Abstract
Use of compost is a common agricultural practice. It improves soil fertility by adding nutrients and plant growth promoting (PGP) microorganisms. The role of bacterial-fungal interactions for compost-driven fertilization, however, is still poorly understood. In this study, we investigated whether putative PGP bacteria associate to and disperse along mycelia of fungal isolates. A ‘Fungal highway column system’ was used to isolate and characterize fungal—bacterial couples derived from commercial compost (C), non-composted bulk soil (BS) and rhizosphere soil with compost application (RSC). Bacterial-fungal couples were identified by 16S and 18S rRNA gene sequencing and isolated bacteria were tested for representative PGP traits. Couples of fungi and associated migrator bacteria were isolated from C and RSC only. They included the fungal genera Aspergillus, Mucor, Ulocladium, Rhizopus and Syncephalastrum, and the bacterial genera Rhodococcus, Bacillus, Pseudomonas, Agrobacterium, Glutamicibacter and Microbacterium. Many of migrator bacteria in RSC and C showed PGP traits (e.g., tryptophane—induced auxin synthesis or phytate mineralizing activity) suggesting that fungi contained in C and RSC allow for dispersal of putative PGP bacteria. Next to being provider of nutrients, compost may therefore be source for PGP bacteria and fungal mycelia serving as networks for their efficient dispersal.
1. Introduction
Compost is considered as an inexpensive and eco-friendly organic fertilizer. Applied to soil it helps to reduce the quantity of chemical fertilizers, to improve soil structure, and to counteract soil erosion and degradation []. In agriculture, there is further an increasing need for sustainable ecofriendly soil fertilization strategies. Efforts should be made to develop new bio-technological processes based on the co-inoculation of bacterial-fungal consortia with weathering abilities to enable the efficient utilization of natural phosphate rocks as alternative to phosphate fertilizers. In this regard, there is a growing interest in certified high-quality compost biofertilizers. Compost is a source of a large number and diversity of beneficial microorganisms, both bacteria and fungi that can be used as inoculum in agroecosystems []. Thus, to know the structure of microbial community as well as the functional capacity of bacterial-fungal interactions (BFI) contained in compost is pivotal if compost shall be applied as an inoculum in agricultural soils, degraded natural ecosystems or for the bioremediaton of contaminated soils []. Up to date a considerable diversity of bacterial species have been isolated from compost. Compost bacteria can act as biocontrol of phytopathogens [,], establish symbioses with different fungal genera or form beneficial associations with soil microorganisms [,]. As example, bacterial genera Rhizobium and Rhodococcus are often found in symbiosis with fungal genus Piriformospora []. Although numerous studies on compost bacteria and fungi have been published, there are only few studies of direct BFI in compost.
BFI are widespread in nature [] including parasitism, competence, symbiosis, mutualism and commensalism in soil [,,]. Knowledge on and targeted management of fungal interactions with beneficial plant-growth promoting (PGP) bacteria hence can be key to promote soil health, fertility, and high crop yields []. If compost shall be used as source microorganisms it is needed that its microbiome can effectively establish and disperse in the soil to where it is applied. Unlike fungi, PGP bacteria do not spread well in air-filled soil, as their dispersal in the soil depends on waterborne transport and/or the presence of continuous water films []. In contrast, the fractal network of fungal mycelia can easily explore the soil space and grow in both water-and air-filled spaces [] because of their ability to overcome air-water interfaces by hydrophobin secretion []. In the mycosphere (i.e., the area surrounded and affected by mycelia) [] and even inside of fungal hyphae, bacteria may find suitable ecological niches in terms of pH, nutrient and water content, and even a possible mean for dispersion in soil [,,].
Bacterial dispersal along mycelia of filamentous fungi (also termed as ‘fungal highways’ [] has received increasing attention, e.g., for the clean-up of contaminated soil [,], the functioning of the oxalate-carbonate pathway [] or in cheese production []. Mycelia thereby constitute a fractal network allowing for bacterial dispersal and increased accessibility of nutrient sources [,]. While recent work describes the effect of arbuscular mycorrhizal fungal effects on bacterial transport and organic phosphorus mineralization of organic phosphorus [] or the colonization of legume roots by a symbiotic nitrogen-fixing bacterium [], effects of compost derived fungi on spread of PGP microorganisms remains unknown. In this study, we investigated whether putative PGP bacteria associate to and disperse along mycelia of fungal isolates derived from compost.
2. Material and Methods
2.1. Compost (C)
Commercial compost (Biofert®; Rosario Co.; Santiago de Chile, Chile) was produced from agro-industrial wastes, mainly consisting of residues from horticulture, food industry and municipal pruning in compliance with Chilean environmental and agricultural regulations (NCh 2880:2015; ISO 9001:2008). The composting process is carried out by pile tumbling technique, which takes approximately one year, until the compost reaches the required grade of stability and maturity. Samples of approximately 500 g were aseptically taken from 12 recently packed Bio-fert® bags. Then, the samples were combined forming 4 composite samples containing material from 3 bags; all the composite samples were transported on ice and stored at (4 °C) prior to treatment. The composite samples were processed in the 3 and 5 days following the sampling.
2.2. Rhizosphere Soil with Compost Application (RSC)
Soil samples were taken from the rhizosphere of a grape plants (Vitis vinifera L.) with a historical Biofert® compost application (>seven years), located in San Esteban, Valparaiso Region, in central Chile (32°48′4.49″ S, 70°34′54.2″ W). Samples of approximately 500 g were aseptically taken from 12 different grape plants (planting distance 3.5 × 2.5 m); samples were then combined to 4 composite samples containing material from 3 plants. As control, composite bulk soil (BS) samples were also taken from grapes crop in area without historical Biofert® compost application. The soil samples were collected between 20 and 40 cm of depth, always looking for the greatest presence of root system of the grape plant. All the composite samples were transported on ice and stored at (4 °C) prior to treatment. The samples were processed in the 3 and 5 days following the sampling.
2.3. Chemical Properties Analysis of Compost and Soil Samples
As reference, the chemical properties was measured in representative composite samples (C, RSC and BS). The pH was measured in a suspension of 1:2.5 (w/w) soil and deionized water suspensions. Available phosphorus (POlsen) was extracted using the bicarbonate method and analyzed using the molybdate-blue method []. Exchangeable cations (K, Ca, Mg, and Na) were extracted with 1 M ammonium acetate at pH 7.0 and analyzed using flame atomic adsorption spectrophotometry (FAAS) [] and exchangeable aluminum was extracted with 1 M KCl and also analyzed by FAAS []. In addition, inorganic nitrogen (N) extracted with 2 M KCl and NO3-N was determined by the Devarda alloy distillation method [] and organic matter (OM) contents were estimated by wet digestion [].
2.4. DNA—Based Profiles of Bacterial Community in Compost and Soil Samples
The profile of total bacterial community was addressed by denaturing gradient gel electrophoresis (DGGE) using bacterial 16S rRNA and fungal 18S rRNA as gene targets for bacteria and fungi, respectively. Total DNA was extracted from 0.5 g of C, RSC and BS samples (in quadruplicate) by using Soil DNA Isolation Kit (Qiagen, Hilden, USA) according to manufacturer instructions. For bacterial community, 16S rRNA genes (regions V6–V8) were amplified by touchdown PCR with primer set EUBf933-GC (5′-GCA CAA GCG GTG GAG CAT GTG GCG CCC GCC GCG CGC GGC GGG CGG GGC GGG GGC ACG GGG G-3′) and EUBr1387 (5′-GCC CGG GAA CGT ATT CAC CG-3′) []. For fungal community, 18S rRNA genes (regions 18S rDNA 400–600 bp) were amplified by nested PCR, initially with primer set EF4 (5′-GTA AAA GTC CTG GTT CCC C-3′) and Fun5 (5′-GGA AGG GRT GTA TTT ATT AG-3′), followed by a second PCR with primer set EF4 and NS2-GC (5′-GGC TGC TGG CAC CAG ACT TGC CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG G-3′) [].
DGGE runs were performed using the DCodeTM universal mutation detection system (Bio-Rad Laboratories Inc., Hercules, CA, USA) and the gels were stained with SYBR Gold (Thermo Fisher Scientific Inc., Waltham, MA, USA) for 30 min and photographed on a UV transilluminator (GelDoc—It TS2 Imager, UVP, Analytik Jena GmbH, Jena, Germany). Clustering of DGGE banding profiles using a dendrogram was carried out with CLIQS 1D Pro software (TotalLab Ltd., Newcastle-Upon-Tyne, UK; http://totallab.com/; accessed on 31 May 2021).
2.5. Community Level Physiological Profile of Bacterial Community in Compost and Soil Samples
In order to elucidate the profile of functional potential of microbial communities present in samples of compost and soils, the community-level physiological profile (CLPP) was determined in triplicate using 96-well Biolog EcoPlatesTM (Biolog Inc., Hayward, CA, USA) containing 31 different carbon sources. Briefly, microbial cells were dislodged from 1.5 g of sample by shaking in 15 mL of phosphate saline buffer solution on an orbital shaker at 25 °C, and then diluted by a factor of 1000 and filtered with a qualitative filter paper (11 µm pore size; Whatman plc, Maidstone, UK) to remove impurities. One hundred μL of the filtered suspension were added to each wells in the microplate and incubated at 25 °C in the dark. Color development in the wells was measured at 590 nm with a Multiskan™ GO Microplate Spectrophotometer (ThermoFisher Scientific, Hercules, CA, USA) every day for one week. Samples for statistical analyses were selected using the average well color development (AWCD) method [], at time points where average OD590 values across plate wells was calculate.
2.6. Isolation of Fungi and Associated Bacteria Dispersing along Their Hyphae
To determine the motility and dispersion of bacterial cells along fungal hyphae in compost and soil samples, fungal highway column system (FHCS) methodology was used [,]. The FHCS is built with sterile plastic materials and permits the growth of fungal mycelia from a substrate (here: RSC, C, and BS) toward a culture medium (here: potato dextrose agar; PDA). Mycelia thereby cross an air-filled volume that is filled with glass beads that only can be passed by migrator bacteria if they disperse along fungal hyphae (Figure 1). Firstly, one kg of either C, RSC or BS was placed in previously alcohol- and UV sterilized plastic boxes (30 × 25 cm), and subsequently, 30 FHCS per box were placed, 3 negative controls (FHCS completely closed), 3 FHCS without glass beads inside, in order to determine if fungi are capable of growing in air-filled space, and 24 FHCS for isolation of fungi and their associated bacteria. At each sampling time (7, 14, 21 and 28 days after placement), 6 FHCS were removed and sacrificed for further analysis. The PDA medium from FHCS was removed from the FHCS and pieces of it transferred on two Petri dishes containing either Luria-Bertani (LB) or PDA agar media. This procedure was carried out in two ways: first placing an undisturbed PDA fragment on the culture media, and second inoculating 100 uL of a suspension of microorganism prepared by washing the other PDA fragment with saline solution (NaCl 0.9%), to disaggregate the microorganisms colonizing the target culture medium, in order to isolate the highest variety of bacteria. Inoculated plates were incubated at 25° for 3 days for bacteria and 10 days for fungi and those showing microbial growth were used to isolate bacterial and fungal strains. Fungal isolates were maintained in PDA—containing Petri dishes and their ability to mineralize Na-phytate (C6H18P6O24·12Na·xH2O) and solubilize Ca-phosphate [Ca3(PO4)2] on agar plates was tested []. Meanwhile the hyphae-associated bacterial isolates were obtained and purified on agar LB plates by streaking and stored at −80 °C in 7:3 LB: glycerol (v/v).
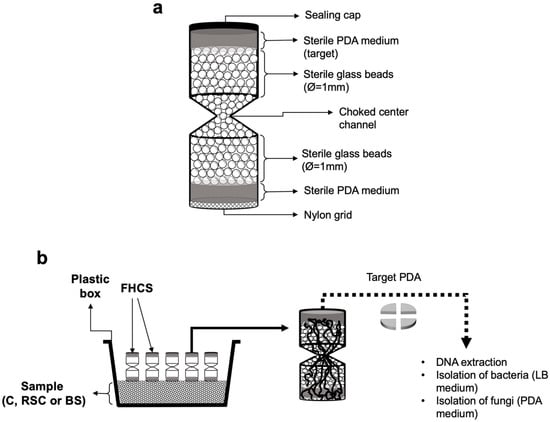
Figure 1.
(a). Schematic of fungal highway column systems (FCHS) used to the isolation of fungal and their hyphae-associated bacterial strains from compost (C), rhizosphere soil with compost application (RSC) and bulk soil (BS) samples. PDA: potato dextrose agar; LB: Luria Bertani agar. (b). Schematic of experimental workflow for isolation and identification of microorganisms from different samples using FCHS.
Then, chromosomal DNA of purified fungal and bacterial isolates was extracted by using UltraClean Microbial DNA Isolation Kit (Qiagen, Hilden, Germany) and DNA extracts were sent to Scientific and Technological Bioresource Nucleus (https://bioren.ufro.cl/) for partial sequencing of 18S and 16S rRNA genes. For bacteria, primer set 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) and 27F (5′-AGA GTT TGA TCC TGG CTC AG3′) were used [], while for fungi, three different primer sets were used: NS1 (5′-GTA GTC ATA TGC TTG TCT C-3′) and NS24 (5′-AAA CCT TGT TAC GAC TTT TA-3′); ITS1 (5′-CTT GGT CAT TTA GAG GAA GTA A-3′) and ITS4 (5′-TCC TCC GCT TAT TG TAT GC-3′); and NS1 (5′-GTA GTC ATA TGC TTG TCT C-3′) and NS8 (5′-TCC GCA GGT TCA CCT ACG GA-3′) [] in order to improve the accuracy of identification. The sequences were then used to determine the taxonomic affiliation of isolates by comparison with those deposited in GenBank by Blastn tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 31 May 2021). Those bacterial strains isolated from the same fungi and showing similar taxonomic affiliation were treated as clones, and only one clone was reported.
The sequences obtained in this study were deposited in the GenBank under accession numbers MW624352 to MW624370 for 18S rRNA gene, and MW624337 to MW624351 for 16S rRNA gene.
2.7. Analysis of Bacterial Dispersal along Mycelia
The dispersion of bacterial cells along fungal hyphae was also confirmed by microscopic observation. To achieve a better observation of the hypha, isloted fungi were grown in Petri dishes and glass slides, both coated with thin layers of PDA culture medium, and observed directly in optical microscope (Olympus BX-41, Tokyo, Japan) at 400× magnification. Additionally, those preparations were also observed in Fluorescent Cell Imager ZOE TM (Bio-Rad Laboratories, Hercules, CA, USA) after staining with LIVE/DEAD® BacLight Bacterial Viability Kits that allow distinguish live and dead bacteria provide a two-color fluorescence assay of mixtures of green-fluorescent nucleic acid stain for living bacterial and the red-fluorescent nucleic acid stain, propidium iodide for dead bacteria. This staining allowed to better visualize bacteria associated to fungal hyphae and determine their viability.
2.8. Screening of Representative Plant Growth‒Promoting Traits in Dispersed Bacterial Isolates
Bacterial isolates showing dispersal along the hyphae were tested for representative PGP traits as follows:
- The production of tryptophan—induced auxin was determined by colorimetrically at 530 nm using Salkowski’s reagent []. The bacterial isolates were incubated on a gyratory shaker (120 rpm) at 30 °C for 3 days in LB broth and LB broth supplemented with tryptophan (1 mg mL−1) as auxin precursor. After incubation, bacterial cells were centrifuged (850× g, at 4 °C for 10 min) and 1 mL of the supernatant was collected and mixed with 2 mL of Salkowski’s reagent [] and incubated for 30 min at room temperature. Standard solution of pure indole acetic acid (Sigma-Aldrich, St. Louis, MO, USA) was used as a positive control. Results are presented qualitatively as positive/negative.
- 1-aminocyclopropane-1-carboxylic acid deaminase (ACCD) activity was measured following the methodology reported by Penrose and Glick []. Briefly, bacterial strains were inoculated in 5 mL tubes containing DF minimum salts medium containing 4.0 g of KH2PO4, 6.0 g of Na2HPO4, 0.2 g of MgSO4·7H2O, 2.0 g of glucose, 2.0 g of gluconic acid and 2.0 g citric acid and trace element: 1 mg of FeSO4·7H2O, 10 µg of H3BO3, 11.19 µg of MnSO4·H2O, 124.6 g of µg ZnSO4·7H2O, 78.22 µg of CuSO4·5H2O, 10 µg of MoO3, pH 7.2, and 2.0 g (NH4)2SO4 as nitrogen source. Cultures were incubated for 48 h at 28 °C. Aliquots (0.1 mL) of each culture were inoculated in tubes containing DF medium supplemented with 3.0 mM of ACC as unique nitrogen source and incubated for 48 h at 28 °C []. Finally, for determining ACCD activity, the amount of α-ketobutyrate produced as ACCD degrades ACC was measured spectrophotometrically in a microplate reader at 540 nm wavelength. An α-ketobutyrate calibration curve in the range from 0.1 to 1.0 µmol was used. Results are presented qualitatively as ACCD positive/negative.
- The ability of bacterial isolates and their associated fungi to mineralize phytate and/or solubilize phosphate was evaluated using the phytate screening medium and National Botanical Research Institute’s phosphate growth medium as reported by Jorquera et al. (2008) []. The P sources used in the media were Na-phytate (C6H18P6O24·2Na·xH2O) and Ca-phosphate [Ca3(PO4)2] analytical grade (pH = 7.0). After incubation at 30 °C for 4 days, the appearance of clear zones around the colonies were considered as a positive result the capacity of phytate mineralization and phosphate solubilization. Results are presented qualitatively as positive/negative.
2.9. Statistical Analyses
Based on the matrix obtained from the banding profiles on DGGE gels, differences between bacterial communities were calculated by similarity profile analysis (SIMPROF test) with Bray-Curtis similarity index, 5% significance level, and 0.1 stress values [] and visualized by nonmetric multidimensional scaling (NMDS) analysis using Primer 6 software (Primer-E Ltd., Auckland, New Zealand; http://www.primer-e.com/; accessed on 31 May 2021).
Based on the matrix obtained from the results of CLPP, differences between bacterial communities were calculated by similarity profile analysis (SIMPROF test) with Bray-Curtis similarity index, 5% significance level, and 0.1 stress values [], and visualized by nonmetric multidimensional scaling (NMDS) analysis using Primer 6 software.
3. Results
3.1. Chemical Properties of Compost and Soil Samples
Differences in chemical properties among representative C, RSC and BS samples were observed (Table 1). Compost contained higher P, K, Ca, Na, and organic matter (OM) levels than B soil. Higher values of extractable potassium (K) and exchangeable cations were observed in C sample whereas RSC sample showed higher values of nitrogen (N). Samples of C and RSC showed similar values of extractable phosphorus (P) and organic matter, but higher to the value observed in BS samples. In contrast, higher values of pH were observed in BS and RSC samples (pH ≈ 7.2) as compared to C (pH ≈ 6.8).

Table 1.
Chemical properties of representative composite samples of compost (C), rhizosphere soil with compost application (RSC) and bulk soil (BS).
3.2. DNA-Based Profiles of Bacterial and Fungal Communities in Compost and Soil Samples
16S rRNA DGGE based analysis comparison of C, RSC and BS showed distinct bacterial communities as visualized by dendrogram and NMDS analyses (Figure 2a,b). NMDS analyses showed three clusters with a clear separation among the bacterial communities at 50% similarity level. Furthermore, highest homogeneity in replicate samples (80% similarity) in C samples as compared to RSC and B was found. Similar results were observed in fungal communities banding profiles obtained from DGGE analysis using 18S rRNA gene as target (Figure 2c,d). Significant differences in fungal communities were also observed between C, RSC and BS samples, where a higher homogeneity was visualized in each sample and a higher separation (70% similarity) between C samples relative to RSC and BS.
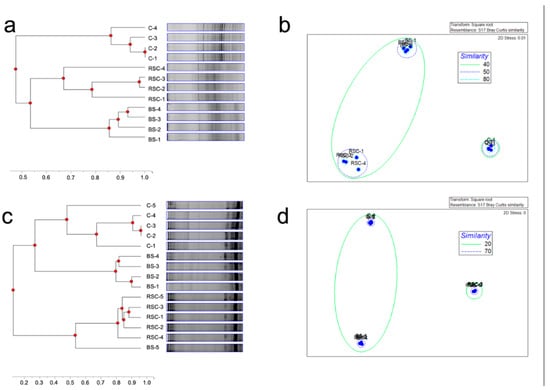
Figure 2.
DNA—based profiles of microbial fingerprints determined by denaturing gradient gel electrophoresis (DGGE). (a) Dendrogram of bacterial community compositions in samples of compost (C), rhizosphere soil with compost application (RSC) and bulk soil (BS) (n = 4) revealed by DGGE using 16S rRNA gene as target. (b) Non-metric multidimensional scaling (NMDS) of bacterial community compositions revealed by DGGE using 16S rRNA gene as target. (c) Dendrogram of fungal community compositions in samples of C, RSC and BS (n = 4) revealed by DGGE using 18S rRNA gene as target. (d) NMDS of fungal community compositions revealed by DGGE using 18S rRNA gene as target.
3.3. Community Level Physiological Profiles of Bacterial Communities in Compost and Soil Samples
The CLPP analysis showed microbial communities in C and RSC of similar metabolic capacity. Both microbial communities were able to catabolize 30 different carbon sources present in the Biolog EcoPlatesTM (Figure 3a). Microbial communities in RSC samples consumed the 30 carbon sources in 48 h of incubation, whereas microbial communities in BS samples consumed a maximum of 19 carbon sources at 120 h of incubation. C and RSC samples showed a higher metabolic diversity as compared to BS samples. While e.g., Tween 40, Tween 80, and D-cellobiose were efficiently consumed by C, RSC, and BS microbial communities, 4-hydroxybenzoic acid, L-arginine, L-threonine, and L-phenylalanine were not consumed by C and BS microbial communities.
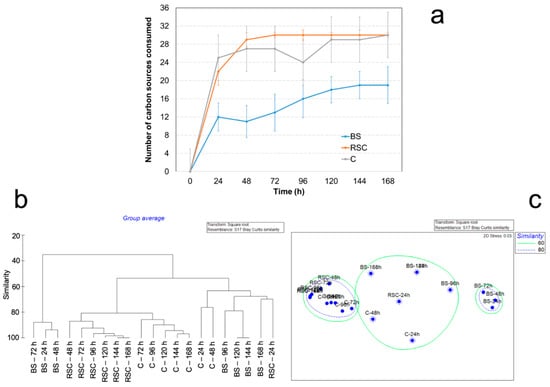
Figure 3.
Physiological profiles of total microbial community determined by community-level physiological profile (CLPP). (a) Number of carbon sources consumed in samples of compost (C), rhizosphere soil with compost application (RSC) and bulk soil (BS) in Biolog EcoPlatesTM after incubation at 25 °C in the dark for one week (168 h). Error bars denote standard deviation (n = 3). Dendrogram (b) and non-metric multidimensional scaling (c) based on CLPP analysis.
Visualization of CLPP with a dendrogram and NMDS analysis (Figure 3b,c) showed three clusters. One cluster with a similarity of 80% was formed by samples of RSC and C from 72 to 168 h of incubation. A second cluster was formed by a lower similarity (60%), including samples of BS (96–168 h incubation) and samples of C (24–48 h incubation) and RSC (48 h incubation). The third cluster with an 80% of similarity was exclusively formed by samples of BS from 24 to 72 h of incubation.
3.4. Characterization of Bacteria Dispersing along Fungal Hyphae
A total of 19 fungi and 10 taxonomically different bacterial isolates were obtained from C and RSC samples by FHCS. No fungi were isolated from BS samples.
Based on partial sequencing of the 18S rRNA genes, the fungal isolates showing hyphae-associated bacteria were identified as members of the Aspergillus, Mucor, Ulocladium, Rhizopus and Syncephalastrum genera (Table 2). All of them (except Ulocladium sp. 2H), showed phytate-mineralizing or phosphate-solubilising capability on agar plates. In contrast, fungal isolates identified as Stereum, Miladina, Aspergillus, Penicillium, Rhizopus, Chaetomium and Lichtheimia genera did not show hyphae-associated bacteria (Table 3).

Table 2.
Taxonomic assignment of fungal isolates with hyphae-associated bacteria using fungal highway column system (FHCS) selection and isolation approach. C and RSC refer to compost and rhizosphere soil with compost application, respectively.

Table 3.
Taxonomic assignment of fungal isolates which did not showed associated bacteria, using the fungal highway column system (FHCS) selection and isolation approach. C and RSC refer to compost and rhizosphere soil with compost application, respectively.
Based on partial sequencing of 16S rRNA gene, the isolated bacteria able to disperse along the fungal hyphae in FHCS were identified as members of Rhodococcus, Bacillus, Pseudomonas, Agrobacterium, Glutamicibacter and Microbacterium genera (Table 4). Isolates belonging to Rhodococcus and Microbacterium genera were exclusively isolated from C samples, while Pseudomonas was found in RSC samples only. Isolates belonging to the genera Bacillus were obtained from both C and RSC samples. Interestingly, Pseudomonas isolates were found associated only with the mycelium of the genus Mucor; while actinobacteria (Rhodococcus, Glutamicibacter and Microbacterium) were isolated in association with the mycelium of the genus Aspergillus. On the other hand, the strains of genus Bacillus (B. subtilis, B. amyloliquefaciens and B. cereus) were found associated with a variety of fungal genera, such as Ulocladium, Rhizopus, Syncephalastrum and Aspergillus.

Table 4.
Taxonomic assignment of bacterial isolates dispersing along of fungal hyphae (host) to the target agar in fungal highway column systems (FHCS). C and RSC refer to compost and rhizosphere soil with compost application, respectively.
Fungal-bacteria associations and bacterial dispersal along hyphae were further confirmed by optical microscopy (Figure 4; Video S1).
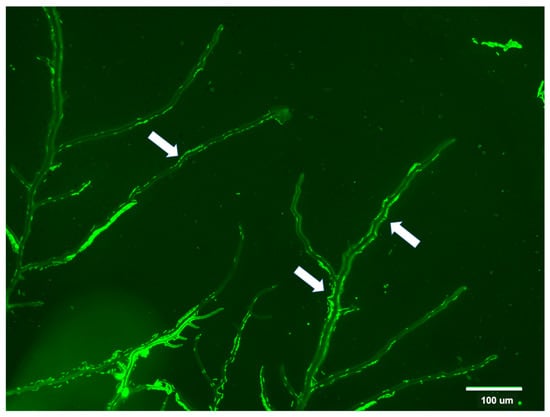
Figure 4.
Optical microscopy photograph (magnification: 400×) showing the association of green‒stained bacterial cells in the liquid layer from hyphae of the fungus contained in the commercial compost and grown on agar plate. Bacterial cells on hyphae are indicated by arrows and they can also be visualized a video in the Supplementary Video S1.
3.5. Screening of Representative Plant Growth‒Promoting Traits in Selected Bacterial Isolates
PGP traits of seven bacterial isolates able to disperse along the fungal hyphae were studied (Table 5). All isolates were able to produce tryptophan-induced auxin, while only three isolates (Pseudomonas sp. 18B and 32B, Glutamicibacter sp. 37B) exhibited ACCD activity. Four isolates (Bacillus sp. 15B, Pseudomonas sp. 18B and 32B, and Rhodococcus sp. 13B) enabled phytate-mineralization. However, no phosphate-solubilizing activity was observed in any of the bacterial isolates, while two fungal strains, both from Aspergillus genus, did showed this capability.

Table 5.
Screening of representative plant growth-promoting traits in selected bacterial isolates dispersed along of fungal hyphae.
4. Discussion
4.1. Microbial Community Composition and Metabolic Activity in C, BS, and RSC
Applying a previously described approach [,,] fungi were isolated from C, RSC and BS and searched for putative PGP bacteria able to disperse along their mycelia. Prior to isolation of the bacterial fungal couples, we characterized the C, RSC, and BS substrata used as inoculum source. BS, RSC and C samples showed distinct differences in their chemical properties (Table 1) with C and RSC exhibiting significantly higher nutrient and OM contents than BS. Not surprisingly, this is in accordance to earlier studies revealing that continuous application of compost induce changes in physical, chemical and microbial traits of agricultural soils [,,]. Observed differences between BS and RSC may however also be driven by grapevine rhizosphere effects, as plants root typically release organic compounds (e.g., aminoacids, sugars, or organic acids) that may select for rhizosphere-specific microbial communities [,,,]. This may explain the more efficient use of amino acids (e.g., L-arginine, L-threonine and L-phenylalanine), carbohydrates (e.g., D-cellobiose and xylose) and organic acids (4-hydroxybenzoic, malic acid and galacturonic acid) by RSC than by BS microbial communities in Biolog® assays. Similar CLPP of C and RSC microbial communities may be explained selective expression of similar catabolic functions [], and/or a functional redundancy of different taxonomic groups in highly diverse RSC and C microbial communities [].
4.2. Isolation of Fungi and Associated Migrator Bacteria
RSC and C samples allowed successful isolation bacterial-fungal associations and bacteria dispersing along mycelia, respectively (Table 2 and Table 4). This finding suggests that RSC and C, likely due to their favorable chemical and physical characteristics (Table 1) allow for manifold bacterial-fungal interfaces []. The high content of lignocellulosic material in plant derived compost favors growth of saprotrophic mycelial fungi, which, by exerting selective pressure, also allow for targeted bacterial communities in their hyphosphere. Aerobic degradation of cellulose and lignin is widespread among compost decomposing fungi (e.g., such as Aspergillus and Mucor [,]) while cellulose degradation by soil bacterial species such as Bacillus, or Pseudomonas (i.e., genera also found in our study) have been described []. In contrast, fungi were not isolated from BS samples. The absence of fungal growth may be related to low abundance of fast-growing saprotrophic fungi (which typically colonize FHCS []) or the fact that FHCS did not provide the appropiate conditions to the colonization and growth of native fungi contained in BS, which was characterized by lower contents of P and organic matter according to Table 1. However, the reasons for which fungi were not isolated from BS are still unclear.
Different ecological relations (e.g., mutualism and antagonism) are established in the bacterial-fungal interface [,,]. In particular, knowledge of beneficial bacteria-fungi interactions is highly relevant as they can be exploited to improve soil fertility and crop yields. For instance, the ability of bacteria to disperse through soil via mycelial networks represents a mechanism that can provide a great thrust to their ecological competence, as it allows them to reach new ressources which confer novel ecological opportunities [,,,]. As an example, genera Bacillus and Pseudomonas are commonly present in bacterial community added to soil with compost application [,,,] however these bacteria often fail to colonize the plant rhizosphere as they fail to compete with the native microbial community due to insufficient access to immobilized ressources pores or poor displacement capability in vadose environments []. Filamentous compost fungi such as Aspergillus and Mucor may play a fundamental role [,] as conduits for efficient colonization of roots and soil habitats or the access to soil nutrients [,]. For instance, co-inoculation of biocontrol bacteria Pseudomonas fluorescens with arbuscular mycorrhizal Glomus mosseae demonstrated that P. fluorescens enabled better mycorrhizal colonization of roots of basil (Ocimum basilicum L.) [].
As migrators along hyphae, bacteria of the genera Bacillus, Pseudomonas, Microbacterium, Glutamicibacter and Rhodococcus were isolated. Members of Bacillus and Pseudomonas have been found in rhizosphere soil amended with compost and are known as PGP bacteria in compost and other organic amendments [,,,]. Six out of 10 bacterial isolates belonged to genus Bacillus and associated to hyphae of Ulocladium, Rhizopus, Syncephalastrum and Aspergillus. Members of genus Bacillus (e.g., B. subtilis and B. amyloliquefaciens) are widely studied and proposed as biocontrol agents for several phytopathogenic fungi, such as Fusarium oxysporum, Ralstonia solanacearum, Rhizoctonia solani and Alternaria tenuissima [,]. On the other hand, there are previous reports of beneficial associations between strains of B. subtilis and fungi of the genus Aspergillus, where co-inoculation of both microorganisms leads to a more efficient biodegradation of contaminants in the soil [], as well as the stimulation of the synthesis of compounds with pharmaceutical application in the fungus []. Also, strains of the genus Bacillus (B. subtilis, B. amyloliquefaciens and B. cereus) and genus of fungi Ulocadium are commonly used in the manufacture of commercial products to control the pathogenic fungus Botrytis cinerea [,]. However, we could not find in these studies reference to formulations of products that included the co-inoculation of both (Bacillus and Ulocladium); taking into account that this fungus can be an effective means for dispersal of these bacteria, it could be expected that a formulation that contains both microorganisms could act more efficiently.
Two bacterial isolates belonged to genus Pseudomonas and associated with Mucor sp. derived from RSC. Member of genus Pseudomonas (e.g., P. fluorescens and P. chlororaphis) are likewise known as biocontrol agents of phytopathogenic fungi, such as Rosellinia necatrix and Pythium aphanidermatum [,]. Recent studies using Pseudomonas (P. fluorescens and P. protegens) showed potential inhibition of mycelial growth and spore fungal germination of several fungal genera including the Mucor genus [,], however the Pseudomonas strain isolated in our study did not show evidence of affecting the growth and germination of this fungus.
The isolates belonging to genera Rhodococcus, Agrobacterium, Glutamicibacter and Microbacterium associated with genus Aspergillus and Rhizopus. Microbacterium humic and Microbacterium agarici have previously been isolated from the fungi Agaricus blazei [], whereas Glutamicibacter arilaitensis has been described to associate with Penicillium [] and to significantly improve the mycelial growth of edible mushroom Pleurotus florida []. Bacteria of the genus Rhodococcus sp. have been isolated from a wide variety of enviroments, including soils and composts [,] and have been found dispersing along the mycelia of the oomycete Pythium ultimum []. Finally, a recent study has also reported the association of Agrobacterium with the fungal mutualist Piriformospora indica (Basidiomycota), establishing a tripartite symbiosis (Sebacinalean) with a broad range of plants [].
4.3. PGP Traits of Bacteria Dispersing along Fungal Mycelia
Anaylsis of putative PGP traits of migrator bacteria revealed that all bacterial isolates were able to synthetise tryptophan-induced auxins (Table 5). Most of PBP bacteria are able to synthesize auxins (e.g., indole-3-acetic acid [IAA]) that stimulate growth and division of plant cells []. Particularly, bacteria belonging to the genera Pseudomonas and Bacillus are commonly reported for their ability to produce and thereby increase IAA levels in stems and leaves of grapvine plants []. Three of our isolates (Pseudomonas spp. and Glutamicibacter sp.) moreover exhibited ACCD activity; i.e., a trait associated to the promotion of plant growth and reduction of plant stress, resp. []. ACCD activity has been described for both bacterial groups, including isolates from compost and organic amendments []. Although we couldn’t observe ACCD activity in our Bacillus sp. isolates, other studies reported on vermicompost-derived Bacillus sp. isolates able to express ACCD activity []. Not surprisingly we observed phytase activity in isolates of the genera Rhodococcus, Bacillus and Pseudomonas as phytate is one of the main organic phosphorus forms in compost and plant-derived residues. Phytase activity has been described for the genera Bacillus, Burkholderia, Enterobacter, Pseudomonas, and Staphylococcus []. Although members of Pseudomonas and Bacillus have been described as phosphate solubilizers [] none of our isolates capable of solubilizing inorganic phosphates. This may be due to the hypothesiszed prevalence of phytate in compost and RSC. Recent studies e.g., have shown that long-term chemical fertilization combined with organic materials can modify the phoD gene expression pattern in soil bacterial communities []; nevertheless, our finding requires further study since the fungi associated with these bacteria showed both phosphate-solubilizing and phytate-mineralizing capabilities. Taking into account that most fungal bacterial associsations are known to share nutrients such as phosphorus [], it seems reasonable to assume that such phenomenon may also occur in the bacterial-fungal associations of this study.
5. Practical Relevance
Our data show that bacteria exibiting PGP traits dispersed along mycelia of fungi presente in compost and RSC. Mycelia-mediated dispersal may allow them to reach plant roots and other PGP microhabitats. Next to being a cheap and readily available fertilizer, compost facilitates the inoculation of complex and adapted bacterial-fungal consortia and their efficient spreading even in vadose environments such as water unsaturated soil. The fractal network of fungal mycelia however has the ability to overcome air-water interfaces and builds a suitable scaffold (‘fungal highway’ []) for efficient dispersion in soil. Such phenomenon may be particularly useful in case compost is solely placed onto agricultural land without further mixing with soil. Inoculation of single isolates with efficient PGP traits often fails, as such strains often fail to compete with soil microbiota leading to poor survival efficiency and root colonization []. Furthermore, bioaugmented PGP bacteria typically do not spread well in air-filled and dry soil, as their dispersal and access to nutrients in the soil depends on waterbone transport and/or the presence of continuous water films []. The high functional and taxonomic diversity of compost in conjunction with its elevated water holding capacity makes compost an interesting seedbank of PGP bacteria and selfpromoted dispersal thereof by mycelial transport [].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agronomy11081567/s1, Video S1: Bacterial cells moving on hyphae of the fungus contained in the commercial compost.
Author Contributions
Conceptualization, S.G.-G., L.Y.W. and M.A.J.; formal analysis, S.G.-G., M.A.-E., M.C., J.J.A. and M.A.J.; investigation, S.G.-G., M.A.-E., M.C., L.Y.W., J.J.A. and M.A.J.; writing—original draft preparation, S.G.-G., L.Y.W. and M.A.J.; writing—review and editing, S.G.-G., L.Y.W., J.J.A. and M.A.J.; funding acquisition, S.G.-G., J.J.A. and M.A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by The National Fund for Scientific and Technological Development (FONDECYT) project no. 1201386 and 1181050 (to M.A.J. and J.J.A.) from Chile’s National Research and Development Agency (ANID), by the National Competition for the Attraction of International Advanced Human Capital, Short Stay Modality (MEC) no. 80180048 (to J.J.A., L.Y.W. and M.A.J.) from International cooperation program (PCI-ANID), by Science and Technology Research Partnership for Sustainable Development (SATREPS; JST/JICA, Japan) project code JPMJSA1705 (to M.A.J. and J.J.A.), by Fund for Scientific and Technological Equipment (FONDEQUIP) code EQM170171 (to M.A.J), by Universidad de La Frontera (DIUFRO) project code DI21-0044, by Ph.D. Scholarship provided by ANID no. 21160935 (to S.G.-G.) and 21151002 (to M.A.-E.), and by the “Apoyo a Profesores Patrocinantes de Alumnos de Pre y Postgrado” program, code DI19-2016, Vicerrectoría de Investigación y Postgrado, Universidad de La Frontera (to S.G.-G. and M.A.-E.).
Data Availability Statement
All sequences reported are available in NCBI GenBank database (http://ncbi.nlm.nih.gov; accessed on 31 May 2021).
Acknowledgments
Authors thank to Jorge Parraguez (Rosario Co.) by provide the samples used in this study.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Palanivell, P.; Susilawati, K.; Ahmed, O.H.; Majid, N.M. Compost and Crude Humic Substances Produced from Selected Wastes and Their Effects onZea maysL. Nutrient Uptake and Growth. Sci. World J. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baliah, T.; Muthulakshmi, P.; Lakshmi, K.; Begum, J. Crop response of different formulations of solubilizing bacteria on cow pea. Int. J. Curr. Adv. Res. 2016, 5, 924–928. [Google Scholar]
- Liu, C.H.; Liu, Y.; Fan, C.; Kuang, S.Z. The effects of composted pineapple residue return on soil properties and the growth and yield of pineapple. J. Soil Sci. Plant Nutr. 2013, 13, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Calderón, C.E.; de Vicente, A.; Cazorla, F.M. Role of 2-hexyl, 5-propyl resorcinol production by Pseudomonas chlororaphis PCL1606 in the multitrophic interactions in the avocado rhizosphere during the biocontrol process. FEMS Microbiol. Ecol. 2014, 89, 20–31. [Google Scholar] [CrossRef] [Green Version]
- Le, X.H.; Ballard, R.A.; Franco, C.M.M. Effects of endophytic Streptomyces and mineral nitrogen on Lucerne (Medicago sativa L.) growth and its symbiosis with rhizobia. Plant Soil 2016, 405, 25–34. [Google Scholar] [CrossRef]
- Yadav, A.; Dubey, R.; Yadav, K. Growth and ectomycorrhization of banj oak plants co-inoculated with Scleroderma bovista and mycorrhizosphere bacteria. J. Appl. Nat. Sci. 2015, 7, 265–272. [Google Scholar] [CrossRef]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Glaeser, S.P.; Alabid, I.; Imani, J.; Haghighi, H.; Kampfer, P.; Kogel, K.-H. The Abundance of Endofungal Bacterium Rhizobium radiobacter (syn. Agrobacterium tumefaciens) Increases in Its Fungal Host Piriformospora indica during the Tripartite Sebacinalean Symbiosis with Higher Plants. Front. Microbiol. 2017, 8, 629. [Google Scholar] [CrossRef]
- Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [Green Version]
- Bonfante, P.; Anca, I.-A. Plants, Mycorrhizal Fungi, and Bacteria: A Network of Interactions. Annu. Rev. Microbiol. 2009, 63, 363–383. [Google Scholar] [CrossRef] [Green Version]
- Nazir, R.; Warmink, J.A.; Boersma, H.; van Elsas, J.D. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 2009, 71, 169–185. [Google Scholar] [CrossRef] [Green Version]
- Carson, J.K.; Gonzalez-Quiñones, V.; Murphy, D.V.; Hinz, C.; Shaw, J.A.; Gleeson, D.B. Low Pore Connectivity In-creases Bacterial Diversity in Soil. Appl. Environ. Microbiol. 2010, 76, 3936–3942. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.-S.; Cao, M.-Q.; Zou, Y.-N.; He, X.-H. Direct and indirect effects of glomalin, mycorrhizal hyphae and roots on aggregate stability in rhizosphere of trifoliate orange. Sci. Rep. 2015, 4, 5823. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.S.; Nene, S.N.; Joshi, K.S. A comparative study of production of hydrophobin like proteins (HYD-LPs) in submerged liquid and solid state fermentation from white rot fungus Pleurotus ostreatus. Biocatal. Agric. Biotechnol. 2020, 23, 101440. [Google Scholar] [CrossRef]
- Worrich, A.; Wick, L.Y.; Banitz, T. Ecology of Contaminant Biotransformation in the Mycosphere: Role of Transport Processes. Adv. Appl. Microbiol. 2008, 104, 93–133. [Google Scholar]
- Van Overbeek, L.S.; Saikkonen, K. Impact of Bacterial-Fungal Interactions on the Colonization of the Endosphere. Trends Plant Sci. 2016, 21, 230–242. [Google Scholar] [CrossRef]
- Kohlmeier, S.; Smits, T.H.; Ford, R.M.; Keel, C.; Harms, H.; Wick, L.Y. Taking the Fungal Highway: Mobilization of Pollutant-Degrading Bacteria by Fungi. Environ. Sci. Technol. 2005, 39, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Genet. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Bravo, D.; Cailleau, G.; Bindschedler, S.; Simon, A.; Job, D.; Verrecchia, E.; Junier, P. Isolation of oxalotrophic bacteria able to disperse on fungal mycelium. FEMS Microbiol. Lett. 2013, 348, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Kastman, E.; Guasto, J.S.; Wolfe, B.E. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Furuno, S.; Foss, S.; Wild, E.; Jones, K.C.; Semple, K.; Harms, H.; Wick, L.Y. Mycelia Promote Active Transport and Spatial Dispersion of Polycyclic Aromatic Hydrocarbons. Environ. Sci. Technol. 2012, 46, 5463–5470. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, L.; Zhou, J.; George, T.S.; Feng, G. Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. New Phytol. 2021, 230, 304–315. [Google Scholar] [CrossRef] [PubMed]
- De Novais, C.B.; Sbrana, C.; Jesus, E.D.C.; Rouws, L.F.M.; Giovannetti, M.; Avio, L.; Siqueira, J.O.; Júnior, O.J.S.; Da Silva, E.M.R.; De Faria, S.M. Mycorrhizal networks facilitate the colonization of legume roots by a symbiotic nitrogen-fixing bacterium. Mycorrhiza 2020, 30, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Warncke, D.; Brown, J.R. Potassium and other basic cations. In Recommended Chemical Soil Test Procedures for the North Central Region; Missouri Agricultural Experiment Station: Columbia, MO, USA, 1998; pp. 31–33. [Google Scholar]
- Bertsch, P.M.; Bloom, P.R. Aluminum. In Methods of Soil Analysis: Chemical Methods; Sparks, D.L., Ed.; Soil Science Society of America, Inc.; American Society of Agronomy, Inc.: Madison, WI, USA, 1996; Pt. 3, pp. 517–550. [Google Scholar]
- Radojevic, M.; Bashkin, V. Practical Environmental Analysis; Royal Society of Chemistry: London, UK, 1999. [Google Scholar]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter and a proposed modi-fication of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Jorquera, M.A.; Inostroza, N.G.; Lagos, L.M.; Barra, P.; Marileo, L.; Rilling, J.I.; Campos, D.C.; Crowley, D.; Richardson, A.E.; Mora, M.L. Bacterial community structure and detection of putative plant growth-promoting rhizobacteria associated with plants grown in Chilean agro-ecosystems and undisturbed ecosystems. Biol. Fertil. Soils 2014, 50, 1141–1153. [Google Scholar] [CrossRef]
- Marshall, M.N.; Cocolin, L.; Mills, D.A.; Vander Gheynst, J.S. Evaluation of PCR primers for denaturing gradient gel elec-trophoresis analysis of fungal communities in compost. J. Appl. Microbiol. 2003, 95, 934–948. [Google Scholar] [CrossRef]
- Choi, K.-H.; Dobbs, F.C. Comparison of two kinds of Biolog microplates (GN and ECO) in their ability to distinguish among aquatic microbial communities. J. Microbiol. Methods 1999, 36, 203–213. [Google Scholar] [CrossRef]
- Simon, A.; Bindschedler, S.; Job, D.; Wick, L.Y.; Filippidou, S.; Kooli, W.M.; Verrecchia, E.P.; Junier, P. Exploiting the fungal highway: Development of a novel tool for the in situ isolation of bacteria migrating along fun-galmycelium. FEMS Microbiol. Ecol. 2015, 91, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junier, P.; Cailleau, G.; Palmieri, I.; Vallotton, C.; Trautschold, O.C.; Junier, T.; Paul, C.; Bregnard, D.; Palmieri, F.; Estoppey, A.; et al. Democratization of fungal highway columns as a tool to investigate bacteria associated with soil fungi. FEMS Microbiol. Ecol. 2021, 97, fiab003. [Google Scholar] [CrossRef] [PubMed]
- Jorquera, M.; Hernández, M.T.; Rengel, Z.; Marschner, P.; Mora, M.D.L.L. Isolation of culturable phosphobacteria with both phytate-mineralization and phosphate-solubilization activity from the rhizosphere of plants grown in a volcanic soil. Biol. Fertil. Soils 2008, 44, 1025–1034. [Google Scholar] [CrossRef]
- Patten, C.; Glick, B. Role of Pseudomonas putida indole acetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [Green Version]
- Acuña, J.J.; Jorquera, M.A.; Martínez, O.A.; Menezes−Blackburn, D.; Fernández, M.T.; Marschner, P.; Greiner, R.; Mora, M.L. Indole acetic acid and phytase activity produced by rhizosphere bacilli as affected by pH and metals. J. Soil Sci. Plant Nutr. 2011, 11, 1–12. [Google Scholar]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Barra, P.; Inostroza, N.G.; Acuña, J.J.; Mora, M.L.; Crowley, D.E.; Jorquera, M. Formulation of bacterial consortia from avocado (Persea americana Mill.) and their effect on growth, biomass and superoxide dismutase activity of wheat seedlings under salt stress. Appl. Soil Ecol. 2016, 102, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R.; Somerfield, P.; Gorley, R.N. Testing of null hypotheses in exploratory community analyses: Similarity profiles and biota-environment linkage. J. Exp. Mar. Biol. Ecol. 2008, 366, 56–69. [Google Scholar] [CrossRef]
- Furuno, S.; Remer, R.; Chatzinotas, A.; Harms, H.; Wick, L.Y. Use of mycelia as paths for the isolation of contaminant-degrading bacteria from soil. Microb. Biotechnol. 2011, 5, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Morra, L.; Pagano, L.; Iovieno, P.; Baldantoni, D.; Alfani, A. Soil and vegetable crop response to addition of different levels of municipal waste compost under Mediterranean greenhouse conditions. Agron. Sustain. Dev. 2010, 30, 701–709. [Google Scholar] [CrossRef] [Green Version]
- Diacono, M.; Montemurro, F. Long-Term Effects of Organic Amendments on Soil Fertility. In Sustainable Agriculture; Springer Science and Business Media: Berlin/Heidelberg, Germany, 2011; Volume 2, pp. 761–786. [Google Scholar]
- Semenov, M.V.; Krasnov, G.S.; Semenov, V.M.; van Bruggen, A.H. Long-term fertilization rather than plant species shapes rhizosphere and bulk soil prokaryotic communities in agroecosystems. Appl. Soil Ecol. 2020, 154, 103641. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Richardson, A.E.; O’Callaghan, M.; de Angelis, K.M.; Jones, E.E.; Stewart, A.; Firestone, M.K.; Condron, L.M. Efects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 2011, 77, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Gianfreda, L. Enzymes of importance to rhizosphere processes. J. Soil Sci. Plant Nutr. 2015, 15, 283–306. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.-W.; Wang, B.; Li, K.; Liu, Z.-D.; Han, X.; Xu, S.-J.; Guo, Y.-S.; Xie, H.-G. Effect of 4-hydroxybenzoic acid on grape (Vitis vinifera L.) soil microbial community structure and functional diversity. Biotechnol. Biotechnol. Equip. 2015, 29, 637–645. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [Green Version]
- Johansson, J.F.; Paul, L.R.; Finlay, R. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 2004, 48, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hasna, N.; Behnam, T.; Touhami, S.; Mohamed, B.A.; Aouatef, M. Isolation, screening and identification of ligno-cellulolytic fungi from northern central Morocco. Biotechnol. Agron. Soc. Environ. Presses Agron. Gembloux 2019, 234, 207–217. [Google Scholar]
- Noreen, N.; Ramzan, N.; Parveen, Z.; Shahzad, S. A comparative study of cow dung compost, goat pellets, poultry waste manure and plant debris for thermophilic, thermotolerant and mesophilic microflora with some new reports from Pakistan. Pak. J. Bot. 2019, 51, 1155–1159. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and bio-technology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [Green Version]
- Simon, A.; Hervé, V.; Al-Dourobi, A.; Verrecchia, E.; Junier, P. Anin situinventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol. Ecol. 2016, 93, fiw217. [Google Scholar] [CrossRef] [Green Version]
- Barea, J.M.; Azcón, R.; Azcón-Aguilar, C. Mycorrhizosphere interactions to improve plant fitness and soil quality. Antonie Van Leeuwenhoek 2002, 81, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Boultermer, J.I.; Trevors, J.T.; Boland, G.J. Microbial studies of compost: Bacterial identification, and their po-tential for turfgrass pathogen suppression. World J. Microbiol. Biotechnol. 2002, 18, 661–671. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Z.; Ling, N.; Yuan, Y.; Zheng, X.; Shen, B.; Shen, Q. Bacillus subtilis SQR9 can control Fusarium wiltin cucumber by colonizing plant roots. Biol. Fertil. Soils 2011, 47, 495–506. [Google Scholar] [CrossRef]
- Escobar, N.; Delgado, J.M.; Romero, N.J. Identificación de poblaciones microbianas en compost de residuos organicos Fincas Cafeteras de Cundinamarca. Boletín Científico. Cent. Museos. Mus. Hist. Nat. 2012, 16, 75–88. [Google Scholar]
- Tecon, R.; Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 2017, 41, 599–623. [Google Scholar] [CrossRef]
- Jangra, E.; Yadav, K.; Aggarwal, A. Impact of arbuscular mycorrhizal fungi and Pseudomonas fluorescens on growth, physiological parameters and essential oil content in Ocimum basilicum L. Eur. J. Environ. Sci. 2019, 9, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Soares, M.A.; Li, H.Y.; Bergen, M.; da Silva, J.M.; Kowalski, K.P.; White, J.F. Functional role of an endophytic Bacillus amyloliquefaciens in enhancing growth and disease protection of invasive English ivy (Hedera helix L.). Plant Soil 2016, 405, 107–123. [Google Scholar] [CrossRef]
- Zang, S.; Lian, B.; Wang, J.; Yang, Y. Biodegradation of 2-naphthol and its metabolites by coupling Aspergillus niger with Bacillus subtilis. J. Environ. Sci. 2010, 22, 669–674. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; Shindia, A.A.; AbouZeid, A.; Koura, A.; Hassanein, S.E.; Ahmed, R.M. Triggering the biosynthetic machinery of Taxol by Aspergillus flavipes via cocultivation with Bacillus subtilis: Proteomic analyses emphasize the chromatin remodeling upon fungal-bacterial interaction. Environ. Sci. Pollut. Res. Int. 2021, 28, 39866–39881. [Google Scholar] [CrossRef]
- Elmer, P.; Reglinski, T. Biosuppression of Botrytis cinerea in grapes. Plant Pathol. 2006, 55, 155–177. [Google Scholar] [CrossRef]
- Bolívar-Anillo, H.J.; Garrido, C.; Collado, I.G. Endophytic microorganisms for biocontrol of the phytopathogenic fungus Botrytis cinerea. Phytochem. Rev. 2019, 19, 721–740. [Google Scholar] [CrossRef]
- Kim, Y.C.; Anderson, A.J. Rhizosphere pseudomonads as probiotics improving plant health. Mol. Plant Pathol. 2018, 19, 2349–2359. [Google Scholar] [CrossRef] [Green Version]
- Abdelwahab, R.; Bensidhoum, L.; Tabli, N.; Bouaoud, Y.; Naili, F.; Cruz, C.; Elhafid, N. A Pseudomonas Protegens with High Antifungal Activity Protects Apple Fruits Against Botrytis Cinerea Gray Mold. Int. J. Sci. Res. Sci. Technol. 2016, 2, 227–237. [Google Scholar]
- Wallace, R.L.; Hirkala, D.L.; Nelson, L.M. Efficacy of Pseudomonas fluorescens for control of Mucor rot of apple during commercial storage and potential modes of action. Can. J. Microbiol. 2018, 64, 420–431. [Google Scholar] [CrossRef]
- Young, C.C.; Busse, H.J.; Langer, S.; Chu, J.N.; Schumann, P.; Arun, A.B. Microbacterium humi sp. nov. and Microbacterium pseudoresistens sp. nov., isolated from the base of the mushroom Agaricus blazei. Int. J. Syst. Evol. Microbiol. 2010, 60, 854–860. [Google Scholar] [CrossRef]
- Cleary, J.L.; Kolachina, S.; Wolfe, B.E.; Sanchez, L.M. Coproporphyrin III produced by the bacterium Glutamicibacter arilaitensis binds zinc and is upregulated by fungi in cheese rinds. mSystems 2018, 3, e00036-18. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Naraian, R. Enhanced growth and yield of oyster mushroom by growth-promoting bacteria Glutamicibacter arilaitensis MRC119. J. Basic Microbiol. 2021, 61, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Kuyukina, M.S.; Ivshina, I.B. Bioremediation of Contaminated Environments Using Rhodococcus. In Probiotics; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2019; Volume 16, pp. 231–270. [Google Scholar]
- Egelkamp, R.; Schneider, D.; Hertel, R.; Daniel, R. Nitrile-Degrading Bacteria Isolated from Compost. Front. Environ. Sci. 2017, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Glick, B.R.; Pasternak, J.J. Molecular Biotechnology: Principles and Application Recombinant DNA Technology, 3rd. ed.; ASM Press: Washington, DC, USA, 2003. [Google Scholar]
- Salomon, M.V.; Bottini, R.; de Souza, G.A.; Cohen, A.C.; Moreno, D.; Gil, M.; Piccoli, P. Bacteria isolated from roots and rhizosphere of Vitis vinífera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 2013, 151, 359–374. [Google Scholar] [CrossRef]
- Glick, B.R. Modulation of plant ethylene levels by the bacterial enzyme ACCdeaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef]
- Nadeem, S.; Imran, M.; Naveed, M.; Khan, M.Y.; Ahmad, M.; Zahird, Z.A.; Crowley, D.E. Synergistic use of biochar, compost and plant growth-promoting rhizobacteria for enhancing cucumber growth under water deficit conditions. J. Sci. Food Agric. 2017, 97, 5139–5145. [Google Scholar] [CrossRef]
- Jayakumar, P.; Natarajan, S. Molecular and functional characterization of bacteria isolated from straw and goat manure based vermicompost. Appl. Soil Ecol. 2013, 70, 33–47. [Google Scholar]
- Hussin, A.S.M.; Farouk, A.E.; Greiner, R.; Salleh, H.M.; Ismail, A.F. Phytate degrading enzyme production by bacteria isolated from Malaysian soil. World J. Microbiol. Biotechnol. 2007, 23, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jianga, N.; Chena, Z.; Tiana, J.; Sunc, N.; Xuc, M.; Chena, L. Response of soil phoD phosphatase gene to long-term combined applications of chemical fertilizers and organic materials. Appl. Soil Ecol. 2017, 119, 197–204. [Google Scholar] [CrossRef]
- Elkoca, E.; Turan, M.; Donmez, M.F. Effects of single, dual and triple inoculations with Bacillus subtilis, Bacillus megaterium and Rhizobium leguminosarum by Phaseoli on nodulation, nutrient uptake, yield and yield parameters of common bean. J. Plant Nutr. 2010, 33, 2104–2119. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).