Transcriptomic Analysis of the Differences in Leaf Color Formation during Stage Transitions in Populus × euramericana ‘Zhonghuahongye’
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sampling
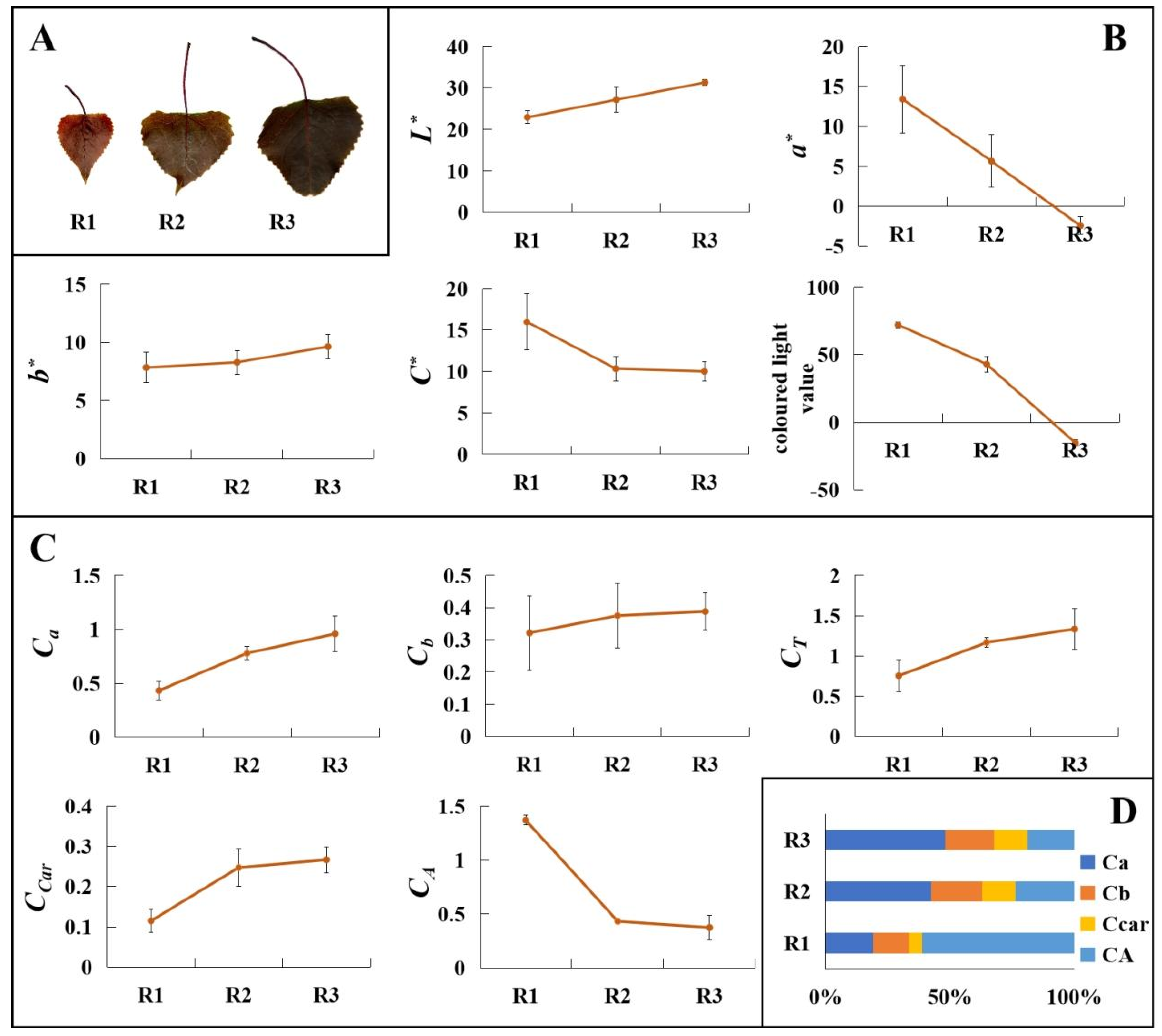
2.2. Leaf Colour Analysis
2.3. Determination of Leaf Pigment Contents
2.4. Total RNA Extraction and Detection
2.5. Library Construction and Sequencing
2.6. Screening and Functional Annotation of DEGs
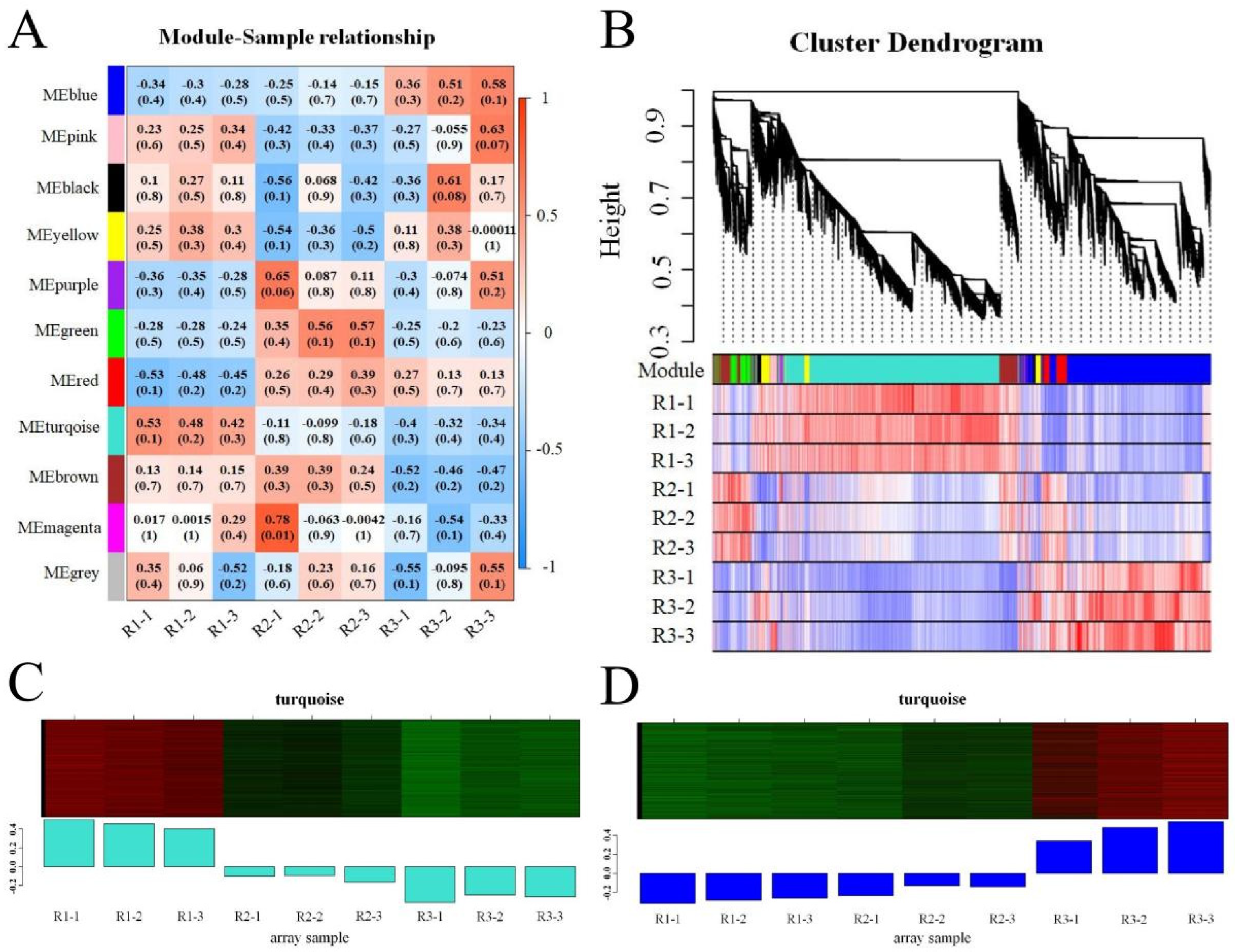
2.7. Construction of a Coexpression Network
2.8. Transcription Factor Analysis
2.9. Quantitative Real-Time PCR (RT-qPCR)
3. Results
3.1. Analysis of Leaf Colour Phenotypes at Different Stages of Leaf Maturation
3.2. Sequencing Data Quality Assessment
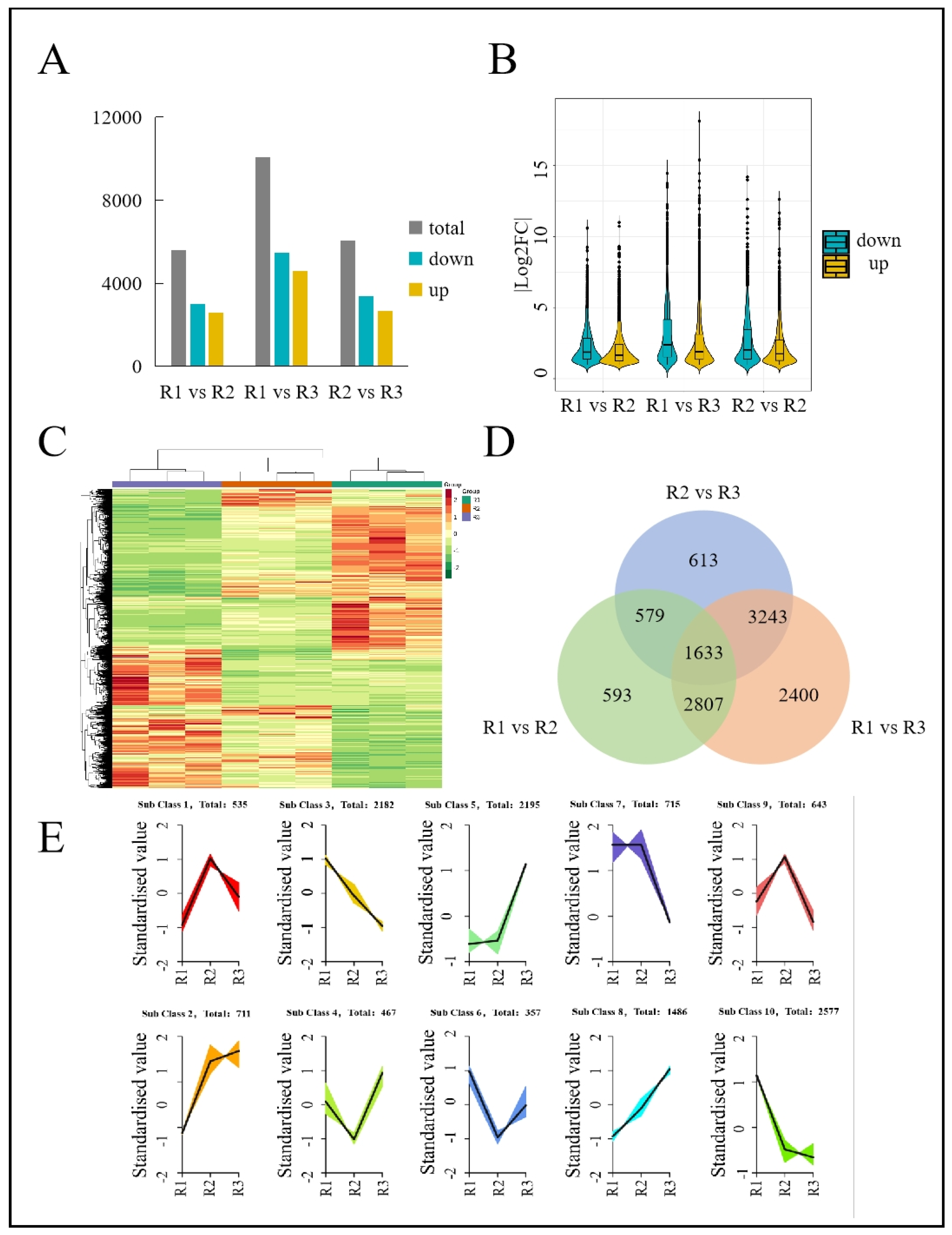
3.3. DEG Analysis
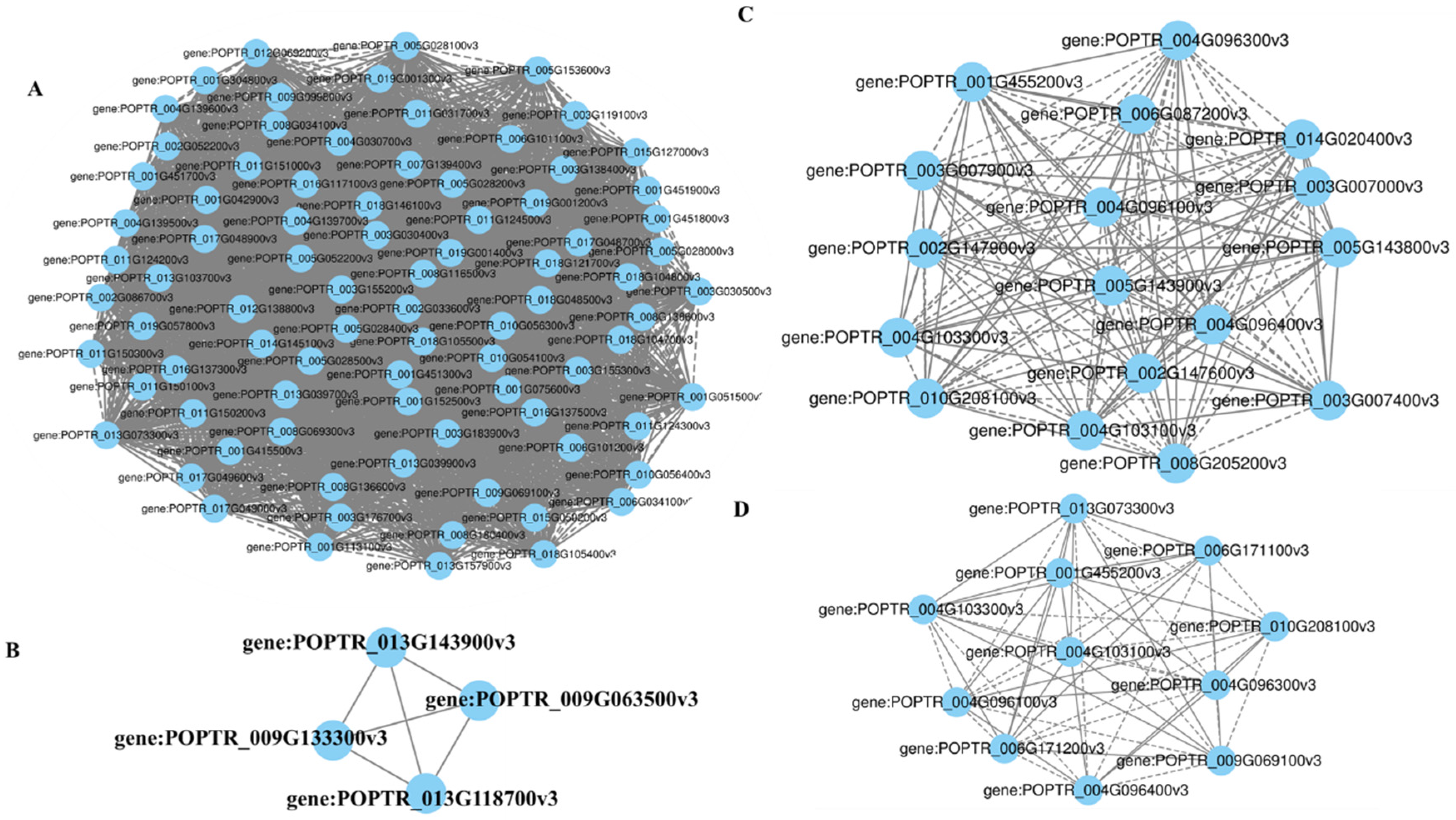
3.4. Coexpression Network Analysis
3.5. Functional Annotation and Enrichment Analysis of DEGs
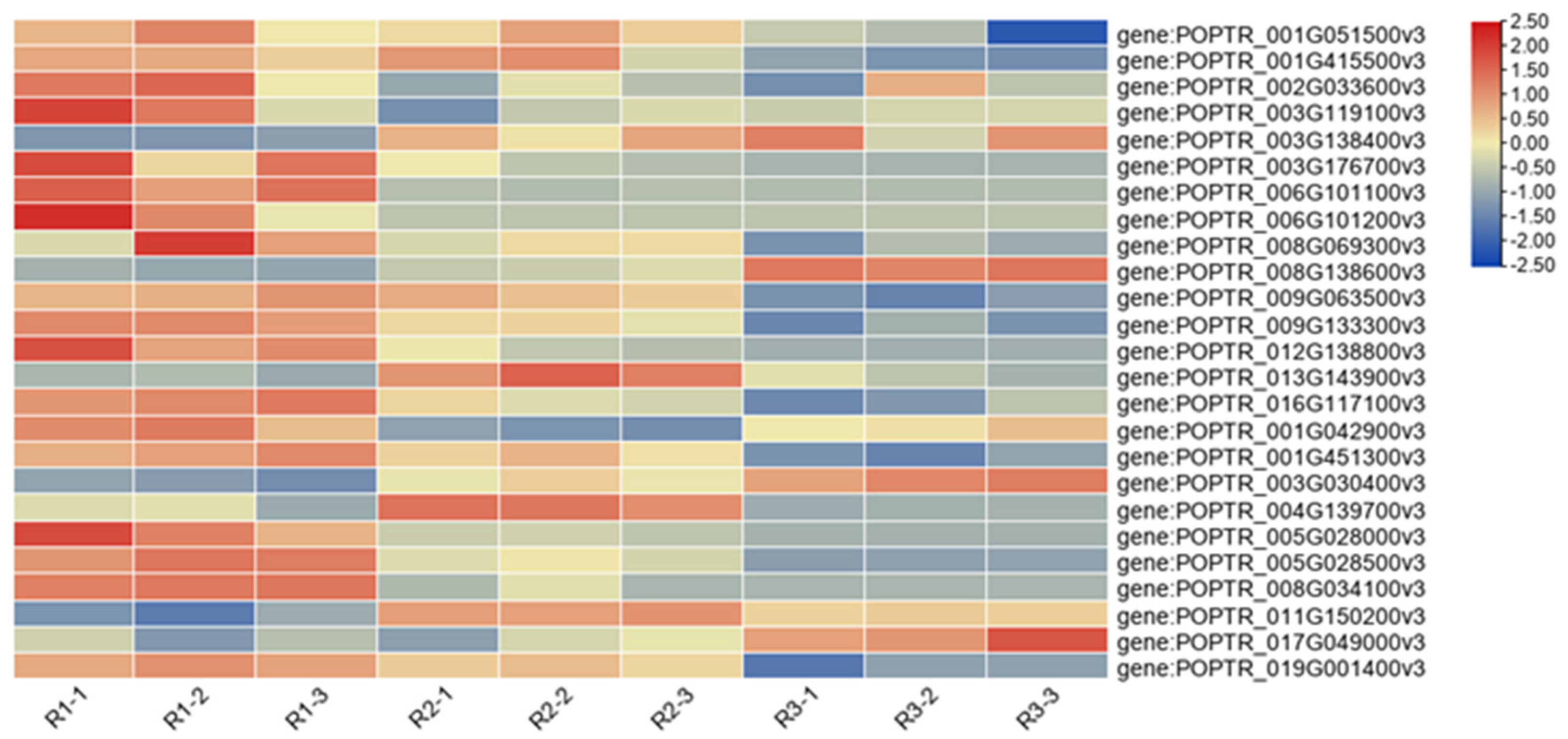
3.6. Analysis of DEGs Related to Anthocyanin Synthesis
3.7. Transcription Factor (TF) Analysis
3.8. qRT-PCR-Based Verification of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, S.; Zhu, Y. Selection and Breeding of Populus Hongye; Northwestern Polytechnical University Press: Xian, China, 2018. [Google Scholar]
- Zhu, Y.; Cheng, X. A new poplar variety ‘Zhonghongyang’. For. Sci. 2008, 10, 173–174. [Google Scholar]
- Chen, X. Research on the Attrition Phenomenon of Red Leaf Poplar; Henan Agricultural University: Zhengzhou, China, 2012. [Google Scholar]
- Yang, S.; Zhu, Y.; Ma, Y.; Yan, Z.; Ding, X. Correlation between leaf color and pigment composition of Poplar in the growing season. J. Northeast For. Univ. 2013, 41, 63–68. [Google Scholar]
- Gentile, D.; Patamia, V.; Fuochi, V.; Furneri, P.M.; Rescifina, A. Analysis of differences in anthocyanin metabolism by transcriptome sequencing of two species of Toona sinensis at different stages. Mol. Plant Breed. 2022, 20, 422–434. [Google Scholar]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Ma, D.; Constabel, C.P. The MYB182 Protein Down-Regulates Proanthocyanidin and Anthocyanin Biosynthesis in Poplar by Repressing Both Structural and Regulatory Flavonoid Genes. Plant Physiol. 2015, 167, 693–710. [Google Scholar] [CrossRef] [Green Version]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.M.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB Proteins SlANT1 and SlAN2: Same Protein Activity, Different Roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef] [Green Version]
- Mazza, G.; Miniati, E. Anthocyanins in Fruits, Vegetables, and Grains, 1st ed.; CRC Press: Boca Raton, FL, USA, 1993; p. 384. [Google Scholar]
- Mori, K.; Goto-Yamamoto, N.; Kitayama, M.; Hashizume, K. Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 2007, 58, 1935–1945. [Google Scholar] [CrossRef]
- Vaknin, H.; Bar-Akiva, A.; Ovadia, R.; Nissim-Levi, A.; Forer, I.; Weiss, D.; Oren-Shamir, M. Active anthocyanin degradation in Brunfelsia calycina (yesterday–today–tomorrow) flowers. Planta 2005, 222, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Gonnet, J.F. Colour effects of co-pigmentation of anthocyanins revisited-1. A colorimetric definition using the CIELAB scale. Food Chem 1998, 63, 409–415. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Lu, Y. Plant color mutants and the anthocyanin pathway. Chin. Bull. Bot. 2016, 51, 107–119. [Google Scholar]
- Liu, X.Y.; Han, H.Q.; Ge, H.Y.; Jiang, M.M.; Chen, H.Y. Cloning, expression and interaction of anthocyaninrelated transcription factors SmTTG1, SmGL3 and SmTT8 in eggplant. Acta Hortic. Sin. 2014, 41, 2241–2249. [Google Scholar]
- Daehwan, K.; Ben, L.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS ONE 2016, 11, e157022. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D822–D827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhao, J.; Wan, X.; Luo, X.; Li, W.; Sun, L.; Chen, Q. From green to red: Large-scale transcriptome comparison of a bud sport in poplar (Populus deltoides). Acta Physiol. Plant. 2016, 38, 244. [Google Scholar] [CrossRef]
- Yang, S.; Liang, K.; Wang, A.; Zhang, M.; Zhang, L. Physiological Characterization and Transcriptome Analysis of Camellia oleifera Abel. During Leaf Senescence. Forests 2020, 11, 812. [Google Scholar] [CrossRef]
- Wang, Z.X.; Yu, Y.F.; Chen, L.; Qin, H.Y.; Liu, Y.X.; Ai, J.; Fan, S.T. Advances in leaf pigment composition, structure and photosynthetic characteristics of colored-leaf plants. Plant Physiol. J. 2016, 52, 1–7. [Google Scholar]
- He, X.; Zhao, X.; Gao, L.; Shi, X.; Dai, X.; Liu, Y.; Xia, T.; Wang, Y. Isolation and characterization of key genes that promote favonoid accumulation in purple-leaf tea (Camellia sinensis L.). Sci. Rep. 2018, 8, 130. [Google Scholar] [CrossRef] [Green Version]
- An, C.H.; Lee, K.W.; Lee, S.H.; Jeong, Y.J.; Woo, S.G.; Chun, H.; Park, Y.I.; Kwak, S.S.; Kim, C.Y. Heterologous expression of IbMYB1a by diferent promoters exhibits diferent patterns of anthocyanin accumulation in tobacco. Plant Physiol. Bioch. 2015, 89, 1–10. [Google Scholar] [CrossRef]
- Cho, J.S.; Nguyen, V.P.; Jeon, H.W.; Kim, M.H.; Eom, S.H.; Lim, Y.J.; Kim, W.C.; Park, E.J.; Choi, Y.I.; Ko, J.H. Overexpression of PtrMYB119, a R2R3-MYB transcription factor from Populus trichocarpa, promotes anthocyanin production in hybrid poplar. Tree Physiol. 2016, 36, 1162–1176. [Google Scholar] [CrossRef] [Green Version]
- Hughes, N.M.; Morley, C.B.; Smith, W.K. Coordination of anthocyanin decline and photosynthetic maturation in juvenile leaves of three deciduous tree species. New Phytologist. 2007, 175, 675–685. [Google Scholar] [CrossRef]
- Wei, L. Cloning and functional study of quinic acid/shikimate-hydroxycinnamoyl transferase gene in tea plants. Henan Agric. Sci. 2016, 45, 35–39. [Google Scholar]
- Huang, X.; Hu, L.; Kong, W.; Yang, C.; Xi, W. Red light-transmittance bagging promotes carotenoid accumulation of grapefruit during ripening. Commun. Biol. 2022, 5, 303. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, S.D.; Di Gaspero, G.; Marconi, R.; Nonis, A.; Peterlunger, E.; Paillard, S.; Adam-Blondon, A.F.; Testolin, R. Colour variation in red grapevines (Vitis vinifera L.): Genomic organisation, expression of flavonoid 3′-hydroxylase, flavonoid 3′,5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genom. 2006, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, Z.; Maximova, S.; Payne, M.J.; Guiltinan, M.J. Proanthocyanidin synthesis in Theobroma cacao: Genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 2013, 13, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quattrocchio, F.R.; Baudry, A.N.; Lepiniec, L.O.; Grotewold, E.R. The Regulation of Flavonoid Biosynthesis. In The Science of Flavonoids; Springer: New York, NY, USA, 2006; pp. 97–122. [Google Scholar]
- Lou, Q.; Liu, Y.; Qi, Y.; Jiao, S.; Tian, F.; Jiang, L.; Wang, Y. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Wu, H.; Ding, Q.; Li, H.; Li, Z.; Ding, J.; Li, Y. The heterologous expression of Arabidopsis PAP2 induces anthocyanin accumulation and inhibits plant growth in tomato. Funct. Integr. Genom. 2018, 18, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, X.; Song, W.; Bao, Y.; Jin, Y.; Jiang, C.; Wang, C.; Li, B.; Zhang, H. PdMYB118, isolated from a red leaf mutant of Populus deltoids, is a new transcription factor regulating anthocyanin biosynthesis in poplar. Plant Cell Rep. 2019, 38, 927–936. [Google Scholar] [CrossRef]
- Piao, W.L.; Kim, S.H.; Lee, B.D.; An, G.; Sakuraba, Y.; Paek, N.C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Li, L.X.; Wang, H.B.; Zhou, H.; Chen, X.S.; Feng, S.Q. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. 2022, 110, 1021–1034. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, Q.; Zeng, D.; Xu, J.; Zhou, H.; Wang, F.; Ma, N.; Li, Y. RhMYB108, an R2R3-MYB transcription factor, is involved in ethylene- and JA-induced petal senescence in rose plants. Hortic Res. 2019, 1, 131. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Wang, C.; Zhang, W.; Liu, S.; Wang, W.; Yu, X.; Song, T.; Yu, M.; Yu, W.; Qu, S. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic Res. 2021, 8, 156. [Google Scholar] [CrossRef]
- Koo, Y.; Poethig, R.S. Expression pattern analysis of three R2R3-MYB transcription factors for the production of anthocyanin in different vegetative stages of Arabidopsis leaves. Appl. Biol. Chem. 2021, 64, 5. [Google Scholar] [CrossRef]
- Jia, J.; Xing, J.H.; Dong, J.G.; Han, J.M.; Liu, J.S. Functional Analysis of MYB73 of Arabidopsis thaliana Against Bipolaris oryzae. Agric. Sci. China 2011, 10, 721–727. [Google Scholar] [CrossRef]
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, A.; Qian, M.; Li, J.; Li, Y.; Shen, J.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef] [Green Version]
- Fan, D.; Wang, X.; Tang, X.; Ye, X.; Ren, S.; Wang, D.; Luo, K. Histone H3K9 demethylase JMJ25 epigenetically modulates anthocyanin biosynthesis in poplar. Plant J. 2018, 96, 1121–1136. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Li, H.; Zhang, W.; Huang, L.; Zhu, J. Transcriptomic Analysis of the Differences in Leaf Color Formation during Stage Transitions in Populus × euramericana ‘Zhonghuahongye’. Agronomy 2022, 12, 2396. https://doi.org/10.3390/agronomy12102396
Chen M, Li H, Zhang W, Huang L, Zhu J. Transcriptomic Analysis of the Differences in Leaf Color Formation during Stage Transitions in Populus × euramericana ‘Zhonghuahongye’. Agronomy. 2022; 12(10):2396. https://doi.org/10.3390/agronomy12102396
Chicago/Turabian StyleChen, Mengjiao, Hui Li, Wan Zhang, Lin Huang, and Jingle Zhu. 2022. "Transcriptomic Analysis of the Differences in Leaf Color Formation during Stage Transitions in Populus × euramericana ‘Zhonghuahongye’" Agronomy 12, no. 10: 2396. https://doi.org/10.3390/agronomy12102396



