Abstract
The recently proposed species Ensifer aridi represents an interesting model to study adaptive mechanisms explaining its maintenance under stressful pedo-climatic conditions. To get insights into functions associated with hyperosmotic stress adaptation in E. aridi, we first performed RNAseq profiling of cells grown under sub-lethal stresses applied by permeating (NaCl) and non-permeating (PEG8000) solutes that were compared to a transcriptome from unstressed bacteria. Then an a priori approach, consisting of targeted mutagenesis of the gene encoding alternative sigma factor (rpoE2), involved in the General Stress Response combined with phenotyping and promoter gfp fusion-based reporter assays of selected genes was carried out to examine the involvement of rpoE2 in symbiosis and stress response. The majority of motility and chemotaxis genes were repressed by both stresses. Results also suggest accumulation of compatible solute trehalose under stress and other metabolisms such as inositol catabolism or the methionine cycling-generating S-adenosyl methionine appears strongly induced notably under salt stress. Interestingly, many functions regulated by salt were shown to favor competitiveness for nodulation in other rhizobia, supporting a role of stress genes for proper symbiosis’ development and functioning. However, despite activation of the general stress response and identification of several genes possibly under its control, our data suggest that rpoE2 was not essential for stress tolerance and symbiosis’ development, indicating that E. aridi possesses alternative regulatory mechanisms to adapt and respond to stressful environments.
1. Introduction
The rhizobium strain LMR001T (=LMG 31426T; =HAMBI 3707T) was isolated from desert sand dune in the Moroccan Merzouga desert [1]. This strain belongs to a new species within the Ensifer genus (formerly Sinorhizobium) that was proposed to be named E. aridi as isolates belonging to this new taxon were primarily isolated from arid or semi-arid environments in Africa, Asia, and America [1,2,3]. These bacteria are gram-negative, rod-shaped, motile, fast growing, mucous and develop a symbiotic interaction with diverse legume species able to survive under environmental constraints such as those found in deserts. More recently, E. aridi strains were recovered in Pakistan from mung bean grown in soils receiving as little as 12 mm of mean rainfall during the crop season [4], and strains have also been recovered from Retama monosperma growing in abandoned lead mine soils in Morocco [5]. The ability to develop the symbiosis with locally growing legume species was proposed to be due to accessory genome variations among strains, which most probably result from large plasmid exchanges through horizontal gene transfer [6].
Studying nitrogen-fixing rhizobia diversity and mechanisms underlying their adaptive responses to environmental constraints such as drought or saline stresses to improve the fertility of soils and arid land resilience was raised decades ago [7]. In a context of climate change, soil degradation, and demographic growth, there is now an emergency to act and propose solutions, as warned by the latest IPCC report on Climate change and land, published in August 2019. Capable to utilize a wide range of carbon sources and to develop the symbiosis with plants adapted to environmental constraints, Ensifer aridi represents an interesting model to study mechanisms involved in the adaptation to aridity.
Microorganisms such as rhizobia are adapted to contrasted growth conditions (soil, rhizosphere, in planta) and may be recovered from contrasted pedo-eco-climatic environments. They may face oxidative, pH, acid, hyperosmotic, and/or thermal stresses during their life cycle and have developed numerous mechanisms to thrive. Among functions associated to environmental stress adaptation, bacteria use various mechanisms including cell membrane structure and composition modifications [8], tuning its permeability and transporter activity to ensure proper nutrition and ionic exchanges. For example, phospholipids and the production of surface polysaccharides may be induced to prevent water loss and maintain a hydrated microenvironment around the cell [9,10,11]. Bacteria may respond by a modification in the transport and/or the metabolism of several molecules such as potassium ions, sugars, or amino acid derivatives that may act as compatible solutes to counteract osmotic pressure and/or protect bacteria against intracellular damages notably in rhizobia [12]. Among them, trehalose, a non-reducing disaccharide, appears promising as it may improve plant stress tolerance through symbiosis-mediated accumulation [13]. Bacteria may also trigger repair mechanisms to protect their macromolecules against damages by reactive oxygen species [14]. Finally, several transcription factors are also involved to modulate gene expression and enable the bacteria to rapidly respond to environmental changes through specific regulatory pathways.
Indeed, when subjected to environmental stressors, alphaproteobacteria, the bacterial class to which rhizobia belong, induce a conserved General Stress Response (GSR) that involves a regulatory system composed of an Extra Cytoplasmic Function (ECF) sigma factor (σECFG), an anti-sigma factor (NepR), and an anti-anti sigma factor (PhyR), whose activity depends on its phosphorylation by the Histidine kinase PhyK [15]. More recently, Lori and colleagues showed the involvement of a single domain response regulator MrrA as an intermediate between histidine kinases and PhyK, suggesting that PhyK may act as a phosphotransferase rather than a phosphate kinase [16]. Inducing molecules interacting with the primary sensors and some regulatory proteins involved in the GSR regulation remain to be determined. However, the phosphorylation of the multi-domain alternative two component system protein PhyR leads to its binding to the anti-sigma factor NepR, which is under normal growth conditions bound to σECFG, thus releasing its interaction with RNA polymerase, which triggers GSR [15].
Here, to explore the regulatory response of the new species E. aridi to salt- or water-induced hyperosmotic stresses, a genome-wide transcriptional study was performed. A mutant in the Ensifer aridi LMR001 σECFG encoding gene (rpoE2) was generated and studied to evaluate its involvement in the GSR-mediated regulation of salt-induced osmotic upshift of selected metabolisms’ transport and regulation or to assess the importance of the GSR to cope with such stress and to develop the symbiosis with compatible hosts.
2. Materials and Methods
2.1. Bacterial Growth
The bacterial strains and plasmids used in this study are listed in Table 1. Ensifer strains and derivatives were cultured in tryptone yeast extract (TY) medium [17] at 28 °C. Ensifer cultures were routinely started from glycerol stocks upon a primary growth on TY agar plates. E. coli strains were grown at 37 °C in Luria-Bertani medium [18]. Solid media contained 2% agar. Gentamycin (Gm, 10 μg mL−1), kanamycin (Km, 50 μg mL−1), ampicillin (Ap, 50 μg mL−1), rifampicin (Rif, 100 μg mL−1), and 5-bromo-4-chloro-3-indolyl-beta-D-galactosidase (X-gal, 80 μg mL−1) were added to the media for selection as required.

Table 1.
Strains, plasmids, and primers used in the study.
2.2. Phenotypic Characterization of the ΔrpoE2 Strain
2.2.1. Plant Nodulation Assay
Seeds of Vachellia tortilis subsp. raddiana and of the endemic Moroccan Vachellia gummifera [19] were scarified and surface sterilized in concentrated sulfuric acid (95%) with agitation for 1 h for V. tortilis and 30 min for V. gummifera and rinsed thoroughly with sterile water. Seeds were then germinated on water agar plates at 25 °C for 5 days in the dark. After germination, seedlings were planted in pots containing sand that was previously sterilized by autoclaving for 1 h at 120 °C. Ensifer aridi LMR001 WT and ∆rpoE2 were pre-grown in TY medium up to an optical density (OD) 600 nm of 0.7 and then washed and re-suspended in sterilized water prior to inoculation using 1 mL of washed cells directly after planting. Six individual plants were inoculated with each rhizobial strain or sterile water for the negative and positive controls. Plants were incubated in a growth chamber at 26 °C under a photoperiod of 16 h day and 8 h night and watered using N-free plant growth BD medium for watering [20] that was supplemented with KNO3 (3 mM) as a source of nitrogen for the positive controls. After 6 months of growth, the plants were harvested, and nodule number, plant shoot, and root fresh weights were recorded.
2.2.2. Motility Assay
LMR001 and ∆rpoE2 were grown at 28 °C in TY broth with shaking at 180 RPM to an OD (600 nm) of 0.7. After incubation, cells were pelleted, washed twice with sterile water, and resuspended in 10 volumes of sterile water. Swimming test assays were performed using 20 mL of TY media that were complemented either with 0, 5.85, or 11.7 g·L−1 NaCl (generating, respectively, ≈0, 0.1, and 0.25 M final NaCl concentrations) and 0.2% agar. Each medium was poured into Petri dishes, upon cooling to 50 °C in a water bath to avoid water loss by evaporation, and allowed to solidify in a horizontal laminar flow for 15 min. Finally, aliquots (5 µL) of washed cells were gently dispensed into the center of the plates and incubated at 28 °C for 5 days and the diameters of motile bacteria were measured after 2, 3, and 5 days of growth. The average diameter and the standard deviations from three biological replicates were calculated.
2.2.3. Salinity Tolerance
Ensifer aridi LMR001 and ∆rpoE2 derivatives were first grown in liquid TY cultures up to an OD (600 nm) of 0.7, and 1 mL was used to inoculate 50 mL of liquid TY medium as well as TY complemented with 14.6, 17.5, or 23.4 g L−1 of NaCl (generating, respectively, 0.25 M, 0.3 M, and 0.4 M final salt concentrations). Cultures were incubated in a rotary shaker (180 RPM) at 28 °C and the OD (600 nm) was recorded after 4, 8, 24, 48, 120, and 144 h incubation. The mean ODs (600 nm) of three independent cultures were calculated for both strains and at each time point with their respective standard deviations.
2.2.4. Sensitivity to Detergent, Acid, and Oxidative Stresses
Sensitivities to oxidative, acid, and detergent stresses induced, respectively, by H2O2, HCl, and SDS were compared between the wild-type LMR001 and the ∆rpoE2 strains. Briefly, 4 mL of bacterial cultures of E. aridi or the ∆rpoE2 strain (pre-grown to OD 600 nm ≈ 0.7 in TY liquid medium) were added to 16 mL of soft TY agar containing 0.7% of agar precooled to 45 °C and poured onto TY agar plates. Once solidified, a paper disc (6-mm diameter) was placed on the plate and 5 µL of H2O2 (2 M), HCl (5.5 M), or SDS (10% w/v) was spotted onto it. Plates were incubated at 28 °C for three days prior to recording the diameters of the inhibition zones, and the average of three replicates were averaged.
2.3. RNAseq Analysis and Characterization of Differentially Regulated Genes in LMR001 upon NaCl- or PEG8000-Induced Hyperosmotic Stresses
For the RNAseq analysis, the method was adapted from previously published work [21]. Briefly, six biological replicates were first grown in TY medium up to an OD (600 nm) of 0.9 (±0.05) in 250 mL Erlenmeyer flasks at 30 °C with 140 rpm. For each replicate, 15 mL were added to an equal volume of pre-warmed TY containing either nothing (Controls), NaCl at 500 mM (Salt treatment, 250 mM final concentration), or PEG8000 at 24% (W:V) (PEG8000 treatment, 12% final concentration), and were further incubated at 30 °C and 140 rpm. After 2 ½ h of incubation, which is less than a generation time, 1/10th volume of stop buffer (ethanol containing phenol at a final concentration of 5%) was added to each culture (6 replicates × 3 treatments) to inhibit RNAses and centrifuged 4 min at 8000× g and 4 °C. The pellets were snap-frozen in liquid nitrogen and kept in the freezer at −80 °C until use.
Total RNAs were purified from each replicate and treated with the RiboPureTM kit (AMBION), which included a DNAse I treatment, according to manufacturer’s recommendations. RNA extracts from the six replicates were then pooled and subjected to two successive rounds of rRNA subtractions with Microbe ExpressTM kit (AMBION), following manufacturer’s recommendations. Purified mRNAs were sent to Montpellier Genomix for sequencing. Three libraries corresponding to the three treatments (Control, Salt, and PEG8000) were prepared, following illumina kit instructions. Briefly, for each library, 100 ng of mRNA were first fragmented chemically, and reverse transcribed using random primers and SuperScript IITM to generate first-strand cDNAs. The second strand was synthesized using DNA polymerase I and RNAse H. The cDNA fragments were then end-repaired, a single ‘A’ base was added, and adapters were ligated. These products were enriched by PCR to select for cDNA fragments containing adaptors and to create the final cDNA library. The efficacy of the library construction was checked in a quality control step that involved measuring the adapter-cDNA size and concentration on an Agilent DNA 1000 chip. Sequencing libraries were denatured with sodium hydroxide and diluted to 6 pM in hybridization buffer for loading onto a single lane of an Illumina HiSeq 2000 flowcell V1.5. Cluster formation, primer hybridization, and single-read, 100 sequencing cycles were performed on cBot and HiSeq2000 (Illumina, San Diego, CA, USA), respectively.
After sequencing, reads that passed the sequencing platform quality filter were imported into CLC Genomics workbench (V5.5.1) and mapped to the RAST-predicted genomic objects [6] including putative protein encoding genes (PEGs) (listed in Table S1) and RNAs from Ensifer aridi LMR001 using 95% of sequence similarity over 70% of read length. Reads that mapped several PEGs were discarded. Finally, the read table obtained upon mapping the three libraries onto PEGs were normalized using a LOESS transformation that appeared to be the most adapted method, as the PEG8000 treatment resulted in an enhanced recovery of the least abundant transcripts as compared to the control or salt treatments (see Section 3). Genes showing an average coverage of 50× and more than 2-fold differences between pairwise comparisons were considered as differentially regulated.
To explore the major changes associated to the transcriptomes, the predicted proteome of the LMR001 strain was functionally annotated using COG and KEGG classifications [22,23]. Differentially regulated genes were then used to estimate major classes and functions associated to salt- and PEG8000-induced hyperosmotic stresses.
2.4. Real-Time PCR on Selected Genes
The list of genes targeted, as well as primers used for real-time PCR experiments, are listed in Table 1. LMR001 was cultivated either in TY or in TY complemented with NaCl at 250 mM, as described for the RNAseq experiment. Three biological replicates for each growth condition were extracted using SV Total RNA Isolation kit (Promega), following manufacturer’s recommendations. RNA samples were treated with Turbo DNAse (Applied Biosystems) to remove any DNA contaminant from RNA samples. For each sample and replicate, 1 μg of total RNA was reverse-transcribed with 400 U of Super-Script II (Invitrogen) and random hexamer primers. The qPCRs were performed on a Strategene MXP3005P system using Power SYBER green master mix (Applied Biosystems). PCR cycling conditions consisted of a primary 10-min incubation at 95 °C, followed by 40 cycles consisting of 15 s at 95 °C and 30 s at 60 °C. Primer specificity and efficiencies and the formation of primer dimers were estimated by dissociation curves. The expression levels of a selection of 12 genes were standardized by using the gene hrcA as reference. PCR efficiency (E) for each amplicon was calculated using the linear regression method on the log (fluorescence)-per-cycle-number data using Stratagene MXpro software. All qPCRs were performed in two technical replicates using total RNA isolated from three biological samples so as to assess biological variability. Data for each sample were expressed relative to the expression levels of hrcA by using the mathematical model described previously, which determines the relative quantification of a target gene in comparison to a reference (ref) gene between treatment and control samples [24]. The relative expression ratio (R) of a target gene is calculated based on efficiency (E) and the threshold cycle (Ct) of an unknown sample versus a control and expression in comparison to a reference gene. The average relative quantity for each gene under study was calculated and log2-transformed. Means and standard deviations of these final log2 ratios were calculated using data from three biological replicates.
2.5. LMR001 ΔrpoE2 Mutant Construction
The regions bordering the rpoE2 gene (PEG2539) were first amplified by PCR with custom-designed primers (see Table 1). The primer pair rpoE2-P2539-A-XI/rpoE2-P2539-B-HIII was used to amplify a 478 bp sequence located upstream of the rpoE2 gene including the first 37 bases of the coding sequence. Furthermore, the primer rpoE2-P2539-B-HIII contained in 5′ a HindIII site as well as 13 bases that were complementary to the 5‘ end of the primer rpoE2-P2539-C-HIII that was used together with rpoE2-P2539-D-XI to amplify a 473 sequence of the 3′ region of the rpoE2 gene including the last 77 nucleotides. The PCR was performed using, as templates, genomic DNA extracts from LMR001 obtained, following the method reported previously [25], using 10 mL of bacterial liquid cultures at early stationary phase. First, PCR reactions were performed to amplify both the 5′ and the 3′ flanking regions using the proof-reading enzyme Phusion (Thermo-Fischer) and recommended chemistry, except for primers (1 µM final concentrations for forward and reverse primers). The cycling conditions included a primary denaturation step of 30 s at 98 °C followed by 35 cycles (98 °C, 10 s/56 °C, 30 s/72 °C, 45 s) and a final elongation step of 7 min at 72 °C. PCR products were purified using Illustra™ GFX PCR DNA and Gel Band purification kits (GE Healthcare), following manufacturer’s recommendations. Then a second PCR reaction was set up, using as primers and matrix, purified PCR products obtained that contain complementary sequences, thus enabling priming of the amplification. The 25-µL reactions contained 5 µL of 5× GoTaq buffer (Promega), 1.25 µL of MgCl2 (25 mM), 2.5 µL of dNTP mix (2.5 mM each), 0.125 µL GoTaq (5 U/µL), 2 µL of each purified PCR products obtained, and nuclease free water. A primary PCR cycling consisting of a denaturation step of 95 °C for 5 min followed by 5 cycles (95 °C, 30 s/50 °C, 45 s/72 °C, 50 s) and 5 min at 72 °C was performed so as to generate chimeric DNA molecules consisting of merged 5′ and 3′ regions. Then, 15 µL of the mix (3 µL of 5× GoTaq buffer, 0.75 µL of MgCl2 (25 mM), 1.5 µL of dNTP mix, 0.125 µL of GoTaq (5 U/µL), 4 µL of external primers rpoE2-P2539-A-XI and rpoE2-P2539-D-XI, and 1.625 µL of nuclease free water) was added to the PCR reaction and subjected to the following cycling conditions: a denaturation step of 95 °C for 5 min followed by 25 cycles (95 °C, 30 s/55 °C, 45 s/72 °C, 50 s) and 5 min at 72 °C. The PCR product was purified from 1% agar gel using promega Wizard®® SV Gel and PCR Clean-Up System, following recommended protocol. As a result of the two amplifications’ process, the PCR-amplified and -purified product was cloned as a fragment into a pGEM-T Easy Vector (Promega kit) and transferred into XL2-Blue Ultra-competent Cells. The resulting positive clones were selected and the plasmid was extracted by using the Wizard®® Plus SV Minipreps DNA Purification System, digested by XbaI enzyme and ligated into suicide vector pJQ200SK [26]. Enzymes were purchased from Promega. The latter plasmid was used to transform Ensifer aridi LMR001 cells by bipartite conjugation method using S17-1 E. coli strains as a donor [27]. Transformants were selected on TY plates containing Rif and 5% sucrose to allow selection of knockout clones that had undergone a double cross-over. Double cross-over gene replacement in mutants was verified by screening for antibiotic resistant/ sensibility phenotypes (Rif and Gm, respectively) and also PCR to verify deletion by using specific external and internal primers.
2.6. Use of Promoter-Gfp Fusions for Transcriptional Analyses in the Wild-Type and the ∆rpoE2 Strains
The promoter region of genes involved in the regulation of the methionine cycle (nesR, PEG5735; sahR, PEG2239), in the regulation of the GSR (rsiA1, PEG2540; rsiB1, PEG2541), in the trehalose transport (thuE,PEG6268) metabolism (otsB, PEG4868; treZ, PEG5323), a glycosyl transferase (PEG4866) located upstream of the otsBA operon in reverse orientation or the regulation of trehalose transport and endogenous utilization (thuR, PEG6269), and in the inositol catabolism (iolC, PEG373) and regulation (iolR, PEG374) was amplified using primers listed in Table 1. PCR products were cleaned up using illustra™ GFX™ PCR DNA and Gel Band Purification Kits (GE Healthcare). The purified PCR products were ligated into TOPO®® TA Cloning®® vector (INVITROGEN), following the manufacturer’s recommendations, and were transferred into E. coli XL2 Blue Ultra-competent Cells. Plasmid DNA was extracted using the Wizard Plus SV Minipreps DNA Purification System kit (Promega), following the manufacturer’s recommendations. Plasmids containing the promoters were sequenced by Genoscreen (Lille, France) to verify sequence integrity. Inserts were excised using the restriction enzyme EcoRI and ligated into a pPROBE NT vector, which allows for transcriptional fusions with the green fluorescent protein (GFP) encoding gene gfp [28], previously digested with EcoRI and dephosphorylated. The ligation mix was transferred into electro-competent E. coli strain S17-1. The selection of clones containing the promoters in the proper orientation was done by using the forward primers of the promoters with the primer gfp-Rev50 located in the reverse orientation of the 5′ end of the gfp gene, which results in positive PCR amplification when correct. The pPROBE NT constructs were transferred into LMR001 RifR or ΔrpoE2 strains by biparental mating. Briefly, Donor S17-1 cells containing the pPROBE NT derivatives were grown on LB agar containing Kanamycine (50 µg mL−1) and the recipient cells were grown onto TY Agar containing Rifampicine (100 µg mL−1). Half a loop of donor cells and 2–3 loops of recipient cells were resuspended in separate Eppendorf tubes containing 1 mL of TY medium each to wash cells from antibiotics. The tubes were mixed by mild vortexing and centrifuged at 7000× g for 7 min and resuspended in 0.5 mL of fresh TY medium. Then, 100 µL of each of the donor and the recipient cells were mixed onto fresh TY plates and grown for 2 days at 28 °C. Half loops of cells were then streaked onto a TY agar plate containing the two antibiotics (Kanamycine and Rifampicine) for selection of transconjugants. Individual colonies were finally isolated and verified by PCR prior conservation in 20% glycerol at −80 °C. When needed, strains carrying the promoter pPROBE constructs were grown on TY agar plates supplemented with appropriate antibiotics at 28 °C. Using sterile toothpicks, fresh colonies were transferred to 10 mL liquid TY containing Kanamycine and grown overnight with shaking at 28 °C. Cells were washed in fresh TY medium and resuspended in an equal volume of fresh TY medium prior to use in transcriptional assays. For each transcriptional assay, the same bacterial inocula (70 μL) were used to inoculate 140 μL TY medium supplemented or not with NaCl (250 mM final concentration) in 96-well plates. The plates were transferred in a spectrofluorometer microplate reader (TECAN Infinite M200) set at 28 °C and subjected to the following cycling conditions. After a preliminary shaking of 900 s using orbital shaking mode with 1-mm amplitude, the mean of three reads of optical density (600 nm) and three reads of fluorescence levels (excitation filter at 485 nm and emission filter at 535 nm) were recorded every 30 min for all wells and at least 9 h. The experiment was performed three times. For the analysis, each plate was treated separately and relative fluorescence values were recorded as follow. First, the fluorescence data were transformed for each time point as a percentage of the maximal fluorescence levels observed in the experiment to enable the comparison between each replicate. The relative fluorescence values were finally obtained by dividing the normalized fluorescence obtained by the corrected OD600 nm measures at each time point. Data from the bacterium containing empty vector were finally subtracted and the means from the three independent repetitions were finally calculated.
3. Results and Discussion
To explore the possible mechanisms that Ensifer aridi possibly uses to face hyperosmotic stresses, a transcriptomic approach was realized using NaCl as a permeating solute and PEG8000 as a non-permeating solute that, respectively, resulted in salt- and water-induced osmotic upshifts. To focus on functions associated with these stresses and disregard mechanisms linked to cell death, mild stresses were triggered using sublethal concentrations for less than a generation time. First, growth kinetics of LMR001 were compared in TY medium containing various concentrations of Salt (NaCl) and PEG8000. Concentration of 250 mM of salt and 12% (w/v) of PEG8000 were chosen, as these concentrations reduced the growth of the bacterium without inhibiting its growth. Indeed, six independent cultures were pre-grown in TY Medium up to an OD (600 nm) of 0.9. Each pre-culture was further used to inoculate an equal volume of either TY or TY complemented with Salt 0.5 M (0.25 M final concentration) or with PEG8000 24% w/v (12% final concentration). After 2 ½ h of incubation, the mean OD (600 nm) of the control rose from 0.52 to 0.94 (ΔOD = 0.42), which was significantly higher than the ΔOD obtained in media containing salt and PEG8000 (−10% in PEG8000 growth medium and −40% in salt containing medium; p-values < 0.05, Student Test), confirming that bacteria were subjected to a mild stress that did not inhibit totally their growth.
All cultures were subjected to a total RNA extraction and purified RNAs were pooled according to the treatments prior to rRNA subtractions. Finally, enriched mRNA samples from controls (TY), salt-stressed bacteria (250 mM NaCl), or Water-stressed bacteria (12% PEG8000) were sent for sequencing at the Montpellier MGX sequencing platform. Sequencing consisted of 100 cycles on Hiseq2000 that generated, respectively, 109.95-, 95.17-, and 105.89-million sequence reads of 100 bp (>31 Gb). For each library, the reads were mapped to the LMR001 RAST Predicted Encoding Genes (PEGs) and rRNAs as well as the spike phiX using CLC genomics Workbench (see Materials and Methods for parameters used). Results of the sequencing and mapping are shown in Figure S1. Even though the number of reads that mapped to rRNAs were high (55.39, 35.1, and 39.23 million reads for control, salt, and PEG libraries, respectively), the procedure enabled us to recover significant numbers of reads that mapped to LMR001-predicted encoding gene sequences (respectively 34.91, 35.12, and 42.79 million reads). If we consider that 95% of the RNAs in cells are corresponding to ribosomal RNAs, while only 5% are mRNAs, the rRNA subtraction procedure enabled the removal of 91.6%, 94.7%, and 95.2% of rRNAs (respectively, for the libraries TY, TY + 250 mM salt, and TY + 12% PEG8000). These rates are comparable to those obtained in Mesorhizobium [21] and indicate that the procedure is also well suited to Ensifer aridi.
Scatter plots showing distributions of reads obtained upon mapping on LMR001 PEGs in control (X Axis) as a function of salt or PEG8000 treatments (Y Axes) (Figure S2A,B, respectively) or M-A plots (showing the logarithmic values of mean read numbers for control and salt or control and PEG8000, as a function of the log2 ratios (Figure S2C,D, respectively)) indicated that while salt and control showed rather centered distributions, the sample PEG8000 returned a strong bias for lowly expressed transcripts that were over-represented in this library as compared with control and salt libraries. This bias may result from the presence of PEG8000 remaining in pelleted cells as these molecules favor precipitation of nucleic acids [29,30]. To estimate whether the presence of PEG8000 in the samples also influenced the recovery of particular size fractions of the RNAs, we compared the relative abundances of read counts in the three libraries upon normalization to the total read numbers mapped to PEGs for all predicted genes and ordering based on PEG size, their mean sequencing coverage (obtained from the three libraries), and, finally, by predicted gene order, which is independent of size or coverage. Figure S3 shows the relative abundances of reads obtained in the three libraries (Y axis) for all genes upon ordering, as described above (X axes), as a piled 100% Surface area graphics. The comparison of profiles confirmed that only a bias toward predicted genes lowly transcribed was found. These results clearly show that the PEG8000 facilitated the recovery of rare transcripts using the method used here, and that this enrichment was higher as the coverage was lowering (Figure S3B).
Furthermore, box plots showing the distributions of raw read counts (as log values) in the three libraries also indicated differences in sequencing medians and quartile values (Figure S2E). To normalize for differences in sequencing depth between the three libraries and obtain statistics on differentially regulated genes without multiple replicates, DESeq can be used [31]. However, given the strong bias obtained from the PEG8000 library that generates incomparable expression levels for the genes that are the least transcribed, we could not use this package, and lack statistics from the RNAseq data. In order to normalize for the bias related to lowly expressed genes’ enrichment in the PEG8000 library and for the sequencing depth differences in the three libraries, we performed locally estimated scatter plot smoothing (LOESS) using a window of 0.1 (Log read counts) for both comparisons, Control vs. Salt or PEG8000. This normalization step enabled us to retrieve expected read counts from all three libraries and to obtain the relative abundances of each gene in the three libraries. As expected, distributions of normalized data presented more centered distributions (Figures S2F–J and S3D–F) even though a slight bias for the least transcribed genes remained visible for genes whose coverage was below 30× mean coverage (Figure S3E). To support the RNAseq data, a selection of genes was analyzed by RT-qPCR using fresh bacterial cultures.
The relative expression of 12 genes showing contrasting expressions levels upon salt stress was monitored using hcrA as normalizer by qPCR assays on samples obtained from three independent samples. This analysis indicated a good correlation with RNAseq data (R2 = 0.917) (Figure S4), which confirms that RNAseq data can be used to explore the major changes associated with stress under study, even though qPCR data indicated lower differential expressions as compared to RNAseq, the latter representing a direct measure of transcript levels.
To explore functions associated to salt- and water-induced hyperosmotic stresses, predicted genes showing 50× of mean coverage between two compared treatments (Salt vs. Control or PEG8000 vs. Control) and presenting fold changes of 2 or greater were first selected. Analyses of COGs and KEGG orthologous assignation of differentially regulated genes enabled us to estimate major metabolic and cellular functional changes associated to stresses. Table S1 shows the entire list of LMR001-predicted encoding genes (PEGs), their predicted annotations (RAST-SEED based, COG classification, and KEGG orthology), and RNAseq data.
Globally, we found that salt stress induced a stronger transcriptional response as compared to PEG8000. Addition of salt in the medium triggered an induction of 473 PEGs and a repression of 708 PEGs, of which 331 and 549, respectively, were classified into at least one COG class. In the PEG8000 treatment, only 75 PEGs presented more than 50× mean coverage and a 2-fold induction, and 237 were repressed, of which, 67 and 122, respectively, were assigned to COG classes. This difference may result from a stronger stress induced by the short-term exposure to salt, as shown by the higher impact salt had on delta growth after 2 ½ h of incubation. It could also possibly be due to distinct global responses induced by the two solutes used to induce osmotic upshifts already shown to be partially conserved in Sphingomonas [32].
The distribution of COGs recovered from regulated genes in relation to the total number of COGs in each class is shown in Figure 1. Besides an upregulation of approximately 30% of genes involved in translation ribosomal structure and biogenesis encoding, for example, ribosomal proteins, we found that a majority of genes involved in chemotaxis and motility were repressed by both stresses. Indeed, nearly 60% of genes with COG assigned to that class were repressed upon stress.
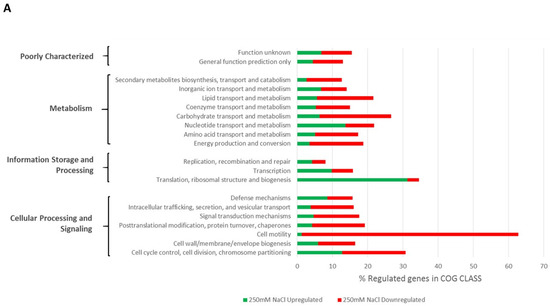
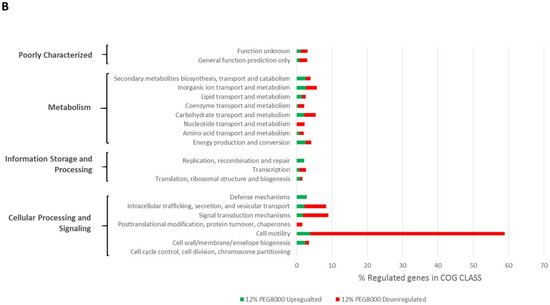
Figure 1.
COG-based functional classification of genes regulated (>50× mean coverage and a minimum of 2-fold change) upon salt (A) or PEG8000 (B) treatments. The horizontal bars represent the % of genes regulated in the corresponding COG Class and, for each bar, the relative proportion of genes that are up- or down-regulated is shown in green and red, respectively. The COG class description and processes to which they belong are shown on the left of graphs.
Hyperosmotic stresses have already been shown to repress motility gene transcription in many bacteria [32,33,34,35,36] including rhizobia [37]. Interestingly the repression of motility is often associated to an induction of genes involved in the synthesis of biofilm components such as surface polysaccharides or proteins [32,35,38,39].
The regulation of motility genes and surface components have been studied in Ensifer meliloti and several transcriptional regulators have been characterized. Regulators such as CbrA, VisN/VisR, and Rem alter the regulation of many genes involved in motility or surface components [40,41], which were shown to be modulated by the ExpR/SinI quorum-sensing system [42], emphasizing the complexity of the regulatory circuits involved. Fine-tuning regulation of motility and cell surface components is of particular importance for bacteria, notably, host-associated ones such as rhizobia that have to adapt to contrasted environments (soil-rhizosphere-nodule) [43]. Among genes that were repressed by the osmotic upshift, VisR/VisN- and Rem-predicted encoding genes (respectively, PEG655/PEG654 and PEG688) were identified. This may explain the downstream down-regulation of motility genes identified here, even though the repression was less marked under PEG80000 condition, with notably visR and visN that were less than 2-fold repressed as compared to the control (see Table S1). Even though the design did not enable us to obtain statistical significance on the RNAseq data set, we also found several genes involved in exopolysaccharides to be induced upon salt stress (exoQ, exoF, exoY exoH, exoO, and exoN, respectively, PEG5904-5906, PEG5914, PEG5919, and PEG5920), supporting a possible developmental switch, as shown in S. meliloti [40].
Among the metabolic pathways known to respond to hyperosmotic stresses, we found that two trehalose de novo biosynthesis pathways were induced. The genes encoding for Alpha,alpha-trehalose-phosphate synthase (otsA, PEG4869) and the Trehalose-6-phosphate phosphatase (otsB, PEG4868) that converts UDP glucose and glucose 6P into trehalose by two successive reactions were induced more strongly under salt stress (fold changes of 3.7 and 5.2, respectively) than upon PEG8000-induced hyperosmotic stresses (fold changes of 2 and 1.6, respectively). The second pathway that converts maltodextrin to trehalose by a Malto-oligosyltrehalose synthase (treY, PEG5229) and a Malto-oligosyltrehalose trehalohydrolase (treZ, PEG5323) appeared, in contrast, more induced under PEG treatment with respective fold changes of 4.9 and 4.3 when these were induced 1.8 and 3.4 times by salt. We also found a strong repression of the trehalose ABC-type import system thuEFGK (PEG6268, 6266, 6265, and 6264) and the trehalose utilization genes thuAB (PEG6263 and 6262), which showed from 9- to 20-fold reduction in expression levels upon salt exposure. These genes were also repressed upon PEG8000 exposure but to a lesser extent (fold changes ranging from 1.5 to 2.7). The induction of trehalose de novo synthesis upon osmotic upshift has been documented in many bacteria, including rhizobia [12,44,45,46]. Ensifer aridi most probably also uses this common non-reducing disaccharide upon osmotic upshift. However, in Ensifer aridi LMR001, we found that the ABC-type transport system encoded by thuEFGK as well as trehalose catabolic genes thuAB [47,48] was strongly repressed, while thuE, thuG, and thuA genes were found induced upon osmotic upshift in Ensifer meliloti [37]. This difference may result from different growth media used and, notably, the presence of trehalose in the medium that appears to induce both the thuEFGK and thuAB genes [47,48]. Interestingly, a repression of the trehalose utilization genes combined with an upregulation of trehalose de novo synthesis should result in increased intracellular trehalose levels that may favor rapid osmo-adaptation to stressful conditions. Interestingly, S. meliloti mutants in the thuAB genes improved the efficiency of nodulation and, more precisely, the early root hair IT-infection competitiveness [48], which may, however, depend on the host genotype [49]. Symbiotic bacteria that are prone to trehalose biosynthesis such as E. aridi may provide advantageous adaptation to plants under abiotic stresses [13].
Among metabolisms that were altered by the salt stress, we found a large locus (PEG2271—PEG2284) shown to be involved in erythritol, adonitol, and L-arabitol transport and catabolism [50]. Predicted genes eryA (PEG2278) and eryB (PEG2275) showed reduced transcription (respectively, 6.7- and 3.7-fold repression) upon salt exposure, while eryC (PEG2271) presented a 2.2-fold repression but a 30× mean coverage. The operon predicted to encode erythritol ABC-type transport system (PEG2283-2279) located just upstream of the catabolic genes was even more strongly repressed by salt (9.5–24-fold repression) when the predicted erythritol transcriptional repressor gene eryD (PEG2284) presented a 10-fold repression, which was surprising, as we would expect an induction. Erythritol transport and catabolism was found to be important for nodule occupancy in Rhizobium leguminosarum [51] but not in Sinorhizobium meliloti, as shown by competitiveness the Triose Phosphate isomerase tpiB mutant that cannot utilize erythritol [52]. Given that the erythritol catabolic genes are induced by the presence of erythritol, the repression observed could be due to an increased and rapid utilization of this polyol under salt stress, leading to reduced external erythritol concentration that may also explain the strong repression observed for thuEFGK and thuAB genes. Other possible explanations may be the involvement of additional transcriptional regulators involved, such as a putative EryR homolog (PEG2270) that was, however, not regulated in the present study or, finally, a possible alternative utilization of an intermediary product of the erythritol catabolism suggested to be involved in its regulation [53].
In contrast to erythritol, inositol transport and metabolism were strongly induced by salt. First a myo-inositol ABC transport system (PEG5305-5307) showed 4.8- to 8.1-fold induction upon salt exposure, as compared to the control treatment. Furthermore, genes encoding 2 myo inositol-2 dehydrogenases idhA (PEG2503) and iolY (PEG375) that covert, respectively, D-chiro inositol and scyllo-inositol into 2-keto myo-inositol presented salt inductions (respectively, 6.7- and 1.8-fold changes). The genes iolE (PEG371), iolD (PEG372), iolB (PEG370), and iolC (PEG373) that are involved in the successive conversion of 2-keto-myo-inositol into 3-D-trihydroxycyclohexane-1,2-dione, 5-deoxy-glucuronic acid, 2-deoxy-5-keto-D-gluconic acid, and 2-deoxy-5-keto-D-gluconic acid 6-phosphate (DKGP) showed 3.6- to 5.6-fold inductions in the salt treatment, as compared to control. These results support active inositol import and catabolism in salt-treated cells that are under the control of the predicted transcriptional repressor IolR (PEG374). Again, iolR was surprisingly also up-regulated by salt with a 2.2-fold induction. It was shown that 2 deoxy 5 keto D gluconic acid 6 P (DKGP), a key product of inositol catabolism, participates in the regulation of many genes involved in this metabolism by interacting with the repressor IolR through competitive binding [54,55], suggesting accumulation of this intermediate product. Interestingly, mutations in genes involved the catabolism of myo-inositol, such as idhA, were shown to negatively impact symbiosis development and, particularly, competitiveness for nodule occupancy with their host plants, as shown in S. meliloti [56], S. fredii [57], and Rhizobium leguminosarum bv. viciae [58], which suggest involvement of inositol derivatives for proper symbiosis establishment and functioning. The induction of inositol catabolism, together with potential trehalose accumulation, as evidenced by trehalose metabolic and transport genes’ expression levels found here upon salt stress together with their respective involvement in symbiosis establishment, further supports a role of such stress-responsive genes in improving symbiosis establishment and functioning [59,60].
We also found an induction by salt stress exposure of genes involved in the methionine cycle. It included the adenosyl homocysteinase ahcY (PE3356, 11.4-fold induction) that converts S-adenosyl-L-homocysteine (SAH) into homocysteine that is, in turn, converted to methionine, either from glycine betaine by a betaine--homocysteine S-methyltransferase BHMT (PEG1969, 18.5-fold induction) or from a sequential conversion of 5, 10-methylene tetrahydrofolate by a reductase encoded by metF (PEG2238, 4.6-fold induction) to methyl tetrahydrofolate that is converted with homocysteine as substrate into methionine and tetrahydrofolate with a 5-methyltetrahydrofolate-homocysteine methyltransferase by metH (PEG3206, 6.7-fold induction). Finally, we found a strong induction (20.5-fold induction by salt) of the S-adenosylmethionine (SAM) synthetase-encoding gene metK (PEG438) that converts methionine into SAM, a universal methyl donor used as cofactor in methyl transferase reactions involved in methylation of nucleic acids and protein or in secondary metabolite biosynthesis such as polyamines. Furthermore, two transcriptional factors previously shown to regulate the methionine cycle were also found differentially regulated. The LuxR-type solo nesR gene (PEG5735), shown to affect the methionine cycle by Patankar and Gonzalez [61], presented a 2-fold induction upon salt stress, while the sahR gene encoding an ArsR-type transcriptional regulator was found 9 times more expressed in the salt condition. SahR was shown to activate several genes of the methionine cycle, including itself upon biding to a SAH, which releases its binding from their promoter, thus enabling their transcriptional activation [62]. An active methionine cycle was shown to be important for osmoprotection in several rhizobia including S. fredii [63] or in S. meliloti [19,61]. Interestingly, a mutation in metH, part of the main pathway for methionine synthesis in S. meliloti strain 102F34, or nesR in Rm2011 reduced competitiveness for nodule occupancy when the mutants were challenged to the wild type [19,61]. Such observation was also observed in a metZ mutant of R. etli impaired for methionine biosynthesis when inoculated to P. vulgaris [64]. Furthermore, a cobO mutant, incapable to biosynthesize the vitamin B12 necessary for MetH-mediated methionine biosynthesis in S. fredii HH103, was severely affected in nodulation. Altogether, these data support a role of the methionine in nodulation competitiveness in the rhizobium legume symbiosis and, again, further suggest a possible improvement of symbiosis performance by rhizobial stress-response genes [60].
Salt stress also induced the transcription of the alternative extracytoplasmic function sigma factor rpoE2 (PEG2539), as well as the anti-sigma factor gene rsiA1, a nepR homolog (PEG2540), and rsiB1, a phyR homolog (PEG2541), that were at least induced by 3 folds. This result indicates that the General Stress Response (GSR) was activated in Ensifer aridi, as already shown in other alphaproteobacteria [65]. The effects of mutations in these genes have been studied in various organisms including rhizobial genera, such as Bradyrhizobium [66,67], Sinorhizobium [68,69], or Rhizobium [70], and have shown their involvement as master regulators of the stress response, which may also be found in Ensifer aridi. Other regulatory systems have been evidenced, such as the lovK-LovR Two-Component System in Caulobacter [71] or in S. meliloti, in which homologs of the rsiA1 and rsiB1 (anti-sigma and anti-anti-sigma factors, respectively) have recently been shown to modulate rpoE2 regulation [68]. This shows that the stress response is complex and finely tuned, which is not surprising, as bacteria have to cope with different but sometimes combined stresses, which may utilize common molecular mechanisms depending on the stress. It is interesting to note that we also found homologs of rsiA1 and rsiB1, named, respectively, rsiA2 (PEG753) and rsiB2 (PEG752), in our strain, which may act similarly as shown for S. meliloti. However, our transcriptomic data showed that the stresses applied here did not affect their transcriptional status, which remained unchanged. Additional studies are required to decipher their regulatory functions. Surprisingly, while bradyrhizobia mutated in genes responsible for the alternative extracytoplasmic function, sigma factor functioning showed strong symbiosis development deficiencies [66,67], symbiosis development and functioning was not impaired by such mutations in Sinorhizobium meliloti [69,72] or Rhizobium etli [70]. Interestingly, Lang and colleagues [72] demonstrated that the absence of such symbiosis deficiency in S. meliloti was not due to extracytoplasmic function sigma factors (ecfG) redundancy. They obtained similar symbiosis phenotypes when all ecfG were mutated and compared to the wild-type strain. Regarding the responses of ecfG mutants to stresses, studies on S. meliloti showed contrasting results. In a primary study, Sauviac and colleagues [69] showed that rpoE2 mutants did not present phenotypic differences with the wild type upon various stresses (heat, salt, acidic pH, or H2O2) in both exponential and stationary growth phases. Later, Flechard and colleagues [45,73] showed that a rpoE2 mutant in a stationary growth phase was sensitive to hydrogen peroxide and hyperosmotic stresses. More recently, Lang and colleagues confirmed Sauviac’s results as no or subtle differences were found in the rpoE2 mutant or a mutant lacking all ECF-like genes, which is surprising, as many stress-responsive genes were identified in the RpoE2 regulon [69,72]. These differences may result from distinct mutation procedures and/or media used in these studies, which included, for example, a deletion of the anti-sigma factors in the study by Lang and colleagues [72]. In Bradyrhizobium, such mutations increased sensitivity of B. japonicum to heat shock and desiccation [66]. Furthermore, recent work on B. diazoefficiens showed that ecfG mutation resulted in increased sensitivity to various stresses including hyperosmotic and ionic stresses [74]. More interestingly, in both Bradyrhzobium diazoefficiens and Sinorhizobium meliloti, a rpoE2 mutation was shown to repress trehalose 6 Phosphate pathway, which was involved in hyperosmotic stresses’ tolerance [45] and was sufficient to impair early symbiotic development in the more sensitive B. diazoefficiens species [74], further supporting that trehalose is a key compatible solute for Bradyrhizobium stress adaptation and nodulation, as shown in B. japonicum [46]. The absence of a symbiotic phenotype observed in the rpoE2 mutant of S. meliloti may result from a high tolerance to hyperosmotic stress in this species as compared to the Bradyrhizobium species. Such stress may be encountered, notably, in infection threads, which is supported by the restoration of symbiosis by addition of exogenous trehalose in B. diazoefficiens ΔotsA strain still capable to import it or in a strain capable to endogenously synthesize other compatible osmoprotectants [74]. To explore the function of RpoE2 and its possible role in the regulation of several functions shown to be transcriptionally regulated by salt stress as well as in the tolerance of the LMR001 bacterium to various stresses and symbiosis development, a deletion mutant of the rpoE2 gene (ΔrpoE2) was generated and phenotypically characterized.
First, the growth of ΔrpoE2 was compared to that of LMR001 in TY medium complemented with salt at 250, 300, and 400 mM final concentrations (Figure 2A). We found that, even though the WT strain reached the stationary phase earlier in TY medium containing 250 mM of NaCl as compared to ΔrpoE2, both strains reached comparable optical densities after 120 h of incubation. Secondly, when we looked at the motility of the two strains, while we found a significant reduction of motility for the two strains upon addition of salt at both concentrations tested (0.1 and 0.25 M), which corroborates transcriptional data observed by RNAseq, no clear differences were observed between the two strains (Figure 2B). Sensitivities of the two strains to detergent, oxidative, and acid stresses were estimated by the inhibitory growth diameter induced by the diffusion in the agar medium of 5 µL concentrated SDS, H2O2, or HCl that were spotted on small paper discs placed in the center of soft agar plates previously inoculated with the strains (see Materials and Methods). The concentration of these molecules decreased as they diffused away from the discs imbibed with the reagents. A measure of the growth inhibition diameter, thus, enabled us to compare tolerance of the strains. We did not find any difference between the wild-type and the ΔrpoE2 strains (Figure 2C), suggesting that the rpoE2 mutation did not alter cell membrane stability under such stresses. Finally, both the LMR001 and ΔrpoE2 strains developed the symbiosis with the tree species V. gummifera and V. tortilis and there were no significant differences associated to plant weights or nodulation (Table 2). These results show that the ECF sigma factor RpoE2 is not essential for motility, symbiosis development, and tolerance to the stresses tested, suggesting that other functions and/or regulators are involved in these processes, as suggested in Sinorhizobium meliloti [69,72].
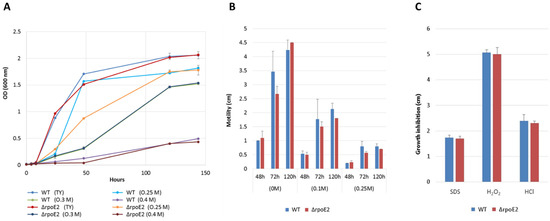
Figure 2.
Effect of the rpoE2 mutation on stress tolerance and motility of LMR001. Ensifer aridi LMR001 (WT) and the LMR001 rpoE2 deletion mutant (ΔrpoE2) growths were compared in liquid TY medium complemented with 0, 0.1, 0.25, 0.3, or 0.4 M of NaCl (A). Their motility was assessed in semi-solid TY complemented with salt at 0, 0.1, or 0.25 M by measurement of the diameter of the swimming colonies after 2, 3, or 5 days of incubation (B). Finally, the diameters of the growth inhibition zones were compared for the two strains inoculated in top agar plates, to the center of which patches imbued with 5 µL of SDS (10% w/v), H2O2 (5.5 M), or HCl (2 M) were placed (C).

Table 2.
Effect of the rpoE2 mutation on symbiosis development with Vachellia.
We further explored the involvement of the extra cytoplasmic function sigma factor in the transcriptional regulation of genes involved in the inositol catabolism (iolC and iolR), methionine cycle regulation (nesR and sahR), trehalose transport, and metabolism (thuR, thuE, otsB, and treZ) including a glycosyl transferase (PEG4866) located upstream of the otsBA operon in reverse orientation or in the regulation of the GSR (rsiA1, rsiB1). Their promoter regions were fused in front of the reporter gene encoding GFP in a mobilizable vector, and resulting constructions were further transferred into LMR001 and ΔrpoE2. Both strains containing the pPROBE derivatives were then grown either in TY medium or in TY complemented with NaCl at a final concentration of 250 mM in 96-microtiter plates and fluorescence with the optical density (600 nm) and were recorded every half hour.
Figure 3 shows the relative expression levels of the GFP under the control of selected promoters recorded every 30 min for 9 h. The relative expression of the GFP varied with the promoter and, for some of them, with the strain. The approach did not allow us to detect differences between salt and control growth conditions for a strain that would require less stable GFP and a more sensitive approach to detect early response to hyperosmotic stress. However, the methodology enabled us to identify transcriptional changes resulting from the mutation. We found that the mutation resulted in an increased transcription of four genes, while it reduced the expression of three genes. An increased transcription of otsB was obtained in the ΔrpoE2 strain, indicating that RpoE2-mediated GSR in E. aridi involves the T6P pathway, as shown in S. meliloti [45] or in B. diazoefficiens [74]. We also found that for rsiA1, the anti-sigma factor that is located upstream of and overlaps the 5′ end of the rpoE2 gene forming an operon was less expressed in the mutant, which suggests, again, that, as shown in other alphaproteobacteria [65], rpoE2 is autoregulated in E. aridi. Given the high similarities, both in terms of syntheny and sequence between S. meliloti and E. aridi for many genes discussed here (for example, 97.8% similarity for RpoE2), we tested whether the same promoter motif GGAACN16-17CGTT that likely represents the −35 and −10 elements of the RpoE2 responsive genes [69,72] could be found in the target promoter sequences of E. aridi genes studied here. This motif was found upstream of rsiA1 and rsiB1, supporting a conserved RpoE2-mediated regulatory mechanism. It was also found twice in the promoter region of the treZ gene, whose transcription was, however, not found significantly different when comparing the WT and ΔrpoE2 strains, and in the range of background values. Regarding the otsB promoter region, we found two motifs that were close to the consensus sequence: (GGAACN17CGAT and AGAACN17CGAT). Finally, one motif with three nucleotide differences as compared to the consensus sequence was found in front of the thuR gene (GGAAAN17AGAT). However, the involvement of these regions requires additional work to assess whether these are directly regulated by RpoE2 and to exclude the possibility that a hidden mutation resulting from the mutagenesis impacted our data. Surprisingly, the most severe transcriptional repression resulting from the mutation was found for thuR, encoding a HTH DNA-Binding LacI-type repressor showing homologies to aglR that may be involved in transport and utilization of trehalose through thuEFGK and thuAB expression [47,48]. It is interesting to note the presence of thuR consensus sequence motifs found among rhizobiales upstream of these operons in LMR001 as well as in front of aglR and aglEFGAK that share partially their motifs [75], which may, however, explain cross-regulation of these transport systems by aglR [47]. Given (1) ThuR homology to AglR, (2) the trehalose transport specificity observed of ThuE, ThuF, ThuG, and ThuK, shown by Jensen and colleagues, and (3) evolutionary and comparative analyses within rhizobiales of LacI-family transcription factors that predicts ThuR as a regulator of two operons (thuEFGKAB involved in trehalose transport and utilization as well as thuR) [47,75], our data further support that ThuR is also autoregulated and involved in the regulation of transport and utilization of trehalose in E. aridi. Nevertheless, additional work is required to characterize the regulatory pathways involved and, notably, whether thuR is directly or indirectly regulated by RpoE2.
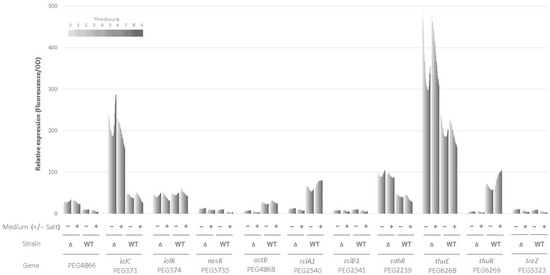
Figure 3.
Effect of the rpoE2 mutation on transcriptional activity of selected genes. The histograms show the relative expression (see Materials and Methods) of gfp under the control of promoters from selected genes mobilized in wild-type Ensifer aridi LMR001 (WT) or the ΔrpoE2 derivative (Δ) strains grown in TY medium containing salt (NaCl at 250 mM) or not (+/−), as indicated at the bottom of the figure. Relative expressions were recorded every 30 min for 9 h and are shown by gray bars of increasing darkness with time (colors indicated on the top left of the figure).
Among genes whose transcriptional activities were increased by the mutation, we found thuE, which may result from the thuR repression in the ΔrpoE2 strain. Interestingly, the glycosyl transferase (PEG4866) located upstream of the otsBA operon in reverse orientation also showed an increased transcriptional activity in ΔrpoE2, which may suggest RpoE2-mediated repression in the wild-type strain. Surprisingly, iolC and sahR genes involved, respectively, in the catabolism of myo-inositol and the regulation of the methionine cycle were also found more expressed in ΔrpoE2 background while they were found induced upon salt stress in the wild-type LMR001 strain. Whether these inductions result from a direct RpoE2-mediated repression remains to be addressed, but our data suggest a complex stress response in E. aridi, which involves RpoE2 that can, nevertheless, be complemented.
4. Conclusions
To explore the mechanisms by which Ensifer aridi LMR001 copes with hyperosmotic stresses, 31 Gb of sequence data were generated by three RNAseq libraries constructed from cells grown either in control medium or upon addition of the permeating solute NaCl at 250 mM or the non-permeating solute PEG8000 at 12% w/v. As shown in other bacteria, hyperosmotic stresses resulted in the repression of motility genes, which was confirmed by phenotypic tests. These effects may be considered for improving the rhizobium–legume symbiosis in soils containing salt, notably, through inoculation of adapted bacteria to ensure contact between non-motile rhizobia and roots. We also found that the extra-cytoplasmic function sigma factor-encoding gene rpoE2 was induced and probably involved, as shown in other rhizobia, in its autoregulation and the induction of trehalose de novo biosynthesis. A concomitant repression of trehalose utilization genes thuAB suggests intracellular accumulation of trehalose in stressed cells. Salt also induced inositol catabolism and the methionine cycle, suggesting a possible role of inositol derivatives and possibly methylation in the early response to such stress. In addition, a combination of directed mutagenesis of rpoE2 and the use of promoter-gfp fusion reporter system suggested that RpoE2 regulates positively thuR, a LacI-type regulator probably involved in trehalose import and utilization. In contrast, the mutation resulted in increased expression of genes involved in inositol catabolism or the regulation of the methionine cycle that, given their induction in the wild-type bacterium upon salt stress, supports the involvement of alternative regulatory circuits. However, additional experiments are required to ascertain that ΔrpoE2 does not carry hidden mutations, which could have impacted our data. Intriguingly, previous studies showed that a mutation of trehalose utilization genes in S. meliloti resulted in improved competitiveness for nodulation, which is also favored with active inositol catabolism and methionine cycle, suggesting that hyperosmotic stress occurs in the early infection of leguminous hosts. The rpoE2 deletion did not result in increased stress sensitivity or symbiosis functioning, suggesting the involvement of alternative regulatory circuits in Ensifer aridi for stress adaptation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agronomy11091787/s1. Figure S1: Sequencing and mapping of the three RNAseq libraries. Figure S2: Distributions of RNAseq data. Figure S3: Effect of PEG order, mean coverage, or size on the relative abundance of reads obtained in the three libraries. Figure S4: Comparison of the relative expression levels of selected genes by RNAseq and qPCR. Table S1: Predicted LMR001-encoding gene annotation and mapping.
Author Contributions
Conceptualization, A.L.Q. and A.F.-M.; methodology, A.L.Q., M.B., E.D., D.S. and H.A.-B.; formal analysis, M.B. and A.L.Q.; investigation, M.B., A.L.Q. and I.S.; resources, E.D. and D.S.; data curation, E.D., D.S. and A.L.Q.; writing—original draft preparation, M.B. and A.L.Q.; writing—review and editing, all authors.; visualization, A.L.Q. and M.B.; supervision, A.L.Q. and A.F.-M.; project administration, A.L.Q. and A.F.-M.; funding acquisition, A.L.Q. and A.F.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the French Institute of Research for the Development (IRD) through the LMI LBMV and an ARTS PhD grant for M.B.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Ensifer aridi LMR001 genome browser, Fasta nucleotide and protein sequences and annotations can be accessed through the RAST server https://rast.nmpdr.org/ (accessed 11 November 2020) using “guest” as login and password under job nb “188440” with ID “6666666.88372”. RNAseq data have been deposited in SRA in NCBI and can be accessed through the Bioproject PRJNA312299.
Acknowledgments
The authors would like to acknowledge Samir El Qaidi for methodological support for the construction of the mutant and Abderrahim Ferradous from the Centre Régional de Recherches Forestière in Marrakech for Vachellia seeds. M.G.X. acknowledges financial support from France Génomique National infrastructure, funded as part of “Investissement d’Avenir” program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Sakrouhi, I.; Belfquih, M.; Sbabou, L.; Moulin, P.; Bena, G.; Filali-Maltouf, A.; Le Quéré, A. Recovery of Symbiotic Nitrogen Fixing Acacia Rhizobia from Merzouga Desert Sand Dunes in South East Morocco—Identification of a Probable New Species of Ensifer Adapted to Stressed Environments. Syst. Appl. Microbiol. 2016, 39, 122–131. [Google Scholar] [CrossRef]
- Rocha, G.; Le Queré, A.; Medina, A.; Cuéllar, A.; Contreras, J.-L.; Carreño, R.; Bustillos, R.; Muñoz-Rojas, J.; Villegas, M. del C.; Chaintreuil, C.; et al. Diversity and Phenotypic Analyses of Salt- and Heat-Tolerant Wild Bean Phaseolus filiformis Rhizobia Native of a Sand Beach in Baja California and Description of Ensifer aridi Sp. Nov. Arch. Microbiol. 2020, 202, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Tak, N.; Awasthi, E.; Bissa, G.; Meghwal, R.R.; James, E.K.; Sprent, J.S.; Gehlot, H.S. Multi Locus Sequence Analysis and Symbiotic Characterization of Novel Ensifer Strains Nodulating Tephrosia Spp. in the Indian Thar Desert. Syst. Appl. Microbiol. 2016, 39, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Hakim, S.; Imran, A.; Mirza, M.S. Phylogenetic Diversity Analysis Reveals Bradyrhizobium yuanmingense and Ensifer aridi as Major Symbionts of Mung Bean (Vigna radiata L.) in Pakistan. Braz. J. Microbiol. 2021, 52, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Lamin, H.; Alami, S.; Bouhnik, O.; ElFaik, S.; Abdelmoumen, H.; Bedmar, E.J.; Missbah-El Idrissi, M. Nodulation of Retama monosperma by Ensifer aridi in an Abandonned Lead Mine Soils in Eastern Morocco. Front. Microbiol. 2019, 10, 1456. [Google Scholar] [CrossRef]
- Le Quéré, A.; Tak, N.; Gehlot, H.S.; Lavire, C.; Meyer, T.; Chapulliot, D.; Rathi, S.; Sakrouhi, I.; Rocha, G.; Rohmer, M.; et al. Genomic Characterization of Ensifer aridi, a Proposed New Species of Nitrogen-Fixing Rhizobium Recovered from Asian, African and American Deserts. BMC Genom. 2017, 18, 85. [Google Scholar] [CrossRef] [PubMed]
- Zahran, H.H. Rhizobium -Legume Symbiosis and Nitrogen Fixation under Severe Conditions and in an Arid Climate. Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef]
- Denich, T.J.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Effect of Selected Environmental and Physico-Chemical Factors on Bacterial Cytoplasmic Membranes. J. Microbiol. Methods 2003, 52, 149–182. [Google Scholar] [CrossRef]
- Chang, W.-S.; van de Mortel, M.; Nielsen, L.; Nino de Guzman, G.; Li, X.; Halverson, L.J. Alginate Production by Pseudomonas putida Creates a Hydrated Microenvironment and Contributes to Biofilm Architecture and Stress Tolerance under Water-Limiting Conditions. J. Bacteriol. 2007, 189, 8290–8299. [Google Scholar] [CrossRef]
- Finn, S.; Condell, O.; McClure, P.; Amézquita, A.; Fanning, S. Mechanisms of Survival, Responses and Sources of Salmonella in Low-Moisture Environments. Front. Microbiol. 2013, 4, 331. [Google Scholar] [CrossRef]
- Lebre, P.H.; De Maayer, P.; Cowan, D.A. Xerotolerant Bacteria: Surviving through a Dry Spell. Nat. Rev. Microbiol. 2017, 15, 285–296. [Google Scholar] [CrossRef]
- Vriezen, J.A.C.; de Bruijn, F.J.; Nüsslein, K. Responses of Rhizobia to Desiccation in Relation to Osmotic Stress, Oxygen, and Temperature. Appl. Environ. Microbiol. 2007, 73, 3451–3459. [Google Scholar] [CrossRef]
- Sharma, M.P.; Grover, M.; Chourasiya, D.; Bharti, A.; Agnihotri, R.; Maheshwari, H.S.; Pareek, A.; Buyer, J.S.; Sharma, S.K.; Schütz, L.; et al. Deciphering the Role of Trehalose in Tripartite Symbiosis Among Rhizobia, Arbuscular Mycorrhizal Fungi, and Legumes for Enhancing Abiotic Stress Tolerance in Crop Plants. Front. Microbiol. 2020, 11, 509919. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, C.Y. Generally Stressed Out Bacteria: Environmental Stress Response Mechanisms in Gram-Positive Bacteria. Integr. Comp. Biol. 2020, 60, 126–133. [Google Scholar] [CrossRef]
- Fiebig, A.; Herrou, J.; Willett, J.; Crosson, S. General Stress Signaling in the Alphaproteobacteria. Annu. Rev. Genet. 2015, 49, 603–625. [Google Scholar] [CrossRef]
- Lori, C.; Kaczmarczyk, A.; de Jong, I.; Jenal, U. A Single-Domain Response Regulator Functions as an Integrating Hub To Coordinate General Stress Response and Development in Alphaproteobacteria. mBio 2018, 9, e00809-18. [Google Scholar] [CrossRef]
- Beringer, J.E. R Factor Transfer in Rhizobium leguminosarum. Microbiology 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor: New York, NY, USA, 1989; Volume 2, ISBN 978-0-87969-309-1. [Google Scholar]
- Barra, L.; Fontenelle, C.; Ermel, G.; Trautwetter, A.; Walker, G.C.; Blanco, C. Interrelations between Glycine Betaine Catabolism and Methionine Biosynthesis in Sinorhizobium meliloti Strain 102F34. J. Bacteriol. 2006, 188, 7195–7204. [Google Scholar] [CrossRef]
- Broughton, W.J.; Dilworth, M.J. Control of Leghaemoglobin Synthesis in Snake Beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Maynaud, G.; Brunel, B.; Mornico, D.; Durot, M.; Severac, D.; Dubois, E.; Navarro, E.; Cleyet-Marel, J.-C.; Le Quéré, A. Genome-Wide Transcriptional Responses of Two Metal-Tolerant Symbiotic Mesorhizobium Isolates to Zinc and Cadmium Exposure. BMC Genom. 2013, 14, 292. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, Information, Knowledge and Principle: Back to Metabolism in KEGG. Nucl. Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef]
- Tatusov, R.L. The COG Database: New Developments in Phylogenetic Classification of Proteins from Complete Genomes. Nucl. Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Kuo, T. A Simple and Rapid Method for the Preparation of Gram-Negative Bacterial Genomic DNA. Nucl. Acids Res. 1993, 21, 2260. [Google Scholar] [CrossRef]
- Quandt, J.; Hynes, M.F. Versatile Suicide Vectors Which Allow Direct Selection for Gene Replacement in Gram-Negative Bacteria. Gene 1993, 127, 15–21. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Miller, W.G.; Leveau, J.H.J.; Lindow, S.E. Improved Gfp and InaZ Broad-Host-Range Promoter-Probe Vectors. Mol. Plant Microbe Interact. 2000, 13, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Wang, Q.; Hu, Y.; Zhao, Y.; Li, Y.; Cheng, H.; Huang, H. Co-Extraction of High-Quality RNA and DNA from Rubber Tree (Hevea brasiliensis). Afr. J. Biotechnol. 2012, 11, 9308–9314. [Google Scholar] [CrossRef]
- Roose-Amsaleg, C.L.; Garnier-Sillam, E.; Harry, M. Extraction and Purification of Microbial DNA from Soil and Sediment Samples. Appl. Soil Ecol. 2001, 18, 47–60. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Alavi, P.; Starcher, M.R.; Zachow, C.; Müller, H.; Berg, G. Root-Microbe Systems: The Effect and Mode of Interaction of Stress Protecting Agent (SPA) Stenotrophomonas rhizophila DSM14405T. Front. Plant Sci. 2013, 4, 141. [Google Scholar] [CrossRef] [PubMed]
- Dressaire, C.; Moreira, R.N.; Barahona, S.; Alves de Matos, A.P.; Arraiano, C.M. BolA Is a Transcriptional Switch That Turns Off Motility and Turns On Biofilm Development. mBio 2015, 6, e02352-14. [Google Scholar] [CrossRef]
- Li, S.; Liang, H.; Wei, Z.; Bai, H.; Li, M.; Li, Q.; Qu, M.; Shen, X.; Wang, Y.; Zhang, L. An Osmoregulatory Mechanism Operating through OmpR and LrhA Controls the Motile-Sessile Switch in the Plant Growth-Promoting Bacterium Pantoea alhagi. Appl. Environ. Microbiol. 2019, 85, e00077-19. [Google Scholar] [CrossRef] [PubMed]
- Steil, L.; Hoffmann, T.; Budde, I.; Völker, U.; Bremer, E. Genome-Wide Transcriptional Profiling Analysis of Adaptation of Bacillus subtilis to High Salinity. J. Bacteriol. 2003, 185, 6358–6370. [Google Scholar] [CrossRef]
- Zhou, A.; Baidoo, E.; He, Z.; Mukhopadhyay, A.; Baumohl, J.K.; Benke, P.; Joachimiak, M.P.; Xie, M.; Song, R.; Arkin, A.P.; et al. Characterization of NaCl Tolerance in Desulfovibrio vulgaris Hildenborough through Experimental Evolution. ISME J. 2013, 7, 1790–1802. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Ferreras, A.; Pérez-Arnedo, R.; Becker, A.; Olivares, J.; Soto, M.J.; Sanjuán, J. Transcriptome Profiling Reveals the Importance of Plasmid PSymB for Osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 7617–7625. [Google Scholar] [CrossRef] [PubMed]
- Bahlawane, C.; McIntosh, M.; Krol, E.; Becker, A. Sinorhizobium meliloti Regulator MucR Couples Exopolysaccharide Synthesis and Motility. Mol. Plant Microbe Interact. 2008, 21, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, W.; Wang, Y.; Wu, L.; Liu, X.; Yan, T.; Alm, E.; Arkin, A.; Thompson, D.K.; Fields, M.W.; et al. Transcriptome Analysis of Shewanella oneidensis MR-1 in Response to Elevated Salt Conditions. J. Bacteriol. 2005, 187, 2501–2507. [Google Scholar] [CrossRef] [PubMed]
- Gibson, K.E.; Barnett, M.J.; Toman, C.J.; Long, S.R.; Walker, G.C. The Symbiosis Regulator CbrA Modulates a Complex Regulatory Network Affecting the Flagellar Apparatus and Cell Envelope Proteins. J. Bacteriol. 2007, 189, 3591–3602. [Google Scholar] [CrossRef]
- Sourjik, V.; Muschler, P.; Scharf, B.; Schmitt, R. VisN and VisR Are Global Regulators of Chemotaxis, Flagellar, and Motility Genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 2000, 182, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Hoang, H.H.; Gurich, N.; González, J.E. Regulation of Motility by the ExpR/Sin Quorum-Sensing System in Sinorhizobium meliloti. J. Bacteriol. 2008, 190, 861–871. [Google Scholar] [CrossRef]
- Janczarek, M. Environmental Signals and Regulatory Pathways That Influence Exopolysaccharide Production in Rhizobia. Int. J. Mol. Sci. 2011, 12, 7898–7933. [Google Scholar] [CrossRef]
- Domínguez-Ferreras, A.; Soto, M.J.; Pérez-Arnedo, R.; Olivares, J.; Sanjuán, J. Importance of Trehalose Biosynthesis for Sinorhizobium meliloti Osmotolerance and Nodulation of Alfalfa Roots. J. Bacteriol. 2009, 191, 7490–7499. [Google Scholar] [CrossRef]
- Flechard, M.; Fontenelle, C.; Blanco, C.; Goude, R.; Ermel, G.; Trautwetter, A. RpoE2 of Sinorhizobium meliloti Is Necessary for Trehalose Synthesis and Growth in Hyperosmotic Media. Microbiology 2010, 156, 1708–1718. [Google Scholar] [CrossRef]
- Sugawara, M.; Cytryn, E.J.; Sadowsky, M.J. Functional Role of Bradyrhizobium Japonicum Trehalose Biosynthesis and Metabolism Genes during Physiological Stress and Nodulation. Appl. Environ. Microbiol. 2010, 76, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.B.; Peters, N.K.; Bhuvaneswari, T.V. Redundancy in Periplasmic Binding Protein-Dependent Transport Systems for Trehalose, Sucrose, and Maltose in Sinorhizobium meliloti. J. Bacteriol. 2002, 184, 2978–2986. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.B.; Ampomah, O.Y.; Darrah, R.; Peters, N.K.; Bhuvaneswari, T.V. Role of Trehalose Transport and Utilization in Sinorhizobium meliloti--Alfalfa Interactions. Mol. Plant Microbe Interact. 2005, 18, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Ampomah, O.Y.; Jensen, J.B.; Bhuvaneswari, T.V. Lack of Trehalose Catabolism in Sinorhizobium Species Increases Their Nodulation Competitiveness on Certain Host Genotypes. New Phytol. 2008, 179, 495–504. [Google Scholar] [CrossRef]
- Geddes, B.A.; Oresnik, I.J. Genetic Characterization of a Complex Locus Necessary for the Transport and Catabolism of Erythritol, Adonitol and l-Arabitol in Sinorhizobium meliloti. Microbiology 2012, 158, 2180–2191. [Google Scholar] [CrossRef] [PubMed]
- Yost, C.K.; Rath, A.M.; Noel, T.C.; Hynes, M.F. Characterization of Genes Involved in Erythritol Catabolism in Rhizobium leguminosarum Bv. viciae. Microbiology 2006, 152, 2061–2074. [Google Scholar] [CrossRef] [PubMed]
- Poysti, N.J.; Oresnik, I.J. Characterization of Sinorhizobium meliloti Triose Phosphate Isomerase Genes. J. Bacteriol. 2007, 189, 3445–3451. [Google Scholar] [CrossRef]
- Geddes, B.A.; Pickering, B.S.; Poysti, N.J.; Collins, H.; Yudistira, H.; Oresnik, I.J. A Locus Necessary for the Transport and Catabolism of Erythritol in Sinorhizobium meliloti. Microbiology 2010, 156, 2970–2981. [Google Scholar] [CrossRef]
- Kohler, P.R.A.; Choong, E.-L.; Rossbach, S. The RpiR-Like Repressor IolR Regulates Inositol Catabolism in Sinorhizobium meliloti. J. Bacteriol. 2011, 193, 5155–5163. [Google Scholar] [CrossRef]
- Herrou, J.; Crosson, S. Myo -Inositol and d-Ribose Ligand Discrimination in an ABC Periplasmic Binding Protein. J. Bacteriol. 2013, 195, 2379–2388. [Google Scholar] [CrossRef] [PubMed]
- Kohler, P.R.A.; Zheng, J.Y.; Schoffers, E.; Rossbach, S. Inositol Catabolism, a Key Pathway in Sinorhizobium meliloti for Competitive Host Nodulation. Appl. Environ. Microbiol. 2010, 76, 7972–7980. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Krishnan, A.H.; Kim, Y.-W.; Wacek, T.J.; Krishnan, H.B. A Functional Myo -Inositol Dehydrogenase Gene Is Required for Efficient Nitrogen Fixation and Competitiveness of Sinorhizobium fredii USDA191 To Nodulate Soybean (Glycine max [L.] Merr.). J. Bacteriol. 2001, 183, 2595–2604. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fry, J.; Wood, M.; Poole, P.S. Investigation of Myo -Inositol Catabolism in Rhizobium leguminosarum Bv. viciae and Its Effect on Nodulation Competitiveness. Mol. Plant Microbe Interact. 2001, 14, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Gourion, B.; Berrabah, F.; Ratet, P.; Stacey, G. Rhizobium–Legume Symbioses: The Crucial Role of Plant Immunity. Trends Plant Sci. 2015, 20, 186–194. [Google Scholar] [CrossRef]
- Rodrigo da-Silva, J.; Alexandre, A.; Brígido, C.; Oliveira, S. Can Stress Response Genes Be Used to Improve the Symbiotic Performance of Rhizobia? AIMS Microbiol. 2017, 3, 365–382. [Google Scholar] [CrossRef]
- Patankar, A.V.; González, J.E. An Orphan LuxR Homolog of Sinorhizobium meliloti Affects Stress Adaptation and Competition for Nodulation. Appl. Environ. Microbiol. 2009, 75, 946–955. [Google Scholar] [CrossRef]
- Novichkov, P.S.; Li, X.; Kuehl, J.V.; Deutschbauer, A.M.; Arkin, A.P.; Price, M.N.; Rodionov, D.A. Control of Methionine Metabolism by the SahR Transcriptional Regulator in Proteobacteria: Control of Methionine Metabolism in Proteobacteria. Environ. Microbiol. 2014, 16, 1–8. [Google Scholar] [CrossRef]
- Jiang, J.Q.; Wei, W.; Du, B.H.; Li, X.H.; Wang, L.; Yang, S.S. Salt-Tolerance Genes Involved in Cation Efflux and Osmoregulation of Sinorhizobium fredii RT19 Detected by Isolation and Characterization of Tn5 Mutants. FEMS Microbiol. Lett. 2004, 239, 139–146. [Google Scholar] [CrossRef][Green Version]
- Taté, R.; Riccio, A.; Caputo, E.; laccarino, M.; Patriarca, E.J. The Rhizobium etli metZ Gene Is Essential for Methionine Biosynthesis and Nodulation of Phaseolus vulgaris. Mol. Plant Microbe Interact. 1999, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Francez-Charlot, A.; Kaczmarczyk, A.; Fischer, H.-M.; Vorholt, J.A. The General Stress Response in Alphaproteobacteria. Trends Microbiol. 2015, 23, 164–171. [Google Scholar] [CrossRef]
- Gourion, B.; Sulser, S.; Frunzke, J.; Francez-Charlot, A.; Stiefel, P.; Pessi, G.; Vorholt, J.A.; Fischer, H.-M. The PhyR-σ EcfG Signalling Cascade Is Involved in Stress Response and Symbiotic Efficiency in Bradyrhizobium japonicum. Mol. Microbiol. 2009, 73, 291–305. [Google Scholar] [CrossRef]
- Ledermann, R.; Bartsch, I.; Müller, B.; Wülser, J.; Fischer, H.-M. A Functional General Stress Response of Bradyrhizobium diazoefficiens Is Required for Early Stages of Host Plant Infection. Mol. Plant Microbe Interact. 2018, 31, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Bastiat, B.; Sauviac, L.; Bruand, C. Dual Control of Sinorhizobium meliloti RpoE2 Sigma Factor Activity by Two PhyR-Type Two-Component Response Regulators. J. Bacteriol. 2010, 192, 2255–2265. [Google Scholar] [CrossRef] [PubMed]
- Sauviac, L.; Philippe, H.; Phok, K.; Bruand, C. An Extracytoplasmic Function Sigma Factor Acts as a General Stress Response Regulator in Sinorhizobium meliloti. J. Bacteriol. 2007, 189, 4204–4216. [Google Scholar] [CrossRef]
- Jans, A.; Vercruysse, M.; Gao, S.; Engelen, K.; Lambrichts, I.; Fauvart, M.; Michiels, J. Canonical and Non-Canonical EcfG Sigma Factors Control the General Stress Response in Rhizobium etli. MicrobiologyOpen 2013, 2, 976–987. [Google Scholar] [CrossRef]
- Foreman, R.; Fiebig, A.; Crosson, S. The LovK-LovR Two-Component System Is a Regulator of the General Stress Pathway in Caulobacter crescentus. J. Bacteriol. 2012, 194, 3038–3049. [Google Scholar] [CrossRef]
- Lang, C.; Barnett, M.J.; Fisher, R.F.; Smith, L.S.; Diodati, M.E.; Long, S.R. Most Sinorhizobium meliloti Extracytoplasmic Function Sigma Factors Control Accessory Functions. mSphere 2018, 3, e00454-18. [Google Scholar] [CrossRef] [PubMed]
- Flechard, M.; Fontenelle, C.; Trautwetter, A.; Ermel, G.; Blanco, C. Sinorhizobium meliloti rpoE2 Is Necessary for H2O2 Stress Resistance during the Stationary Growth Phase. FEMS Microbiol. Lett. 2009, 290, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, R.; Emmenegger, B.; Couzigou, J.-M.; Zamboni, N.; Kiefer, P.; Vorholt, J.A.; Fischer, H.-M. Bradyrhizobium diazoefficiens Requires Chemical Chaperones To Cope with Osmotic Stress during Soybean Infection. mBio 2021, 12, e00390-21. [Google Scholar] [CrossRef] [PubMed]
- Ravcheev, D.A.; Khoroshkin, M.S.; Laikova, O.N.; Tsoy, O.V.; Sernova, N.V.; Petrova, S.A.; Rakhmaninova, A.B.; Novichkov, P.S.; Gelfand, M.S.; Rodionov, D.A. Comparative Genomics and Evolution of Regulons of the LacI-Family Transcription Factors. Front. Microbiol. 2014, 5, 294. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).