ab | 15.8 c | 16.5 c | 16.2 c | 16.1 c | 25.6 bc | 26.0 cd | 25.9 d | 25.8 bc |
| Maruku | 40.1 a | 57.6 ab | 79.6 ab | 57.2 ab | 72.2 b | 66.0 a | 67.3 ab | 69.9 ab | 19.2 de | 20.8 f | 20.5 f | 20.1 f | 23.2 ab | 25.3 bc | 25.3 b | 24.5 b |
| Mitarula | 53.8 ab | 68.4 ab | 81.4 ab | 66.5 ab | 73.1 b | 71.1 ab | 72.4 ab | 72.9 b | 12.8 a | 13.5 a | 13.9 ab | 13.4 a | 23.0 a | 24.4 b | 24.0 a | 23.6 ab |
| Source | Coefficient Value | Standard Error | T (Standardized Coefficients) | p Value | Coefficient of Determination (R2) |
|---|---|---|---|---|---|
| Intercept | −2676.347 | 730.904 | −3.662 | 0.000 | 0.457 |
| Rainfall | −0.038 | 0.024 | −1.555 | 0.121 | |
| RH | 19.139 | 6.241 | 3.067 | 0.002 | |
| Tmin | −7.982 | 3.873 | −2.061 | 0.040 | |
| Tmax | 66.975 | 10.343 | 6.476 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kimunye, J.; Jomanga, K.; Tazuba, A.F.; Were, E.; Viljoen, A.; Swennen, R.; Mahuku, G. Genotype X Environment Response of ‘Matooke’ Hybrids (Naritas) to Pseudocercospora fijiensis, the Cause of Black Sigatoka in Banana. Agronomy 2021, 11, 1145. https://doi.org/10.3390/agronomy11061145
Kimunye J, Jomanga K, Tazuba AF, Were E, Viljoen A, Swennen R, Mahuku G. Genotype X Environment Response of ‘Matooke’ Hybrids (Naritas) to Pseudocercospora fijiensis, the Cause of Black Sigatoka in Banana. Agronomy. 2021; 11(6):1145. https://doi.org/10.3390/agronomy11061145
Chicago/Turabian StyleKimunye, Janet, Kennedy Jomanga, Anthony Fredrick Tazuba, Evans Were, Altus Viljoen, Rony Swennen, and George Mahuku. 2021. "Genotype X Environment Response of ‘Matooke’ Hybrids (Naritas) to Pseudocercospora fijiensis, the Cause of Black Sigatoka in Banana" Agronomy 11, no. 6: 1145. https://doi.org/10.3390/agronomy11061145
APA StyleKimunye, J., Jomanga, K., Tazuba, A. F., Were, E., Viljoen, A., Swennen, R., & Mahuku, G. (2021). Genotype X Environment Response of ‘Matooke’ Hybrids (Naritas) to Pseudocercospora fijiensis, the Cause of Black Sigatoka in Banana. Agronomy, 11(6), 1145. https://doi.org/10.3390/agronomy11061145






