Research Trends and Challenges of Using CRISPR/Cas9 for Improving Rice Productivity
Abstract
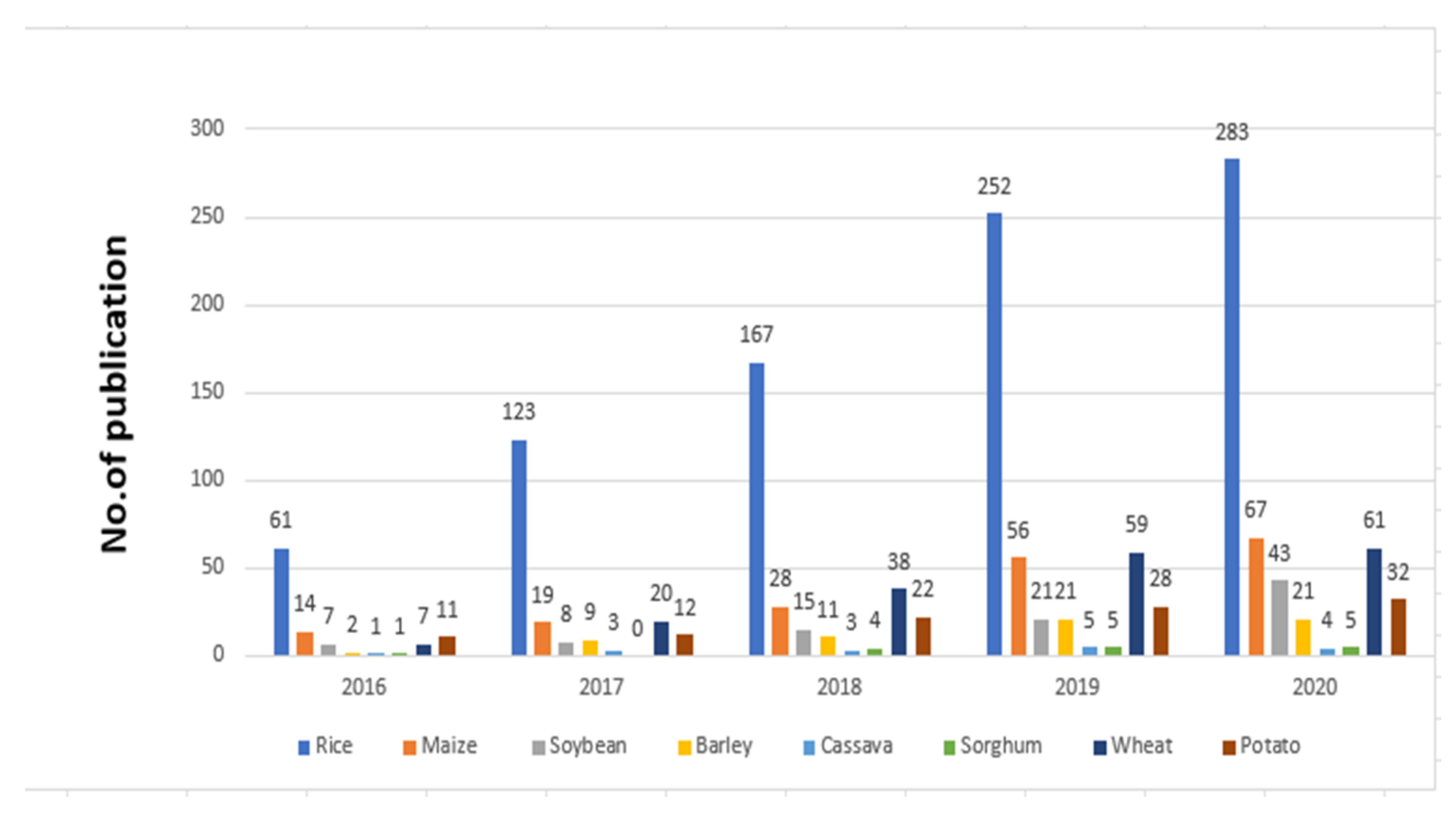
1. Introduction
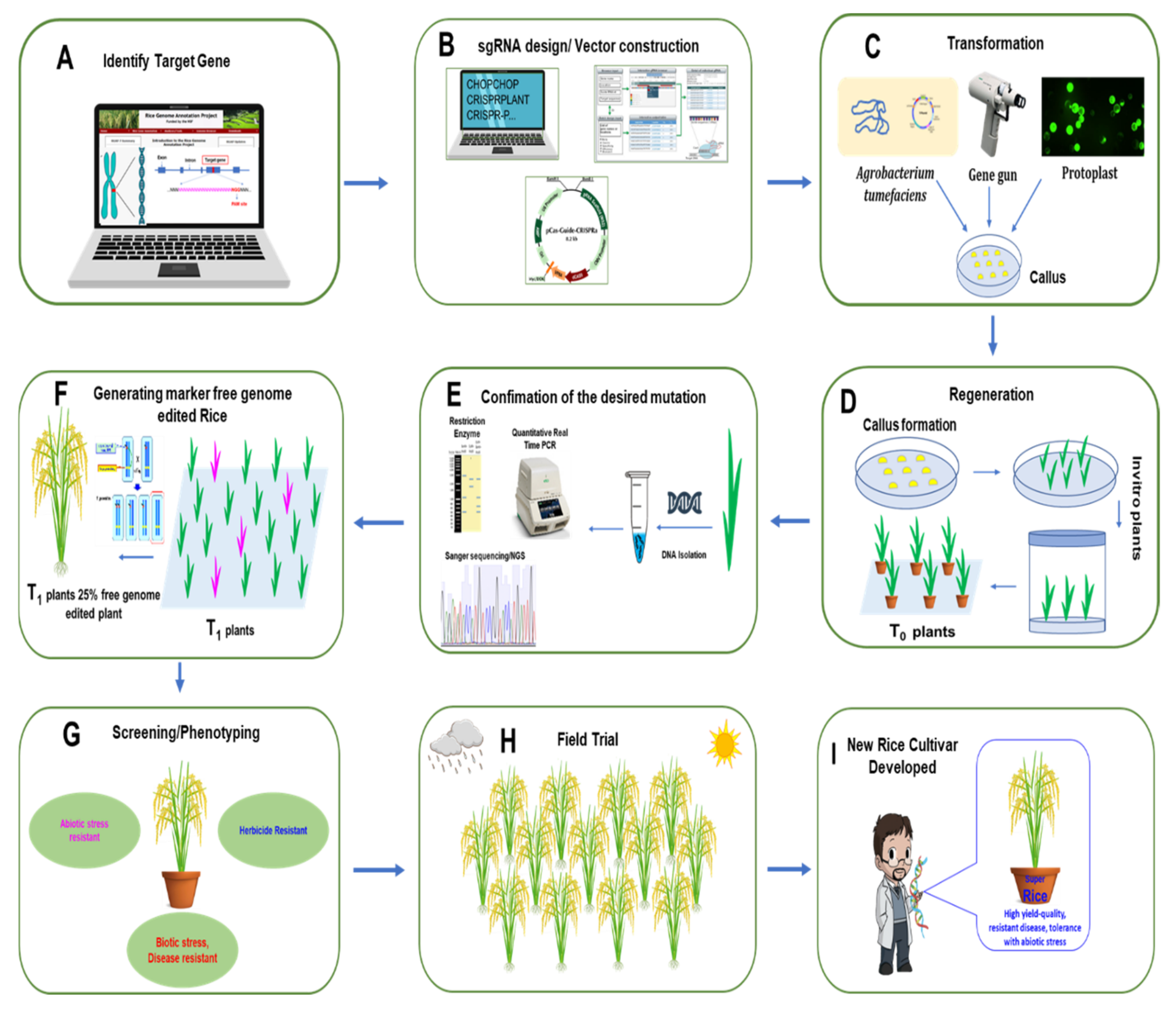
2. CRISPR/Cas9 System and How It Works
3. Research Trends of Major Traits
3.1. Rice Improvement via CRISPR/Cas9 System
3.1.1. Grain Yield Increase
3.1.2. High Quality and Nutrition Fortification
| Target Gene | Gene ID | Cas9 Promoter | sgRNA Promoter | Method of Delivery | Mutation Type | Function | Reference |
|---|---|---|---|---|---|---|---|
| OsIPA1 | LOC_Os08g39890 | p35S, ZmUBI | OsU6, OsU3 | Agrobacterium-mediated | NHEJ | Enhanced grain number, Grain yield. | [45] |
| OsGn1a | LOC_Os09g31310 | ||||||
| OsGS3 | LOC_Os12g34380 | 35S pro Pubi | U6a, U6b | Agrobacterium-mediated | NHEJ | Grain size | [46] |
| OsGW2, OsGW6 | LOC_Os02g14720 LOC_Os06g15620 | pUBQ | OsU3, OsU6, TaU3 | Agrobacterium-mediated | NHEJ | Grain yield, grain size | [47] |
| OsGA20ox2 | LOC_Os01g66100 | Pubi-H | OsU6a, OsU6b | Agrobacterium-mediated | NHEJ | Grain yield, plant architecture | [48] |
| OsGW2 | LOC_Os02g14720 | p35S | OsU6, OsU3 | Agrobacterium-mediated | NHEJ | Grain yield, grain size, grain weight | [49] |
| OsPAO5 | LOC_Os04g57560 | CaMV 35S | U6 | Rice protoplasts | NHEJ | Increase grain weight, grain number | [51] |
| OsBADH2 | LOC_Os08g32870 | 35S/ubi | U3m, U6a | Agrobacterium-mediated | NHEJ | Grain yield, grain size, aromatic | [52] |
| OsPIN5b OsMYB30 | LOC_Os08g41720 LOC_Os02g41510 | 2 × 35S pro Pubi-H | OsU6a | Agrobacterium-mediated | NHEJ | Grain yield, grain size Cold tolerance | [53] |
| OsFWL4 | LOC_Os03g61440 | Maize Ubi1 | OsU6 | Agrobacterium-mediated | NHEJ | Grain yield, plant architecture | [54] |
| OsTB1/ OsFC1 | LOC_Os03g49880 | 2 × 35S pro CaMV | gRNA1,2,3 gRNA4,5,6 | Agrobacterium-mediated | NHEJ | Plant architecture, number of tillers | [55] |
| OsSPL16/ qGW8 | LOC_Os08g41940 | 2 × 35S pro Pubi | OsU6a | Agrobacterium-mediated | NHEJ | Grain yield, grain weight, grain size | [56] |
| OsPYL1, OsPYL12 | LOC_Os10g42280 LOC_Os02g15620 | Maize Ubi1 | OsU6, OsU3 | Agrobacterium-mediated | NHEJ | Number of grains, grain yield | [57] |
| OsWaxy | LOC_Os06g04200 | CaMV 35S | U6 | Agrobacterium-mediated | NHEJ | Decrease amylose content | [65] |
| OsBEI, | LOC_Os06g51084 | pCXUN | OsU3 | Agrobacterium-mediated | NHEJ | High amylose content | [67] |
| OsISA1 | LOC_Os08g40930 | CaMV 35S | OsU6 | Agrobacterium-mediated | NHEJ | Reduced amylose content | [69] |
| OsBADH2 | LOC_Os08g32870 | OsUbi | OsU6a | Agrobacterium-mediated | NHEJ | Enhanced fragrance | [70] |
| OsFAD2-1/ OsFAD2 | LOC_Os02g48560 | pZH 2 × 35S | U3 and U6 OsU6a, OsU6b | Agrobacterium-mediated Rice protoplast | NHEJ | High oleic/low linoleic | [72] [73] |
| OsAAP6, OsAAP10 | LOC_Os01g65670 LOC_Os02g49060 | CaMV35S | OsU3 | Agrobacterium-mediated | NHEJ | Improved eating and cooking quality | [74] |
| OsGS9 | LOC_Os09g27590 | OsUb | OsU6a | Agrobacterium-mediated | NHEJ | Slender grain shape, less chalkiness | [75] |
| OsNramp5 | LOC_Os07g15370 | ZmUBI | OsU6a | Agrobacterium-mediated | NHEJ | Grain low Cd accumulation | [76] |
| OsOr | LOC_Os02g43500 | 2 × 35S | OsU6-2 | Rice Protoplast | NHEJ | Increased β-carotene | [77] |
| SSU-crtI, ZmPsy | - | Ubi1 | OsU6 | Gene gun | NHEJ | Increased β-carotene | [78] |
| OsALS | LOC_Os02g30630 | ZmUBI | U3 | Agrobacterium-mediated | HDR | Herbicide resistance | [79] |
| OsAOX1a, OsAOX1b, OsAOX1c | LOC_Os04g51150 | - | OsU3 | Agrobacterium-mediated | NHEJ | Fertility, Male sterility | [80] |
| OsEPSPS | LOC_Os06g04280 | - | OsU3 | Agrobacterium-mediated | NHEJ, HDR | Glyphosate resistance, high grain yield | [81] |
| OsHd2 | LOC_Os06g16370 | 35S | U3, U6a, U6c | Protoplast | NHEJ | Rice early maturing | [82] |
| OsHd4, OsHd5 | - | - | - | Agrobacterium-mediated | NHEJ | Rice early maturing | [83] |
| OsCIPK3 | LOC_Os07g48760 | CaMV 35S | LacZa | Agrobacterium-mediated | NHEJ | Long day flowering | [84] |
| OsMYB1 | LOC_Os05g35500 | CaMV 35S | OsU6-2 | Agrobacterium-mediated | NHEJ | Seed maturation | [85] |
| OsHAK-1 | LOC_Os04g32920 | - | - | Agrobacterium-mediated | NHEJ | Low cesium accumulation | [86] |
| OsPRX2 | LOC_Os02g33450 | CaMV 35S | OsU3t | Agrobacterium-mediated | NHEJ | Potassium deficiency tolerance | [87] |
3.1.3. Developing Rice Variety with Herbicide Tolerance
3.1.4. Male Sterile Lines for Hybrid Breeding
3.1.5. Rice Early Maturing and Long Day Flowering
3.1.6. Abiotic Stress Resistance
Salinity Tolerance
Drought Tolerance
Cold Tolerance
Heat Stress Tolerance
| Target Gene | Gene ID | Cas9 Promoter | sgRNA Promoter | Method of Delivery | Mutation Type | Functional | Reference |
|---|---|---|---|---|---|---|---|
| OsRAV2 | LOC_Os01g04800 | CaMV 35S | U6 | Agrobacterium mediated | NHEJ | Salt induction | [95] |
| OsRR22 | LOC_Os06g08440 | CaMV35S (2 × 35S) | OsU6a | Agrobacterium -mediated | NHEJ | Salinity tolerance | [96] |
| OsERA1 | LOC_Os01g53600 | - | - | Agrobacterium mediated | NHEJ | Drought tolerance | [100] |
| OsSAPK2 | LOC_Os07g42940 | Pubi-H | U3 | Agrobacterium mediated | NHEJ | Reduced salinity, Drought tolerance | [101] |
| OsDST | LOC_Os03g57240 | OsUBQ | OsU3 | Agrobacterium mediated | NHEJ | Salinity tolerance, osmotic tolerance | [102] |
| OsmiR535 (OsSPL7) | LOC_Os04g46580 | UBI 35S-P | OsU3, OsU6 | Agrobacterium mediated | NHEJ | Drought tolerance | [103] |
| OsPYL9 | LOC_Os06g33690 | PubiH | OsU6a, OsU6b | Agrobacterium mediated | NHEJ | Drought tolerance, Grain yield | [104] |
| OsSRL1, OsSRL2 | LOC_Os01g54390 | Pubi-H | U6a, U6b, U6c, U3m | Agrobacterium mediated | NHEJ | Improved drought tolerance | [105] |
| OsAnn3 | LOC_Os07g46550 | CaMV 35S | U3 | Agrobacterium mediated | NHEJ | Cold stress tolerance | [108] |
| OsPDS | LOC_Os03g08570 | 35S-P | U3 | Gene gun | NHEJ | Heat shock resistant | [111] |
| OsSub1 | LOC_Os01g17160.1 | CaMV 35S | OsU6 | Agrobacterium mediated | NHEJ | Submergence tolerance | [112] |
| OsERF922 | LOC_Os01g54890 | 2 × 35S pro Pubi-H | OsU6a | Agrobacterium -mediated | NHEJ | Rice blast resistance | [113] |
| OsALB1, OsRSY1 | - | TrpC, TEF1 | SNR52, U6-1, U6-2 | Agrobacterium -mediated | NHEJ | Rice blast resistance | [114] |
| OsPi21 | LOC_Os04g32850 | PubiH | OsU6a, OsU3 | Agrobacterium -mediated | NHEJ | Rice blast resistance | [115] |
| OsSEC3A | LOC_Os03g427500 | OsU3 | OsU3 | Rice Protoplast | NHEJ | Rice blast resistance | [116] |
| OsMPK5, MPK2,MPK5, MPK6 | LOC_Os06g06090 | UBI | OsU3 | Agrobacterium -mediated | NHEJ | Disease resistance | [117] |
| OsSWEET14 | LOC_Os08g42350 | 35S, Ubi | U3, U6a | Agrobacterium -mediated | NHEJ | Resistance to Bacterial leaf blight | [118] |
| OsSWEET11, OsSWEET13, OsSWEET14 | LOC_Os11g31190 | CaMV 35S | U6 | Agrobacterium -mediated | NHEJ | Resistance to Bacterial leaf blight | [119] |
| Os8N3 | LOC_Os08g42350 | 35S-p | OsU6a | Agrobacterium -mediated | NHEJ | Resistance to Bacterial leaf blight | [120] |
| OseIF4G | - | ZmUBI1, CaMV35S | TaU6 | Agrobacterium -mediated | NHEJ | Rice tungro virus resistant | [121] |
Submergence Tolerance
3.1.7. Biotic Stress: Disease Resistance
3.1.8. Biotic Stress: Insect Resistance
3.2. Rice Improvement by Homology-Directed Repair (HDR)
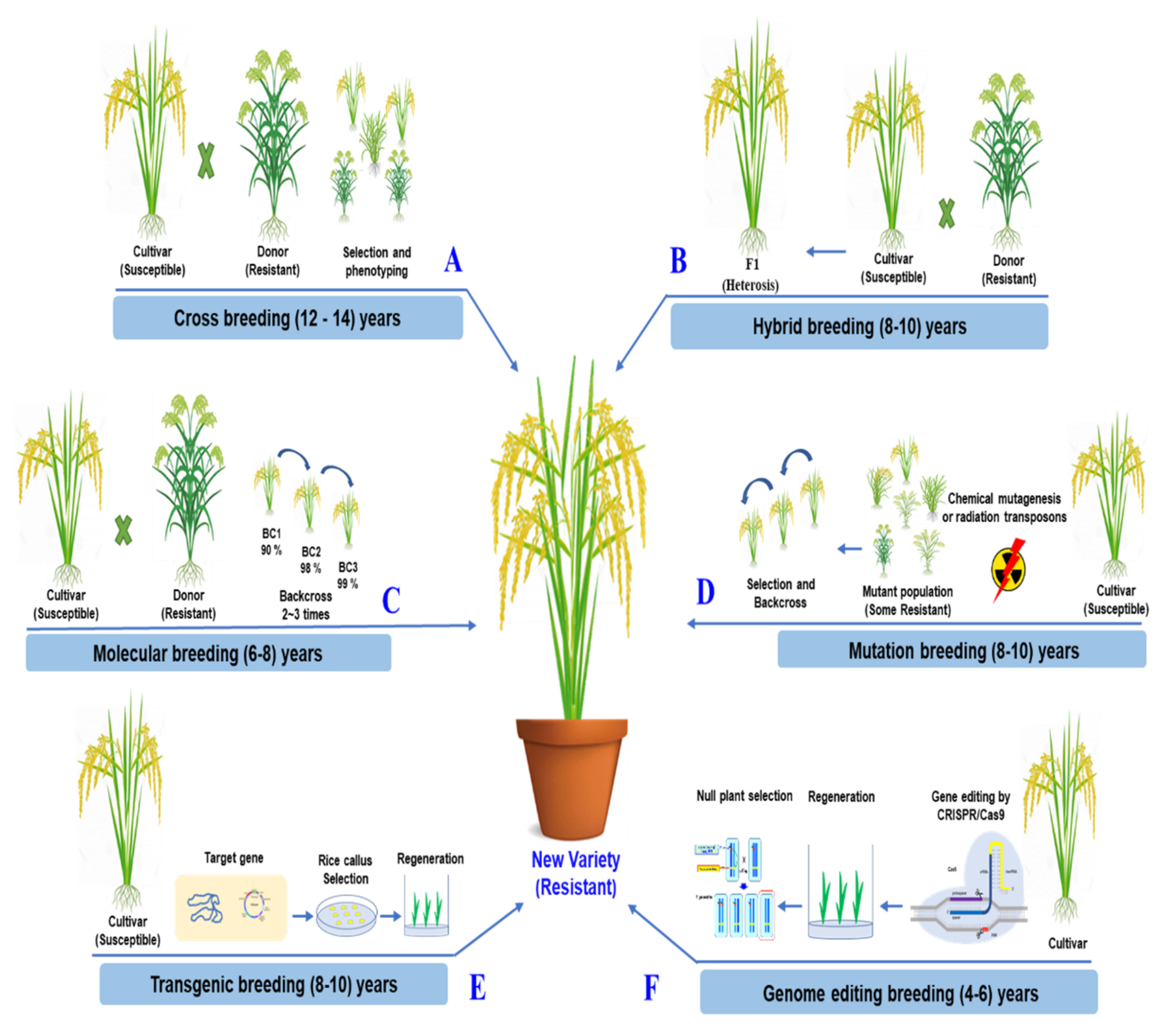
4. Challenge and Future Prospects
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gross, B.L.; Zhao, Z. Archaeological and genetic insights into the origins of domesticated rice. Proc. Natl. Acad. Sci. USA 2014, 111, 6190–6197. [Google Scholar] [CrossRef]
- FAO. Available online: https://www.fao.org/faostat/en/#home (accessed on 16 August 2021).
- FAO Statistics. Available online: https://www.un.org/development/desa/en/news/population/world-population-prospects-2017.html (accessed on 16 August 2021).
- Hunter, M.C.; Smith, R.G.; Schipanski, M.E.; Atwood, L.W.; Mortensen, D.A. Agriculture in 2050: Recalibrating Targets for Sustainable Intensification. BioScience 2017, 67, 386–391. [Google Scholar] [CrossRef]
- Bastin, J.F.; Clark, E.; Elliott, T.; Hart, S.; van den Hoogen, J.; Hordijk, I.; Ma, H.; Majumder, S.; Manoli, G.; Maschler, J.; et al. Correction: Understanding climate change from a global analysis of city analogues. PLoS ONE 2019, 14, e0224120. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar]
- Ma, N.L.; Che Lah, W.A.; Abd Kadir, N.; Mustaqim, M.; Rahmat, Z.; Ahmad, A.; Lam, S.D.; Ismail, M.R. Susceptibility and tolerance of rice crop to salt threat: Physiological and metabolic inspections. PLoS ONE 2018, 13, e0192732. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- Pingali, P.L. Green revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Wolter, F.; Schindele, P.; Puchta, H. Plant breeding at the speed of light: The power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant. Biol. 2019, 19, 176. [Google Scholar] [CrossRef]
- Mishra, R.; Zheng, W.; Joshi, R.K.; Kaijun, Z. Genome Editing Strategies Towards Enhancement of Rice Disease Resistance. Rice Sci. 2021, 28, 133–145. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR/Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Majumder, R.R.; Sakhale, S.; Yadav, S.; Sandhu, N.; Hassan, L.; Hossain, M.A.; Kumar, A. Molecular Breeding for Improving Drought Tolerance in Rice: Recent Progress and Future Perspectives. Mol. Breed. Rice Abiotic Stress Toler. Nutr. Qual. 2021, 2, 5374. [Google Scholar]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Capistrano-Gossmann, G.; Braatz, J.; Sashidhar, N.; Melzer, S. Recent developments in genome editing and applications in plant breeding. Plant. Breed. 2018, 137, 1–9. [Google Scholar] [CrossRef]
- Shino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar]
- Mojica, F.J.; Díez-Villaseñor, C.; Soria, E.; Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Researc. 2007, 35, 52–57. [Google Scholar] [CrossRef]
- Boyaval, P.; Moineau, S.; Romero, D.; Horvath, P. Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Thurtle-Schmidt, D.M.; Lo, T.W. Molecular biology at the cutting edge: A review on CRISPR/CAS9 gene editing for undergraduates. Biochem. Mol. Biol. Educ. 2018, 46, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR/Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S. The Heroes of CRISPR. Cell 2016, 164, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Kuang, Y.; Ren, B.; Yan, D.; Yan, F.; Spetz, C.; Sun, W.; Wang, G.; Zhou, X.; Zhou, H. SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol. 2021, 22, 6. [Google Scholar] [CrossRef]
- Gao, L.; Cox, D.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Lieber, M.R. The Mechanism of DSB Repair by the NHEJ. Annu. Rev. Biochem. 2011, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2018, 77, 229–257. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar]
- Oerke, E.C. Crop Losses to Pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.G.; Kim, S.T.; Choe, S.; Kim, J.S. DNA-free genome editing in plants with preassembled CRISPR/Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Um, T.; Park, T.; Shim, J.S.; Kim, Y.S.; Lee, G.S.; Choi, I.Y.; Kim, J.K.; Seo, J.S.; Park, S.C. Application of Upstream Open Reading Frames (uORFs) Editing for the Development of Stress-Tolerant Crops. Int. J. Mol. Sci. 2021, 22, 3743. [Google Scholar] [CrossRef]
- Wade, M. High-Throughput Silencing Using the CRISPR/Cas9 System: A Review of the Benefits and Challenges. J. Biomol. Screen. 2015, 20, 1027–1039. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Dar, Z.; Dar, S. Breeding Strategies for Improving Rice Yield—A Review. Agric. Sci. 2015, 6, 467–478. [Google Scholar] [CrossRef][Green Version]
- Cho, Y.; Kang, H.; Lee, J.; Lee, Y.; Lim, S.; Gauch, H.G.; Eun, M.Y.; Mccouch, S. Identification of Quantitative Trait Loci in Rice for Yield, Yield Components, and Agronomic Traits across Years and Locations. Crop. Sci. 2007, 47, 2403–2417. [Google Scholar] [CrossRef]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant. Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant. Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Usman, B.; Zhao, N.; Nawaz, G.; Qin, B.; Liu, F.; Liu, Y.; Li, R. CRISPR/Cas9 Guided Mutagenesis of Grain Size 3 Confers Increased Rice (Oryza sativa L.) Grain Length by Regulating Cysteine Proteinase Inhibitor and Ubiquitin-Related Proteins. Int. J. Mol. Sci. 2021, 22, 3225. [Google Scholar] [CrossRef]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. = Yi Chuan Xue Bao 2016, 43, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Teng, K.; Nawaz, G.; Feng, X.; Usman, B.; Wang, X.; Luo, L.; Zhao, N.; Liu, Y.; Li, R. Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations. 3 Biotech 2019, 9, 387. [Google Scholar] [CrossRef]
- Zhou, J.; Xin, X.; He, Y.; Chen, H.; Li, Q.; Tang, X.; Zhong, Z.; Deng, K.; Zheng, X.; Akher, S.A.; et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant. Cell Rep. 2019, 38, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Lacchini, E.; Kiegle, E.; Castellani, M.; Adam, H.; Jouannic, S.; Gregis, V.; Kater, M.M. CRISPR-mediated accelerated domestication of African rice landraces. PLoS ONE 2020, 15, e0229782. [Google Scholar] [CrossRef]
- Lv, Y.; Shao, G.; Jiao, G.; Sheng, Z.; Xie, L.; Hu, S.; Tang, S.; Wei, X.; Hu, P. Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol. Plant. 2021, 14, 344–351. [Google Scholar] [CrossRef]
- Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. Generation of High Yielding and Fragrant Rice (Oryza sativa L.) Lines by CRISPR/Cas9 Targeted Mutagenesis of Three Homoeologs of Cytochrome P450 Gene Family and OsBADH2 and Transcriptome and Proteome Profiling of Revealed Changes Triggered by Mutations. Plants 2020, 9, 788. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR/Cas9 System. Front. Plant. Sci. 2020, 10, 1663. [Google Scholar] [CrossRef]
- Gao, Q.; Li, G.; Sun, H.; Xu, M.; Wang, H.; Ji, J.; Wang, D.; Yuan, C.; Zhao, X. Targeted Mutagenesis of the Rice FW 2.2-Like Gene Family Using the CRISPR/Cas9 System Reveals OsFWL4 as a Regulator of Tiller Number and Plant Yield in Rice. Int. J. Mol. Sci. 2020, 21, 809. [Google Scholar] [CrossRef]
- Cui, Y.; Hu, X.; Liang, G.; Feng, A.; Wang, F.; Ruan, S.; Dong, G.; Shen, L.; Zhang, B.; Chen, D.; et al. Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9. Plant. Biotechnol. J. 2020, 18, 1987–1989. [Google Scholar] [CrossRef]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Qin, B.; Liu, F.; Liu, Y.; Li, R. Programmed Editing of Rice (Oryza sativa L.) OsSPL16 Gene Using CRISPR/Cas9 Improves Grain Yield by Modulating the Expression of Pyruvate Enzymes and Cell Cycle Proteins. Int. J. Mol. Sci. 2020, 22, 249. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Cuevas, R.P.; Fitzgerald, M.A. Genetic Diversity of Rice Grain Quality. Genet. Divers. Plants 2012. [Google Scholar] [CrossRef][Green Version]
- Custodio, M.C.; Cuevas, R.P.; Ynion, J.; Laborte, A.G.; Velasco, M.L.; Demont, M. Rice quality: How is it defined by consumers, industry, food scientists, and geneticists? Trends Food Sci. Technol. 2019, 92, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xia, D.; He, Y. Rice grain quality—Traditional traits for high quality rice and health-plus substances. Mol. Breed. 2019, 40, 1–17. [Google Scholar] [CrossRef]
- Sharma, N.; Khanna, R. Rice Grain Quality: Current Developments and Future Prospects. In Recent Advances in Grain Crops Research; IntechOpen: London, UK, 2019; pp. 1–17. [Google Scholar]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 System for Rice Grain Quality Improvement: Perspectives and Opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Chen, X.; Cai, X.; Tang, S.; Yu, H.; Zhang, J.; Hong, M.; Gu, M. Stable inheritance of the antisense Waxy gene in transgenic rice with reduced amylose level and improved quality. Transgenic Res. 2003, 12, 71–82. [Google Scholar] [CrossRef]
- Tian, Z.; Qian, Q.; Liu, Q.; Yan, M.; Liu, X.; Yan, C.; Liu, G.; Gao, Z.; Tang, S.; Zeng, D.; et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 2009, 106, 21760–21765. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Botella, J.R.; Zhu, J.K. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant. Biol. 2018, 60, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant. 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of High-Amylose Rice through CRISPR/Cas9-Mediated Targeted Mutagenesis of Starch Branching Enzymes. Front. Plant. Sci. 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, J.; Ahmad, S.; Hussain, B.; Mawia, A.M.; Zeb, A.; Ju, L. Applications and Potential of Genome-Editing Systems in Rice Improvement: Current and Future Perspectives. Agronomy 2021, 11, 1359. [Google Scholar] [CrossRef]
- Chao, S.; Cai, Y.; Feng, B.; Jiao, G.; Sheng, Z.; Luo, J.; Tang, S.; Wang, J.; Hu, P.; Wei, X. Editing of Rice Isoamylase Gene ISA1 Provides Insights into Its Function in Starch Formation. Rice Sci. 2019, 26, 77–87. [Google Scholar]
- Ashokkumar, S.; Jaganathan, D.; Ramanathan, V.; Rahman, H.; Palaniswamy, R.; Kambale, R.; Muthurajan, R. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing. PLoS ONE 2020, 15, e0237018. [Google Scholar] [CrossRef]
- Zaplin, E.S.; Liu, Q.; Li, Z.; Butardo, V.M.; Blanchard, C.L.; Rahman, S. Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene. Funct. Plant. Biol. 2013, 40, 996–1004. [Google Scholar] [CrossRef]
- Abe, K.; Araki, E.; Suzuki, Y.; Toki, S.; Saika, H. Production of high oleic/low linoleic rice by genome editing. Plant. Physiol. Biochem. 2018, 131, 58–62. [Google Scholar] [CrossRef]
- Bahariah, B.; Masani, M.; Rasid, O.A.; Parveez, G. Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: A model genome editing system for oil palm. J. Genet. Eng. Biotechnol. 2021, 19, 86. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Y.; Guo, M.; Zhong, C.; Yan, C.; Sun, S. Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop. J. 2020, 8, 457–464. [Google Scholar] [CrossRef]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Songmei, L.; Jie, J.; Yang, L.; Jun, M.; Shouling, X.; Yuanyuan, T.; Youfa, L.; Qingyao, S.; Jianzhong, H. Characterization and Evaluation of OsLCT1 and OsNramp5 Mutants Generated Through CRISPR/Cas9-Mediated Mutagenesis for Breeding Low Cd Rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef]
- Endo, A.; Saika, H.; Takemura, M.; Misawa, N.; Toki, S. A novel approach to carotenoid accumulation in rice callus by mimicking the cauliflower Orange mutation via genome editing. Rice 2019, 12, 81. [Google Scholar] [CrossRef]
- Dong, O.X.; Yu, S.; Jain, R.; Zhang, N.; Duong, P.Q.; Butler, C.; Li, Y.; Lipzen, A.; Martin, J.A.; Barry, K.W.; et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 2020, 11, 1178. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering Herbicide-Resistant Rice Plants through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Mol. Plant. 2016, 9, 628–631. [Google Scholar] [CrossRef]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Reddy, G.M.; Singh, J.; Vani, K.; Vijayalakshmi, M.; Kaul, T.; Reddy, M.K. Development of Transgenic Rice Harbouring Mutated Rice 5-Enolpyruvylshikimate 3-Phosphate Synthase (Os-mEPSPS) and Allium sativum Leaf Agglutinin (ASAL) Genes Conferring Tolerance to Herbicides and Sap-Sucking Insects. Plant. Mol. Biol. Report. 2014, 32, 1146–1157. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Zhang, Y.; He, M.; Li, R.; Meng, W.; Wang, Z.; Li, X.; Bu, Q. Fine-tuning Flowering Time via Genome Editing of Upstream Open Reading Frames of Heading Date 2 in Rice. Rice 2021, 14, 59. [Google Scholar] [CrossRef]
- Li, X.; Zhou, W.; Ren, Y.; Tian, X.; Lv, T.; Wang, Z.; Fang, J.; Chu, C.; Yang, J.; Bu, Q. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genom. 2017, 44, 175–178. [Google Scholar] [CrossRef]
- Peng, Q.; Zhu, C.; Liu, T.; Zhang, S.; Feng, S.; Wu, C. Phosphorylation of OsFD1 by OsCIPK3 promotes the formation of RFT1-containing florigen activation complex for long-day flowering in rice. Mol. Plant. 2021, 14, 1135–1148. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.K. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant. 2013, 6, 2008–2011. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A.A. Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR-Cas system. Plant. J. Cell Mol. Biol. 2017, 92, 43–56. [Google Scholar] [CrossRef]
- Mao, X.; Zheng, Y.; Xiao, K.; Wei, Y.; Zhu, Y.; Cai, Q.; Chen, L.; Xie, H.; Zhang, J. OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem. Biophys. Res. Commun. 2018, 495, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Huang, Y.; Wang, K. The Development of Herbicide Resistance Crop Plants Using CRISPR/Cas9-Mediated Gene Editing. Genes 2021, 12, 912. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.X.; Lin, C.; Shen, Z. Development of Transgenic Glyphosate-Resistant Rice with G6 Gene Encoding 5-Enolpyruvylshikimate-3-Phosphate Synthase. Agric. Sci. China 2011, 10, 1307–1312. [Google Scholar] [CrossRef]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef] [PubMed]
- Xu Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B.; et al. Development of Commercial Thermo-sensitive Genic Male Sterile Rice Accelerates Hybrid Rice Breeding Using the CRISPR/Cas9-mediated TMS5 Editing System. Sci. Rep. 2016, 6, 37395. [Google Scholar] [CrossRef]
- Gu, W.; Zhang, D.; Qi, Y.; Yuan, Z. Generating Photoperiod-Sensitive Genic Male Sterile Rice Lines with CRISPR/Cas9. Methods Mol. Biol. 2019, 1917, 97–107. [Google Scholar]
- Lan, S.; Guojun, D.; Yu, Z.; Guocheng, H.; Qiang, Z.; Guanglian, H.; Bo, X.; Deyong, R.; Jiang, H.; Li, Z.; et al. Rapid Creation of New Photoperiod-/Thermo-Sensitive Genic Male-Sterile Rice Materials by CRISPR/Cas9 System. Rice Sci. 2019, 26, 129–132. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant. Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant. Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. New Strateg. Plant. Improv. 2019, 39, 47. [Google Scholar] [CrossRef]
- Mitra, J. Genetics and genetic improvement of drought resistance in crop plants. Curr. Sci. 2001, 80, 758–763. [Google Scholar]
- Babu, R.C.; Zhang, J.; Blum, A.; Ho, T.D.; Wu, R.; Nguyen, H.T. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant. Sci. 2004, 166, 855–862. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Basra, S.M.A. and Islam-ud-Din. Improving Water Relations and Gas Exchange with Brassinosteroids in Rice under Drought Stress. J. Agron. Crop. Sci. 2009, 195, 262–269. [Google Scholar] [CrossRef]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 2020, 15, e0243376. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant. Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR/Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants Int. J. Funct. Plant. Biol. 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Yue, E.; Cao, H.; Liu, B. OsmiR535, a Potential Genetic Editing Target for Drought and Salinity Stress Tolerance in Oryza sativa. Plants 2020, 9, 1337. [Google Scholar] [CrossRef] [PubMed]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Liu, Y.; Li, R. Precise Editing of the OsPYL9 Gene by RNA-Guided Cas9 Nuclease Confers Enhanced Drought Tolerance and Grain Yield in Rice (Oryza sativa L.) by Regulating Circadian Rhythm and Abiotic Stress Responsive Proteins. Int. J. Mol. Sci. 2020, 21, 7854. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef]
- Shakiba, E.; Edwards, J.D.; Jodari, F.; Duke, S.E.; Baldo, A.M.; Korniliev, P.; McCouch, S.R.; Eizenga, G.C. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 2017, 12, e0172133. [Google Scholar] [CrossRef] [PubMed]
- Koseki, M.; Kitazawa, N.; Yonebayashi, S.; Maehara, Y.; Wang, Z.X.; Minobe, Y. Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol. Genet. Genom. 2010, 284, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2016, 60, 539–547. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chu, C.; Yao, S. The impact of high-temperature stress on rice: Challenges and solutions. Crop. J. 2021, 9, 963–976. [Google Scholar] [CrossRef]
- Nandy, S.; Pathak, B.; Zhao, S.; Srivastava, V. Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant. Direct 2019, 3, e00145. [Google Scholar] [CrossRef]
- Liang, Y.; Biswas, S.; Kim, B.; Bailey-Serres, J.; Septiningsih, E.M. Improved Transformation and Regeneration of Indica Rice: Disruption of SUB1A as a Test Case via CRISPR-Cas9. Int. J. Mol. Sci. 2021, 22, 6989. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Foster, A.J.; Martin-Urdiroz, M.; Yan, X.; Wright, H.S.; Soanes, D.M.; Talbot, N.J. CRISPR/Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 2018, 8, 14355. [Google Scholar] [CrossRef]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, K.; Wang, Y. Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant. Biol. 2019, 61, 1201–1205. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z.; Sodmergen. Disruption of OsSEC3A Increases the Content of Salicylic Acid and Induces Plant Defense Responses in Rice. J. Exp. Bot. 2018, 69, 1051–1064. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Zeng, X.; Luo, Y.; Vu, N.; Shen, S.; Xia, K.; Zhang, M. CRISPR/Cas9-mediated mutation of OsSWEET14 in rice cv. Zhonghua11 confers resistance to Xanthomonas oryzae pv. oryzae without yield penalty. BMC Plant. Biol. 2020, 20, 313. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef]
- Kim, Y.A.; Moon, H.; Park, C.J. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 2019, 12, 67. [Google Scholar] [CrossRef]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant. Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Wassmann, R.; Jagadish, S.V.; Sumfleth, K.; Pathak, H.; Howell, G.; Ismail, A.M.; Serraj, R.; Redoña, E.D.; Singh, R.K.; Heuer, S. Chapter 3 Regional Vulnerability of Climate Change Impacts on Asian Rice Production and Scope for Adaptation. Adv. Agron. 2009, 102, 91–133. [Google Scholar]
- Xu, J.; Xing, Y.; Xu, Y.; Wan, J. Breeding by design for future rice: Genes and genome technologies. Crop. J. 2021, 9, 491–496. [Google Scholar] [CrossRef]
- Yin, J.; Zou, L.; Zhu, X.; Cao, Y.; He, M.; Chen, X. Fighting the enemy: How rice survives the blast pathogen’s attack. Crop. J. 2021, 9, 543–552. [Google Scholar] [CrossRef]
- Nalley, L.; Tsiboe, F.; Durand-Morat, A.; Shew, A.; Thoma, G. Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLoS ONE 2016, 11, e0167295. [Google Scholar]
- Tran, T.T.; Pérez-Quintero, A.L.; Wonni, I.; Carpenter, S.; Yu, Y.; Wang, L.; Leach, J.E.; Verdier, V.; Cunnac, S.; Bogdanove, A.J.; et al. Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. PLoS Pathog. 2018, 14, e1007092. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Torollo, G.; Angeles-Shim, R.B.; Cabunagan, R.C.; Choi, I.R.; Yeo, U.S.; Ha, W.G. Rice tungro spherical virus resistance into photoperiod-insensitive japonica rice by marker-assisted selection. Breed. Sci. 2015, 65, 345–351. [Google Scholar] [CrossRef][Green Version]
- Yarasi, B.; Sadumpati, V.; Immanni, C.P.; Vudem, D.R.; Khareedu, V.R. Transgenic rice expressing Allium sativum leaf agglutinin (ASAL) exhibits high-level resistance against major sap-sucking pests. BMC Plant. Biol. 2008, 8, 102. [Google Scholar] [CrossRef]
- Douglas, A.E. Strategies for Enhanced Crop Resistance to Insect Pests. Annu. Rev. Plant. Biol. 2018, 69, 637–660. [Google Scholar] [CrossRef]
- Van, F.K. Cross-order and cross-phylum activity of Bacillus thuringiensis pesticidal proteins. J. Invertebr. Pathol. 2013, 114, 76–85. [Google Scholar]
- Paul, S.; Das, S. Natural insecticidal proteins, the promising bio-control compounds for future crop protection. Nucleus 2021, 64, 7–20. [Google Scholar] [CrossRef]
- Rajadurai, G.; Kalaivani, A.; Varanavasiyappan, S.; Balakrishnan, N.; Udayasuriyan, V.; Sudhakar, D.; Natarajan, N. Generation of insect resistant marker-free transgenic rice with a novel cry2AX1 gene. Electron. J. Plant. Breed. 2018, 9, 723. [Google Scholar] [CrossRef]
- Qiu, L.; Fan, J.; Zhang, B.; Liu, L.; Wang, X.; Lei, C.; Lin, Y.; Ma, W. RNA interference knockdown of aminopeptidase N genes decrease the susceptibility of Chilo suppressalis larvae to Cry1Ab/Cry1Ac and Cry1Ca-expressing transgenic rice. J. Invertebr. Pathol. 2017, 145, 9–12. [Google Scholar] [CrossRef]
- Zuo, Y.; Xue, Y.; Lu, W.; Ma, H.; Chen, M.; Wu, Y.; Yang, Y.; Hu, Z. Functional validation of nicotinic acetylcholine receptor (nAChR) α6 as a target of spinosyns in Spodoptera exigua utilizing the CRISPR/Cas9 system. Pest. Manag. Sci. 2020, 76, 2415–2422. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Huang, J.; Jin, W.; Yang, Y.; Wu, Y. CRISPR-Mediated Knockout of the ABCC2 Gene in Ostrinia furnacalis Confers High-Level Resistance to the Bacillus thuringiensis Cry1Fa Toxin. Toxins 2020, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Liu, S.; Liu, L.; Tay, W.T.; Walsh, T.K.; Yang, Y.; Wu, Y. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 2017, 87, 147–153. [Google Scholar] [CrossRef]
- Gheraoing, H.; Piganeau, M.; Renouf, B.; Renaud, J.B.; Sallmyr, A.; Ruis, B.; Oh, S.; Tomkinson, A.E.; Hendrickson, E.A.; Giovannangeli, C.; et al. Chromosomal Translocations in Human Cells Are Generated by Canonical Nonhomologous End-Joining. Mol. Cell 2014, 55, 829–842. [Google Scholar]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J.K. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol Plant. 2017, 10, 1007–1010. [Google Scholar] [CrossRef]
- Ali, Z.; Shami, A.; Sedeek, K.; Kamel, R.; Alhabsi, A.; Tehseen, M.; Hassan, N.; Butt, H.; Kababji, A.; Hamdan, S.M.; et al. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun. Biol. 2020, 3, 44. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Hashimoto, R.; Ueta, R.; Abe, C.; Osakabe, Y.; Osakabe, K. Efficient Multiplex Genome Editing Induces Precise, and Self-Ligated Type Mutations in Tomato Plants. Front. Plant. Sci. 2018, 9, 916. [Google Scholar] [CrossRef]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular therapy. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.K. Genome Engineering in Rice Using Cas9 Variants that Recognize NG PAM Sequences. Mol. Plant. 2019, 12, 1003–1014. [Google Scholar] [CrossRef]
- Chen, W.; McKenna, A.; Schreiber, J.; Haeussler, M.; Yin, Y.; Agarwal, V.; Noble, W.S.; Shendure, J. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019, 47, 7989–8003. [Google Scholar] [CrossRef]
- Hahn, F.; Nekrasov, V. CRISPR/Cas precision: Do we need to worry about off-targeting in plants? Plant. Cell Rep. 2019, 38, 437–441. [Google Scholar] [CrossRef]
- Razzaq, A.; Saleem, F.; Kanwal, M.; Mustafa, G.; Yousaf, S.; Imran Arshad, H.M.; Hameed, M.K.; Khan, M.S.; Joyia, F.A. Modern trends in plant genome editing: An inclusive review of the CRISPR/Cas9 Toolbox. Int. J. Mol. Sci. 2019, 20, 4045. [Google Scholar] [CrossRef]
- Koo, T.; Lee, J.; Kim, J.-S. Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Mol. Cells 2015, 38, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Botella, J.R.; Liu, Y.; Zhu, J.K. Gene editing in plants: Progress and challenges. Natl. Sci. Rev. 2019, 6, 421–437. [Google Scholar] [CrossRef]
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant. Sci. 2020, 11, 56. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Belisle, M.; Frommer, W.B. The evolving landscape around genome editing in agriculture: Many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO Rep. 2020, 21, e50680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rymarquis, L.A.; Ezura, H.; Nekrasov, V. Editorial: CRISPR-Cas in Agriculture: Opportunities and Challenges. Front. Plant. Sci. 2021, 12, 672329. [Google Scholar] [CrossRef] [PubMed]
- Romero, F.M.; Gatica-Arias, A. CRISPR/Cas9: Development and Application in Rice Breeding. Rice Sci. 2019, 26, 265–281. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, V.T.; Kim, M.-S.; Jung, Y.-J.; Kang, K.-K.; Cho, Y.-G. Research Trends and Challenges of Using CRISPR/Cas9 for Improving Rice Productivity. Agronomy 2022, 12, 164. https://doi.org/10.3390/agronomy12010164
Le VT, Kim M-S, Jung Y-J, Kang K-K, Cho Y-G. Research Trends and Challenges of Using CRISPR/Cas9 for Improving Rice Productivity. Agronomy. 2022; 12(1):164. https://doi.org/10.3390/agronomy12010164
Chicago/Turabian StyleLe, Van Trang, Me-Sun Kim, Yu-Jin Jung, Kwon-Kyoo Kang, and Yong-Gu Cho. 2022. "Research Trends and Challenges of Using CRISPR/Cas9 for Improving Rice Productivity" Agronomy 12, no. 1: 164. https://doi.org/10.3390/agronomy12010164
APA StyleLe, V. T., Kim, M.-S., Jung, Y.-J., Kang, K.-K., & Cho, Y.-G. (2022). Research Trends and Challenges of Using CRISPR/Cas9 for Improving Rice Productivity. Agronomy, 12(1), 164. https://doi.org/10.3390/agronomy12010164










