Molecular Manipulation of the MiR396/GRF Expression Module Alters the Salt Stress Response of Arabidopsis thaliana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Expression Vector Construction
2.2. Plant Material Preparation for Agrobacterium-Mediated Transformation
2.3. Application of the 7-Day Salt Stress Treatment Regime
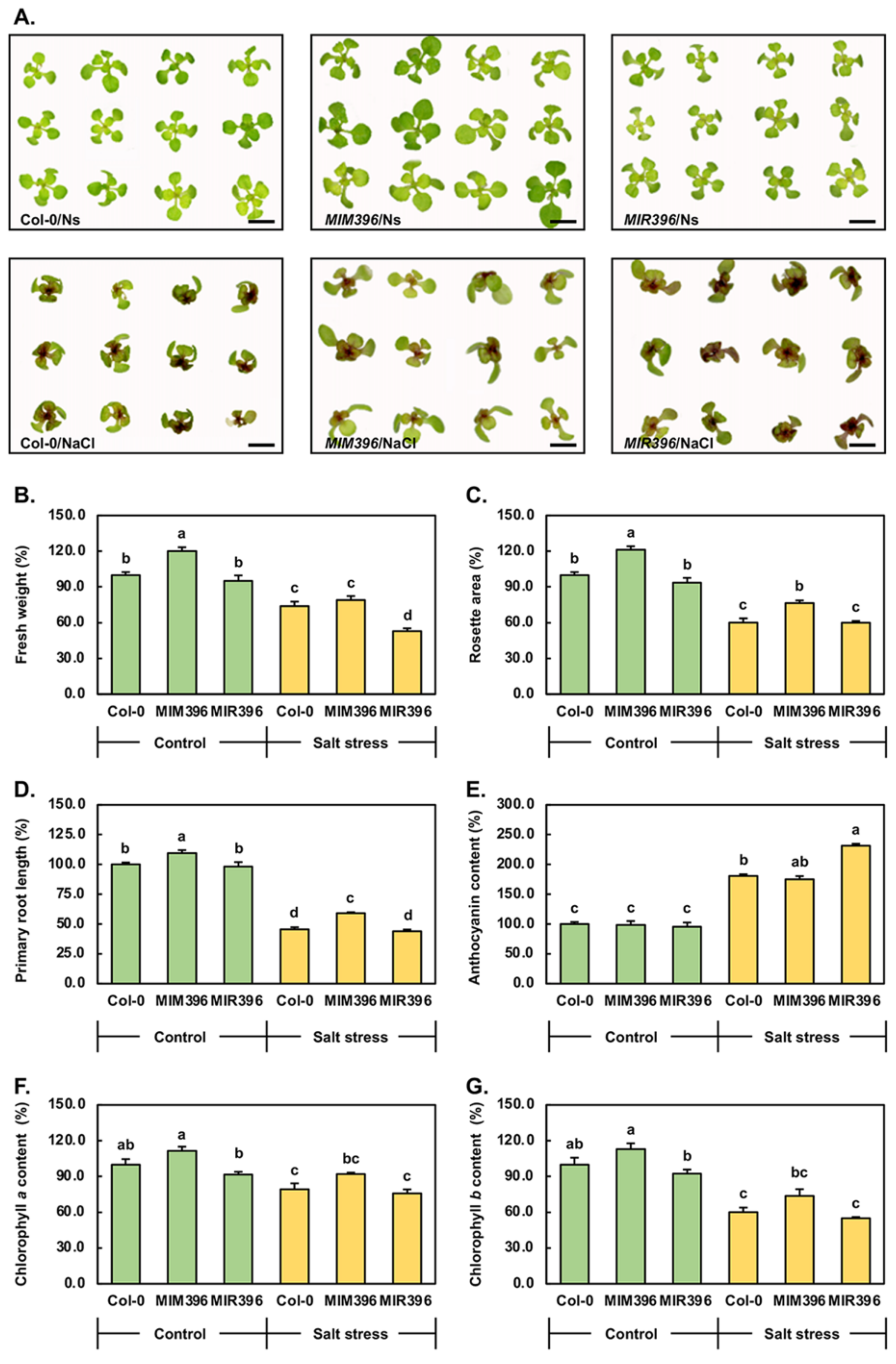
2.4. Phenotypic and Physiological Assessment of 15-Day-Old Control and Salt-Stressed Col-0, MIM396, and MIR396 Seedlings
2.5. RT-qPCR Assessment of miR396 Abundance and Target Gene Expression in 15-Day-Old Control and Salt-Stressed Col-0, MIM396, and MIR396 Seedlings
2.6. Statistical Analysis
3. Results
3.1. Phenotypic and Molecular Assessment of the MIM396 and MIR396 Transformant Lines
3.2. Phenotypic and Physiological Analysis of 15-Day-Old Control and Salt-Stressed Col-0, MIM396, and MIR396 Plants
3.3. Molecular Profiling of the miR396/GRF Expression Module in 15-Day-Old Control and Salt-Stressed Col-0, MIM396, and MIR396 Plants
3.4. Molecular Profiling of the Expression Trends of Three Additional Putative Target Genes of miR396-Directed Expression Regulation in Col-0, MIM396, and MIR396 Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Casati, P. Analysis of UV-B regulated miRNAs and their targets in maize leaves. Plant Signal. Behav. 2013, 8, e26758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Zhang, D.F.; Li, B.; Jia, G.Q.; Zhang, T.F.; Dai, J.R.; Li, J.S.; Wang, S.C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Baum, T.J. Complex feedback regulations govern the expression of miRNA396 and its GRF target genes. Plant Signal. Behav. 2012, 7, 749–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [Green Version]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Zhao, M. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar]
- Gao, F.; Wang, K.; Liu, Y.; Chen, Y.; Chen, P.; Shi, Z.; Li, S. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2015, 2, 15196. [Google Scholar] [CrossRef]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Han, N. miR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajoro, A.; Madrigal, P.; Muiño, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Ó’Maoiléidigh, D.S. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, R41. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014, 164, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, P.M.Y.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Yoshizumi, T. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid–and osmotic stress–responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Xie, K.; Xiong, L. Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol. Genet. Genom. 2010, 284, 477–488. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Profiling the abiotic stress responsive microRNA landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef] [Green Version]
- Hamza, N.B.; Sharma, N.; Tripathi, A.; Sanan-Mishra, N. MicroRNA expression profiles in response to drought stress in Sorghum bicolor. Gene Expr. Patterns 2016, 20, 88–98. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, J.; Li, Z.; Hu, Q.; Yuan, N.; Zhou, M.; Xia, X.; Noorai, R.; Saski, C.; Li, S.; et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 2019, 6, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Li, Y.; Cai, H.; Ji, W.; Guo, D.; Zhu, Y. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 2010, 231, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef]
- Chen, L.; Luan, Y.; Zhai, J. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 2015, 34, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Pegler, J.L.; Grof, C.P.L.; Eamens, A.L. Profiling of the differential abundance of drought and salt stress responsive microRNAs across grass crop and genetic model plant species. Agronomy 2018, 8, 118. [Google Scholar] [CrossRef] [Green Version]
- Pandey, G.; Yadav, C.B.; Sahu, P.P.; Muthamilarasan, M.; Prasad, M. Salinity induced differential methylation patterns in contrasting cultivars of foxtail millet (Setaria italica L.). Plant Cell Rep. 2017, 36, 759–772. [Google Scholar] [CrossRef]
- Puranik, S.; Bahadur, R.P.; Srivastava, P.S.; Prasad, M. Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet (Setaria italica (L.) P. Beauv). Mol. Biotechnol. 2011, 49, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.A.; Zaman, M.; Heng, L. Soil Salinity: Historical Perspectives and a World Overview of the Problem; Springer: Cham, Switzerland, 2018; pp. 43–53. [Google Scholar]
- Hossain, M. Present scenario of global salt affected soils, its management and importance of salinity research. Int. Res. J. Biol. Sci. 2019, 1, 1–3. [Google Scholar]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J.; et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 2020, 246–247, 153142. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.T.; Feng, S.J.; Li, H.; Faust, F.; Kleine, T.; Li, L.N.; Yang, Z.M. Salt stress-induced FERROCHELATASE1 improves resistance to salt stress by limiting sodium accumulation in Arabidopsis thaliana. Sci. Rep. 2017, 7, 14737–14753. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Yang, Z.; Liu, J.; Zhu, T.; Wei, X.; Fan, H.; Wang, B. Adaptation mechanism of salt excluders under saline conditions and its applications. Int. J. Mol. Sci. 2018, 19, 3668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Manoj, V.M.; Anunanthini, P.; Swathik, P.C.; Dharshini, S.; Ashwin Narayan, J.; Manickavasagam, M.; Sathishkumar, R.; Suresha, G.S.; Hemaprabha, G.; Ram, B.; et al. Comparative analysis of glyoxalase pathway genes in Erianthus arundinaceus and commercial sugarcane hybrid under salinity and drought conditions. BMC Genom. 2019, 19, 986. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Yang, Z.; Liu, M.; Wang, B. Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genom. 2015, 16, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Yue, J.; Wu, X.; Xu, C.; Yu, J. An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 2014, 65, 5415–5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lata, C.; Mishra, A.K.; Muthamilarasan, M.; Bonthala, V.S.; Khan, Y.; Prasad, M. Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e113092. [Google Scholar] [CrossRef] [Green Version]
- Pegler, J.L.; Nguyen, D.Q.; Grof, C.P.L.; Eamens, A.L. Profiling of the salt stress responsive microRNA landscape of C4 genetic model species Setaria viridis (L.) Beauv. Agronomy 2020, 10, 837. [Google Scholar] [CrossRef]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of salt tolerance related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.; Stewart, C.N. Jr.; Taki, F.A.; He, Q.; Liu, H.; Zhang, B. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol. J. 2014, 12, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Covarrubias, A.A.; Reyes, J.L. Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ. 2010, 33, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laby, R.J.; Kincaid, M.S.; Kim, D.; Gibson, S.I. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000, 23, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Eamens, A.L.; Smith, N.A.; Curtin, S.J.; Wang, M.B.; Waterhouse, P.M. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 2009, 15, 2219–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eamens, A.L.; Kim, K.W.; Curtin, S.J.; Waterhouse, P.M. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS ONE 2012, 7, e35933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mecchia, M.A.; Debernardi, J.M.; Rodriguez, R.E.; Schommer, C.; Palatnik, J.F. MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech. Dev. 2013, 130, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhou, Y.; Li, X.; Wang, X.; Dong, Y.; Wang, N.; Liu, X.; Chen, H.; Yao, N.; Cui, X.; et al. Tissue-specific regulation of Gma-miR396 family on coordinating development and low water availability responses. Front. Plant Sci. 2017, 8, 1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casadevall, R.; Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F.; Casati, P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 2013, 25, 3570–3583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto-Suárez, M.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; San Segundo, B. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci. Rep. 2017, 7, 44898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, Z.; Wang, J.; Yang, X.; Lu, H.; Miao, X.; Shi, Z. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3 8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Wang, J.; Ju, Z.; Liu, Q.; Li, S.; Tian, H.; Fu, D.; Zhu, H.; Luo, Y.; Zhu, B. Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. Plant Sci. 2016, 247, 1–12. [Google Scholar] [CrossRef]
- Gao, J.; Xiao, Q.; Yin, L.; He, G. Isolation of cDNA clones for genes up-regulated in drought-treated Alternanthera philoxeroides root. Mol. Biol. Rep. 2008, 35, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R.; Porcel, R.; Ruiz-Lozano, J.M. Regulation of root water uptake under abiotic stress conditions. J. Exp. Bot. 2012, 63, 43–57. [Google Scholar] [CrossRef]
- Liu, D.; Yu, D. MicroRNA (miR396) negatively regulates expression of ceramidase-like genes in Arabidopsis. Prog. Nat. Sci. 2009, 19, 781–785. [Google Scholar] [CrossRef]
- Wang, L.; Tian, Y.; Shi, W.; Yu, P.; Hu, Y.; Lv, J.; Fu, C.; Fan, M.; Bai, M.Y. The miR396-GRFs module mediates the prevention of photo-oxidative damage by brassinosteroids during seedling de-etiolation in Arabidopsis. Plant Cell. 2020, 32, 2525–2542. [Google Scholar] [CrossRef]
- Zheng, X.; Li, H.; Chen, M.; Zhang, J.; Tan, R.; Zhao, S.; Wang, Z. smi-miR396b targeted SmGRFs, SmHDT1, and SmMYB37/4 synergistically regulates cell growth and active ingredient accumulation in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2020, 39, 1263–1283. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants 2015, 1, 14027. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. MicroRNA regulatory mechanisms play different roles in Arabidopsis. J. Proteome Res. 2015, 14, 4743–4751. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [Green Version]
- Niemeier, S.; Alves Junior, L.; Merkle, T. Improvement of the design and generation of highly specific plant knockdown lines using primary synthetic microRNAs (pri-smiRNAs). BMC Res. Notes 2010, 3, 59. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.J.; Reyes, J.L.; Chua, N.H.; Gaasterland, T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004, 5, R65. [Google Scholar]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pegler, J.L.; Nguyen, D.Q.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Molecular Manipulation of the MiR396/GRF Expression Module Alters the Salt Stress Response of Arabidopsis thaliana. Agronomy 2021, 11, 1751. https://doi.org/10.3390/agronomy11091751
Pegler JL, Nguyen DQ, Oultram JMJ, Grof CPL, Eamens AL. Molecular Manipulation of the MiR396/GRF Expression Module Alters the Salt Stress Response of Arabidopsis thaliana. Agronomy. 2021; 11(9):1751. https://doi.org/10.3390/agronomy11091751
Chicago/Turabian StylePegler, Joseph L., Duc Quan Nguyen, Jackson M.J. Oultram, Christopher P.L. Grof, and Andrew L. Eamens. 2021. "Molecular Manipulation of the MiR396/GRF Expression Module Alters the Salt Stress Response of Arabidopsis thaliana" Agronomy 11, no. 9: 1751. https://doi.org/10.3390/agronomy11091751





