Characterization of Leaf Transcriptome of Grafted Tomato Seedlings after Rhizospheric Inoculation with Azospirillum baldaniorum or Paraburkholderia graminis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions and Microbial Inoculation
2.2. Rhizobacteria Rhizosphere Colonization
2.3. RNA Extraction and Illumina Sequencing
2.4. RNA-Seq Data Handling and Differential Gene Expression Analysis
2.5. GO Annotation and Enrichment
2.6. Quantitative RT-Real Time PCR Validation
3. Results
3.1. Colonization by Culturable Aerobic Rhizobacteria
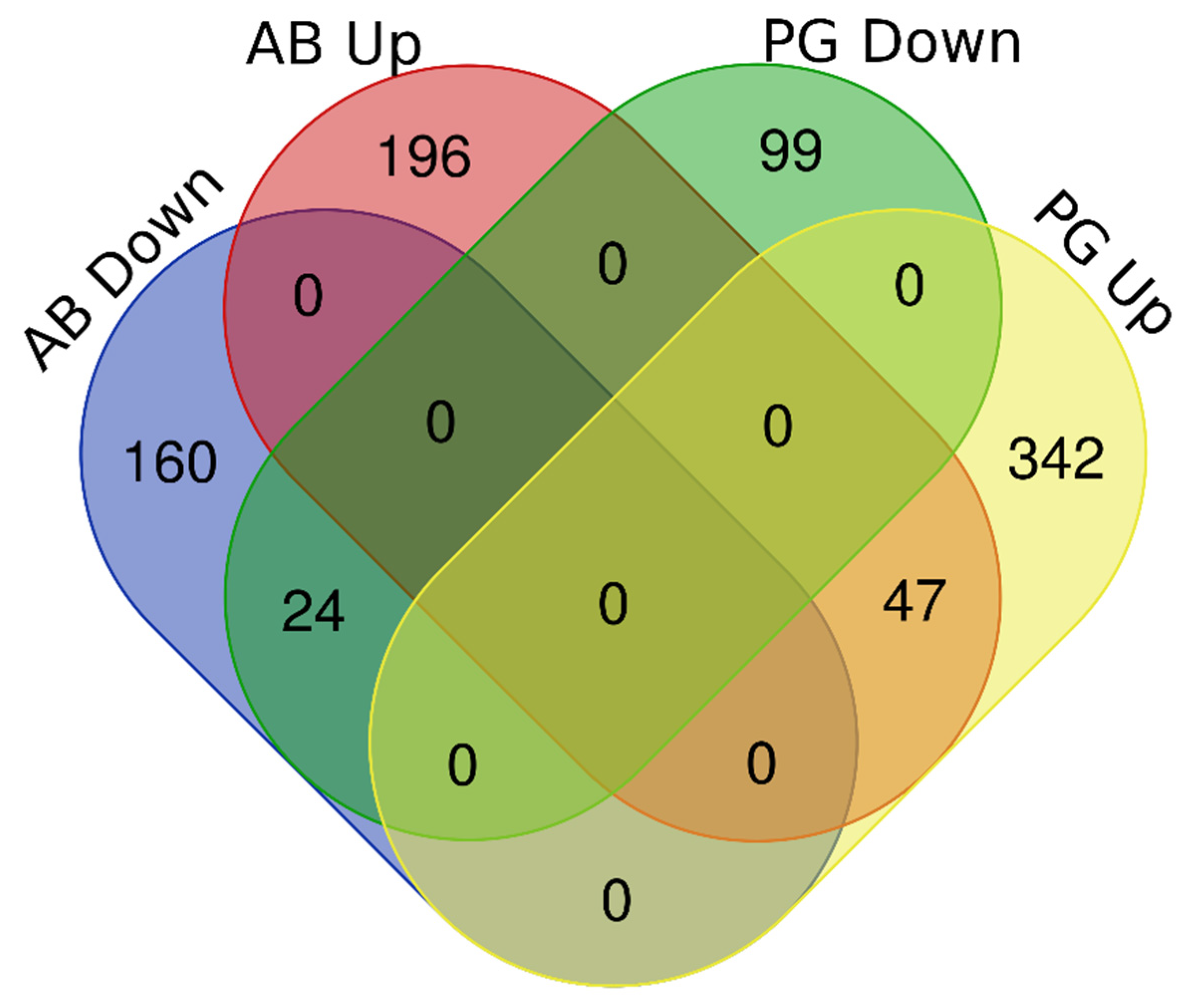
3.2. RNA Sequencing and Differential Expression Analyses
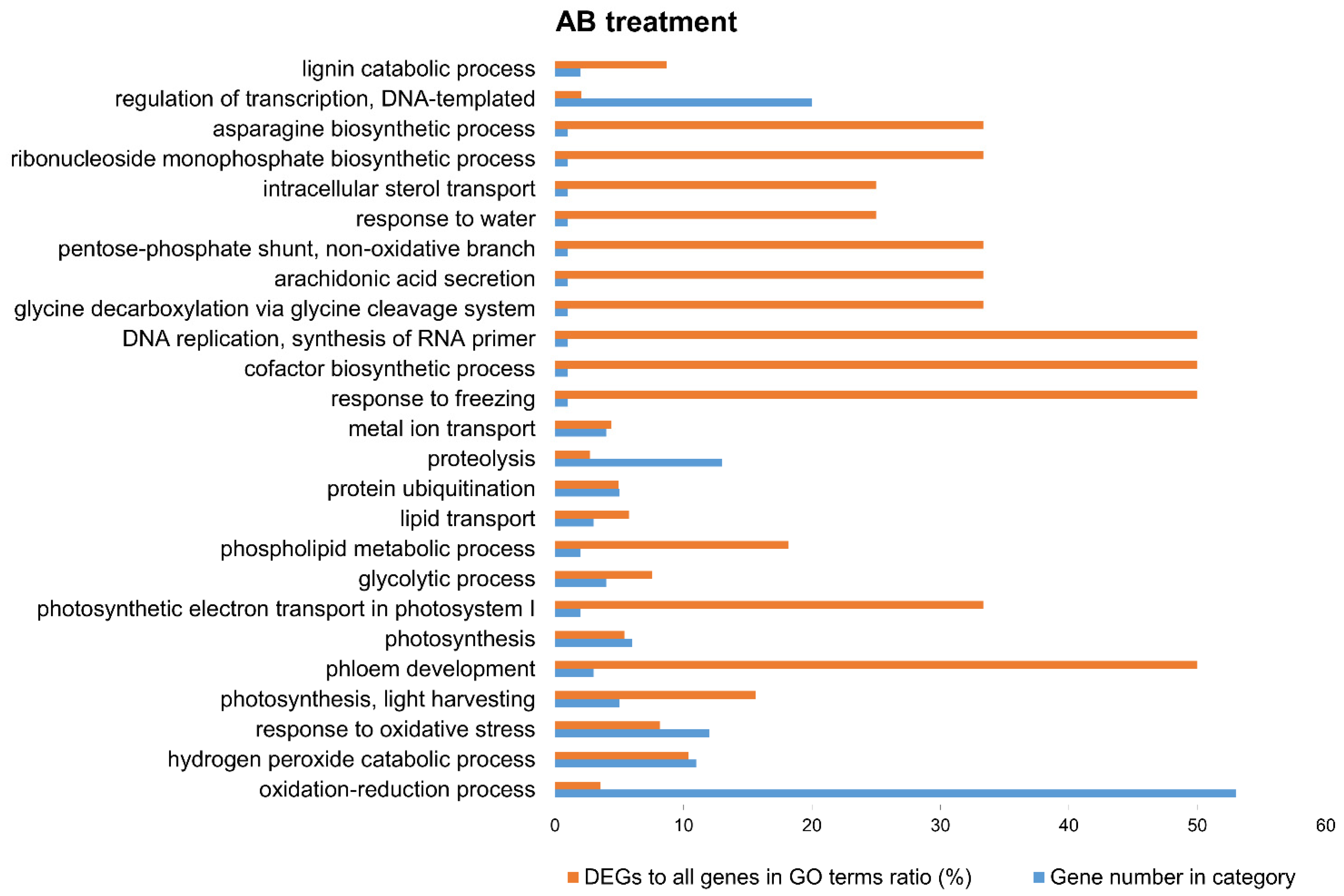
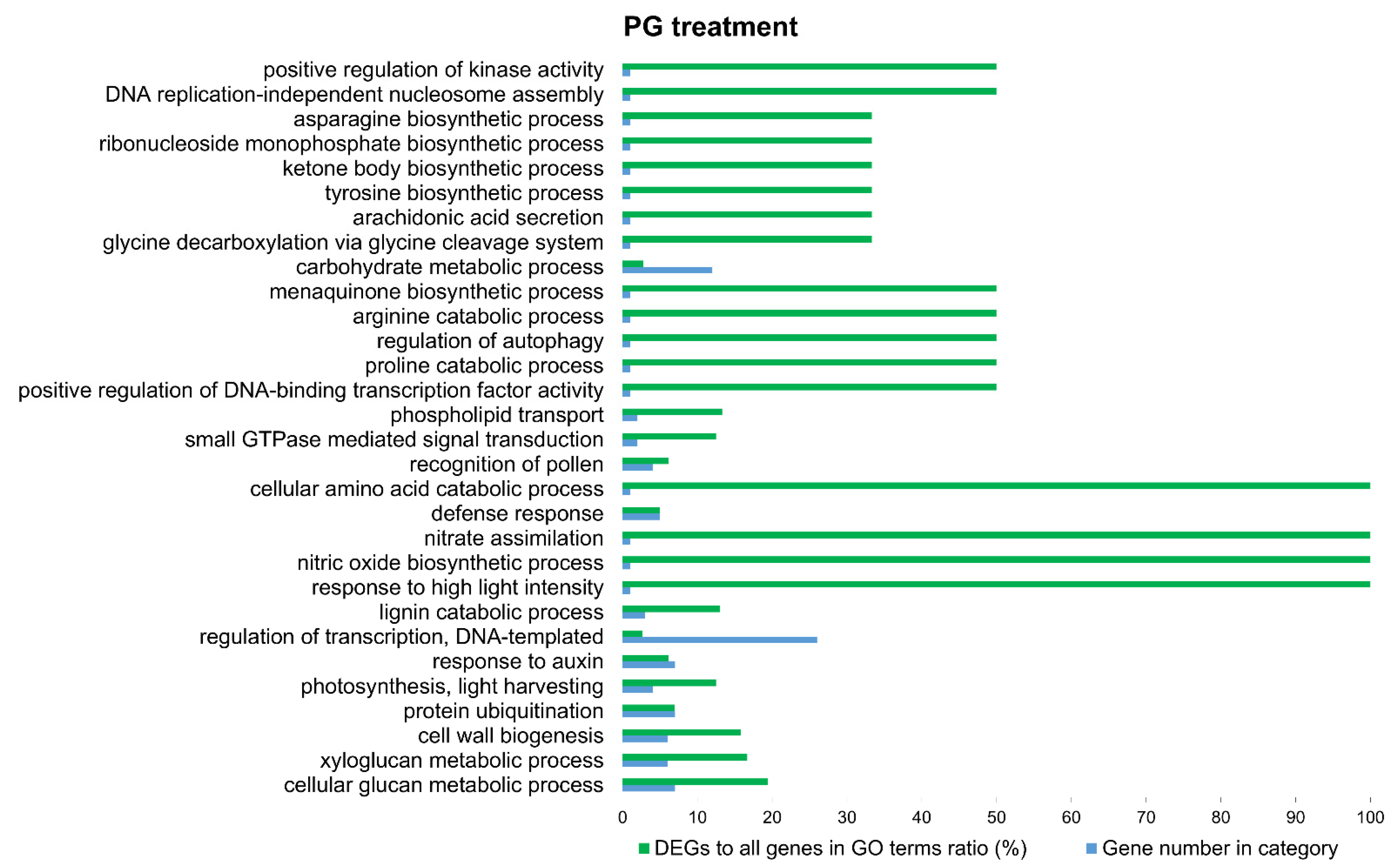
3.3. Functional Annotation and GO-Enrichment Analyses
3.4. Validation of Nine DEGs Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations FAOSTAT 2022. Available online: https://www.fao.org/faostat/en/#data (accessed on 10 January 2022).
- Storniolo, C.E.; Sacanella, I.; Lamuela-Raventos, R.M.; Moreno, J.J. Bioactive Compounds of Mediterranean Cooked Tomato Sauce (Sofrito) Modulate Intestinal Epithelial Cancer Cell Growth Through Oxidative Stress/Arachidonic Acid Cascade Regulation. ACS Omega 2020, 5, 17071–17077. [Google Scholar] [CrossRef] [PubMed]
- Foolad, M.R. Genome Mapping and Molecular Breeding of Tomato. Int. J. Plant Genom. 2007, 2007, 64358. [Google Scholar] [CrossRef] [Green Version]
- Cammarano, D.; Ronga, D.; Di Mola, I.; Mori, M.; Parisi, M. Impact of Climate Change on Water and Nitrogen Use Efficiencies of Processing Tomato Cultivated in Italy. Agric. Water Manag. 2020, 241, 106336. [Google Scholar] [CrossRef]
- Raklami, A.; Bechtaoui, N.; Tahiri, A.; Anli, M.; Meddich, A.; Oufdou, K. Use of Rhizobacteria and Mycorrhizae Consortium in the Open Field as a Strategy for Improving Crop Nutrition, Productivity and Soil Fertility. Front. Microbiol. 2019, 10, 1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; De Deyn, G.B.; Bezemer, T.M. Separating Effects of Soil Microorganisms and Nematodes on Plant Community Dynamics. Plant Soil 2019, 441, 455–467. [Google Scholar] [CrossRef] [Green Version]
- Dimkpa, C.; Weinand, T.; Asch, F. Plant-Rhizobacteria Interactions Alleviate Abiotic Stress Conditions: Plant-Rhizobacteria Interactions. Plant Cell Environ. 2009, 32, 1682–1694. [Google Scholar] [CrossRef]
- Vessey, J.K. Plant Growth Promoting Rhizobacteria as Biofertilizers. Plan Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Rothballer, M.; Schmid, M.; Fekete, A.; Hartmann, A. Comparative in Situ Analysis of IpdC-Gfpmut3 Promoter Fusions of Azospirillum Brasilense Strains Sp7 and Sp245. Environ. Microbiol. 2005, 7, 1839–1846. [Google Scholar] [CrossRef]
- Mariotti, L.; Picciarelli, P.; Lombardi, L.; Ceccarelli, N. Fruit-Set and Early Fruit Growth in Tomato Are Associated with Increases in Indoleacetic Acid, Cytokinin, and Bioactive Gibberellin Contents. J. Plant Growth Regul. 2011, 30, 405–415. [Google Scholar] [CrossRef]
- Caradonia, F.; Francia, E.; Morcia, C.; Ghizzoni, R.; Moulin, L.; Terzi, V.; Ronga, D. Arbuscular Mycorrhizal Fungi and Plant Growth Promoting Rhizobacteria Avoid Processing Tomato Leaf Damage during Chilling Stress. Agronomy 2019, 9, 299. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Saa, S. Biostimulants in Agriculture. Front. Plant Sci. 2015, 6, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yakhin, O.I.; Lubyanov, A.A.; Yakhin, I.A.; Brown, P.H. Biostimulants in Plant Science: A Global Perspective. Front. Plant Sci. 2017, 7, 2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naidoo, S.; Visser, E.A.; Zwart, L.; du Toit, Y.; Bhadauria, V.; Shuey, L.S. Dual RNA-Sequencing to Elucidate the Plant-Pathogen Duel. Curr. Issues Mol. Biol. 2018, 27, 127–142. [Google Scholar] [CrossRef]
- Zanardo, L.G.; de Souza, G.B.; Alves, M.S. Transcriptomics of Plant–Virus Interactions: A Review. Theor. Exp. Plant Physiol. 2019, 31, 103–125. [Google Scholar] [CrossRef]
- Milc, J.; Bagnaresi, P.; Aragona, M.; Valente, M.T.; Biselli, C.; Infantino, A.; Francia, E.; Pecchioni, N. Comparative Transcriptome Profiling of the Response to Pyrenochaeta Lycopersici in Resistant Tomato Cultivar Mogeor and Its Background Genotype—Susceptible Moneymaker. Funct. Integr. Genom. 2019, 19, 811–826. [Google Scholar] [CrossRef]
- Tan, G.; Liu, K.; Kang, J.; Xu, K.; Zhang, Y.; Hu, L.; Zhang, J.; Li, C. Transcriptome Analysis of the Compatible Interaction of Tomato with Verticillium Dahliae Using RNA-Sequencing. Front. Plant Sci. 2015, 6, 428. [Google Scholar] [CrossRef] [Green Version]
- Zouari, I.; Salvioli, A.; Chialva, M.; Novero, M.; Miozzi, L.; Tenore, G.C.; Bagnaresi, P.; Bonfante, P. From Root to Fruit: RNA-Seq Analysis Shows That Arbuscular Mycorrhizal Symbiosis May Affect Tomato Fruit Metabolism. BMC Genom. 2014, 15, 221. [Google Scholar] [CrossRef] [Green Version]
- Caradonia, F.; Ronga, D.; Flore, A.; Barbieri, R.; Moulin, L.; Terzi, V.; Francia, E. Biostimulants and Cherry Rootstock Increased Tomato Fruit Yield and Quality in Sustainable Farming Systems. Ital. J. Agron. 2020, 15, 121–131. [Google Scholar] [CrossRef]
- Jeon, J.-S.; Carreno-Quintero, N.; van Eekelen, H.D.L.M.; De Vos, R.C.H.; Raaijmakers, J.M.; Etalo, D.W. Impact of Root-Associated Strains of Three Paraburkholderia Species on Primary and Secondary Metabolism of Brassica Oleracea. Sci. Rep. 2021, 11, 2781. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 April 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An Improved de Novo Assembly and Annotation of the Tomato Reference Genome Using Single-Molecule Sequencing, Hi-C Proximity Ligation and Optical Maps. bioRxiv 2019, 767764. [Google Scholar] [CrossRef]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The Tomato Genome Sequence Provides Insights into Fleshy Fruit Evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLOS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference Gene Selection for Quantitative Real-Time PCR Normalization in Tomato Subjected to Nitrogen, Cold, and Light Stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Salvioli, A.; Zouari, I.; Chalot, M.; Bonfante, P. The Arbuscular Mycorrhizal Status Has an Impact on the Transcriptome Profile and Amino Acid Composition of Tomato Fruit. BMC Plant Biol. 2012, 12, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Hamid, B.; Zaman, M.; Farooq, S.; Fatima, S.; Sayyed, R.Z.; Baba, Z.A.; Sheikh, T.A.; Reddy, M.S.; El Enshasy, H.; Gafur, A.; et al. Bacterial Plant Biostimulants: A Sustainable Way towards Improving Growth, Productivity, and Health of Crops. Sustainability 2021, 13, 2856. [Google Scholar] [CrossRef]
- Cervantes-Gámez, R.G.; Bueno-Ibarra, M.A.; Cruz-Mendívil, A.; Calderón-Vázquez, C.L.; Ramírez-Douriet, C.M.; Maldonado-Mendoza, I.E.; Villalobos-López, M.Á.; Valdez-Ortíz, Á.; López-Meyer, M. Arbuscular Mycorrhizal Symbiosis-Induced Expression Changes in Solanum Lycopersicum Leaves Revealed by RNA-Seq Analysis. Plant Mol. Biol. Report. 2016, 34, 89–102. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vannini, C.; Ortolani, F.; Garcia-Seco, D.; Chiapello, M.; Novero, M.; Domingo, G.; Terzi, V.; Morcia, C.; Bagnaresi, P.; et al. Omics Approaches Revealed How Arbuscular Mycorrhizal Symbiosis Enhances Yield and Resistance to Leaf Pathogen in Wheat. Sci. Rep. 2018, 8, 9625. [Google Scholar] [CrossRef] [Green Version]
- Spanò, R.; Ferrara, M.; Montemurro, C.; Mulè, G.; Gallitelli, D.; Mascia, T. Grafting Alters Tomato Transcriptome and Enhances Tolerance to an Airborne Virus Infection. Sci. Rep. 2020, 10, 2538. [Google Scholar] [CrossRef] [Green Version]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Prigent-Combaret, C. Plant Secondary Metabolite Profiling Evidences Strain-Dependent Effect in the Azospirillum–Oryza Sativa Association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef]
- Walker, V.; Bertrand, C.; Bellvert, F.; Moënne-Loccoz, Y.; Bally, R.; Comte, G. Host Plant Secondary Metabolite Profiling Shows a Complex, Strain-dependent Response of Maize to Plant Growth-promoting Rhizobacteria of the Genus Azospirillum. New Phytol. 2011, 189, 494–506. [Google Scholar] [CrossRef]
- Hu, H.; Wang, C.; Li, X.; Tang, Y.; Wang, Y.; Chen, S.; Yan, S. RNA-Seq Identification of Candidate Defense Genes Targeted by Endophytic Bacillus Cereus -Mediated Induced Systemic Resistance against Meloidogyne Incognita in Tomato: RNA-Seq Identification of Candidate Defense Genes Targeted by Endophytic Bacillus Cereus. Pest Manag. Sci. 2018, 74, 2793–2805. [Google Scholar] [CrossRef]
- Zhou, C.; Zhu, J.; Qian, N.; Guo, J.; Yan, C. Bacillus Subtilis SL18r Induces Tomato Resistance Against Botrytis Cinerea, Involving Activation of Long Non-Coding RNA, MSTRG18363, to Decoy MiR1918. Front. Plant Sci. 2021, 11, 634819. [Google Scholar] [CrossRef]
- Balestrini, R.; Rosso, L.C.; Veronico, P.; Melillo, M.T.; De Luca, F.; Fanelli, E.; Colagiero, M.; di Fossalunga, A.S.; Ciancio, A.; Pentimone, I. Transcriptomic Responses to Water Deficit and Nematode Infection in Mycorrhizal Tomato Roots. Front. Microbiol. 2019, 10, 1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Maldonado-Mendoza, I.; Lopez-Meyer, M.; Cheung, F.; Town, C.D.; Harrison, M.J. Arbuscular Mycorrhizal Symbiosis Is Accompanied by Local and Systemic Alterations in Gene Expression and an Increase in Disease Resistance in the Shoots: Local and Systemic Alterations in Transcript Profiles in AM Symbiosis. Plant J. 2007, 50, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Gaufichon, L.; Reisdorf-Cren, M.; Rothstein, S.J.; Chardon, F.; Suzuki, A. Biological Functions of Asparagine Synthetase in Plants. Plant Sci. 2010, 179, 141–153. [Google Scholar] [CrossRef]
- Nasr Esfahani, M.; Inoue, K.; Nguyen, K.H.; Chu, H.D.; Watanabe, Y.; Kanatani, A.; Burritt, D.J.; Mochida, K.; Tran, L.P. Phosphate or Nitrate Imbalance Induces Stronger Molecular Responses than Combined Nutrient Deprivation in Roots and Leaves of Chickpea Plants. Plant Cell Environ. 2021, 44, 574–597. [Google Scholar] [CrossRef] [PubMed]
- Hagassou, D.; Francia, E.; Ronga, D.; Buti, M. Blossom End-Rot in Tomato (Solanum Lycopersicum L.): A Multi-Disciplinary Overview of Inducing Factors and Control Strategies. Sci. Hortic. 2019, 249, 49–58. [Google Scholar] [CrossRef]
- Chaumont, F.; Moshelion, M.; Daniels, M.J. Regulation of Plant Aquaporin Activity. Biol. Cell 2005, 97, 749–764. [Google Scholar] [CrossRef] [Green Version]
- Reuscher, S.; Akiyama, M.; Mori, C.; Aoki, K.; Shibata, D.; Shiratake, K. Genome-Wide Identification and Expression Analysis of Aquaporins in Tomato. PLoS ONE 2013, 8, e79052. [Google Scholar] [CrossRef] [Green Version]
- Roychoudhury, A.; Nayek, S. Structural Aspects and Functional Regulation of Late Embryogeniesis Abundant (LEA) Genes and Proteins Conferring Abiotic Stress Tolerance in Plants. In Abiotic Stress: Role in Sustainable Agriculture, Detrimental Effects and Management Strategies; Nova Publishers: New York, NY, USA, 2014; pp. 43–109. [Google Scholar]
- Lim, J.-H.; Kim, S.-D. Induction of Drought Stress Resistance by Multi-Functional PGPR Bacillus Licheniformis K11 in Pepper. Plant Pathol. J. 2013, 29, 201–208. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone Crosstalk in Plant Disease and Defense: More Than Just JASMONATE-SALICYLATE Antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Lushchak, V.I. Adaptive Response to Oxidative Stress: Bacteria, Fungi, Plants and Animals. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 153, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Hossain, M.A.; da Silva, J.A.T.; Fujita, M. Plant Response and Tolerance to Abiotic Oxidative Stress: Antioxidant Defense Is a Key Factor. In Crop Stress and its Management: Perspectives and Strategies; Venkateswarlu, B., Shanker, A.K., Shanker, C., Maheswari, M., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 261–315. ISBN 978-94-007-2219-4. [Google Scholar]
- Pignocchi, C.; Kiddle, G.; Hernández, I.; Foster, S.J.; Asensi, A.; Taybi, T.; Barnes, J.; Foyer, C.H. Ascorbate Oxidase-Dependent Changes in the Redox State of the Apoplast Modulate Gene Transcript Accumulation Leading to Modified Hormone Signaling and Orchestration of Defense Processes in Tobacco. Plant Physiol. 2006, 141, 423–435. [Google Scholar] [CrossRef] [Green Version]
- De Tullio, M.; Guether, M.; Balestrini, R. Ascorbate Oxidase Is the Potential Conductor of a Symphony of Signaling Pathways. Plant Signal. Behav. 2013, 8, e23213. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Steffens, J. Overexpression of Polyphenol Oxidase in Transgenic Tomato Plants Results in Enhanced Bacterial Disease Resistance. Planta 2002, 215, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Thipyapong, P.; Hunt, M.D.; Steffens, J.C. Antisense Downregulation of Polyphenol Oxidase Results in Enhanced Disease Susceptibility. Planta 2004, 220, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Narendra Babu, A.; Jogaiah, S.; Ito, S.; Kestur Nagaraj, A.; Tran, L.-S.P. Improvement of Growth, Fruit Weight and Early Blight Disease Protection of Tomato Plants by Rhizosphere Bacteria Is Correlated with Their Beneficial Traits and Induced Biosynthesis of Antioxidant Peroxidase and Polyphenol Oxidase. Plant Sci. 2015, 231, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Hwang, J.; Kim, K.; Kang, J.; Kim, B.; Xu, S.; Ahn, Y. Development of the Gene-Based SCARs for the Ph-3 Locus, Which Confers Late Blight Resistance in Tomato. Sci. Hortic. 2013, 164, 9–16. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Zhou, X.; Hou, X.; Yang, G.; Meng, J.; Luan, Y. Tomato MYB49 Enhances Resistance to Phytophthora Infestans and Tolerance to Water Deficit and Salt Stress. Planta 2018, 248, 1487–1503. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-Wide Analysis of WRKY Transcription Factors in Solanum Lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Wang, Y.; Zhuang, K.; Meng, Q.; Meng, C. Characterization and Analysis of Some Chilling-Response WRKY Transcription Factors in Tomato; Research Square: Durham, NC, USA, 2020. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-Wide Identification and Characterization of R2R3MYB Family in Solanum Lycopersicum. Mol. Genet. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef]
- Campobenedetto, C.; Mannino, G.; Beekwilder, J.; Contartese, V.; Karlova, R.; Bertea, C.M. The Application of a Biostimulant Based on Tannins Affects Root Architecture and Improves Tolerance to Salinity in Tomato Plants. Sci. Rep. 2021, 11, 354. [Google Scholar] [CrossRef]
- Buoso, S.; Pagliari, L.; Musetti, R.; Martini, M.; Marroni, F.; Schmidt, W.; Santi, S. ‘Candidatus Phytoplasma Solani’ Interferes with the Distribution and Uptake of Iron in Tomato. BMC Genom. 2019, 20, 703. [Google Scholar] [CrossRef]
- Puga-Freitas, R.; Blouin, M. A Review of the Effects of Soil Organisms on Plant Hormone Signalling Pathways. Environ. Exp. Bot. 2015, 114, 104–116. [Google Scholar] [CrossRef]
- Zhang, S.; Gu, X.; Shao, J.; Hu, Z.; Yang, W.; Wang, L.; Su, H.; Zhu, L. Auxin Metabolism Is Involved in Fruit Set and Early Fruit Development in the Parthenocarpic Tomato “R35-P”. Front. Plant Sci. 2021, 12, 671713. [Google Scholar] [CrossRef]
- Liu, M.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.E.P.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.-P.; Bouzayen, M.; et al. Comprehensive Profiling of Ethylene Response Factor Expression Identifies Ripening-Associated ERF Genes and Their Link to Key Regulators of Fruit Ripening in Tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.A.; Khan, M.I.R.; Ferrante, A.; Poor, P. Editorial: Ethylene: A Key Regulatory Molecule in Plants. Front. Plant Sci. 2017, 8, 1782. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Seymour, G.B.; Lu, C.; Hu, Z.; Chen, X.; Chen, G. An Ethylene Response Factor (ERF5) Promoting Adaptation to Drought and Salt Tolerance in Tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Soni, D.K.; Singh, R.; Dwivedi, U.N.; Pathre, U.V.; Nath, P.; Sane, A.P. SlERF36, an EAR-Motif-Containing ERF Gene from Tomato, Alters Stomatal Density and Modulates Photosynthesis and Growth. J. Exp. Bot. 2013, 64, 3237–3247. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, D.; Chen, J.; Yang, Y.; Huang, Z.; Huang, D.; Wang, X.-C.; Huang, R. Tomato Stress-Responsive Factor TSRF1 Interacts with Ethylene Responsive Element GCC Box and Regulates Pathogen Resistance to Ralstonia Solanacearum. Plant Mol. Biol. 2004, 55, 825–834. [Google Scholar] [CrossRef]
| Sample Properties | Trimming and Filtering | Aligning to Genome | |||||
|---|---|---|---|---|---|---|---|
| Sample ID | Treatment 1 | Replicate | Paired-End Reads | No Pairs Survived | % Pairs Survived | No Aligned Pairs | % Aligned Pairs |
| T25l_CON | Ctrl | 1 | 26,783,825 | 25,548,670 | 95.39% | 24,277,920 | 95.03% |
| T26l_CON | Ctrl | 2 | 24,651,592 | 23,369,549 | 94.80% | 21,413,685 | 91.63% |
| T27l_CON | Ctrl | 3 | 23,637,298 | 22,874,761 | 96.77% | 20,835,340 | 91.08% |
| T22l_ABA | AB | 1 | 22,012,255 | 21,065,854 | 95.70% | 19,617,111 | 93.12% |
| T23l_ABB | AB | 2 | 27,225,211 | 26,424,621 | 97.06% | 24,341,237 | 92.12% |
| T24l_ABC | AB | 3 | 23,976,755 | 22,934,673 | 95.65% | 21,820,168 | 95.14% |
| T19l_PGA | PG | 1 | 23,263,479 | 22,361,982 | 96.12% | 21,024,789 | 94.02% |
| T20l_PGB | PG | 2 | 26,445,081 | 25,487,216 | 96.38% | 24,121,624 | 94.64% |
| T21l_PGC | PG | 3 | 23,896,293 | 23,217,583 | 97.16% | 22,004,594 | 94.78% |
| DE Analysis 1 | Down-Regulated Transcripts | Up-Regulated Transcripts | Total No DE Transcripts |
|---|---|---|---|
| AB vs. Ctrl | 184 | 243 | 427 |
| PG vs. Ctrl | 123 | 389 | 512 |
| Treatment 1 | Molecular FUNCTION | Cellular Component | Biological Process | Total No GO Terms |
|---|---|---|---|---|
| AB | 33 | 7 | 25 | 65 |
| PG | 24 | 6 | 30 | 60 |
| Gene ID 1 | Chr 1 | Description 1 | AB | PG | ||
|---|---|---|---|---|---|---|
| RNA-Seq | RT q-PCR | RNA -Seq | RT q-PCR | |||
| Solyc10g076240.3.1 | 10 | Cationic peroxidase 1-like | 6.06 | 3.55 | −0.33 | −0.22 |
| Solyc02g078650.4.1 | 2 | Polyphenol oxidase, chloroplastic-like | 5.33 | 2.78 | 1.16 | 0.74 |
| Solyc01g106605.1.1 | 1 | Basic form of pathogenesis-related protein 1-like | 8.24 | 2.11 | 0.15 | |
| Solyc02g062390.3.1 | 2 | Dehydrin DHN2 | 5.05 | 4.27 | 2.47 | 1.65 |
| Solyc06g075650.3.1 | 6 | Aquaporin TIP1-3-like | 3.18 | 2.48 | 2.29 | 1.52 |
| Solyc03g026280.3.1 | 3 | AP2 domain CBF protein | 0.55 | −0.58 | 4.41 | 3.13 |
| Solyc07g056000.2.1 | 7 | Xyloglucan Endotransglucosylase/hydrolase 2 | 2.19 | 1.81 | 2.61 | 2 |
| Solyc05g052040.1.1 | 5 | Ethylene-responsive transcription factor 5 | 0.43 | 0.03 | 5.56 | 1.44 |
| Solyc12g009240.1.1 | 12 | Ethylene-responsive transcription factor ERF017 | 1.95 | −0.22 | 4.72 | 2.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caradonia, F.; Buti, M.; Flore, A.; Gatti, R.; Morcia, C.; Terzi, V.; Ronga, D.; Moulin, L.; Francia, E.; Milc, J.A. Characterization of Leaf Transcriptome of Grafted Tomato Seedlings after Rhizospheric Inoculation with Azospirillum baldaniorum or Paraburkholderia graminis. Agronomy 2022, 12, 2537. https://doi.org/10.3390/agronomy12102537
Caradonia F, Buti M, Flore A, Gatti R, Morcia C, Terzi V, Ronga D, Moulin L, Francia E, Milc JA. Characterization of Leaf Transcriptome of Grafted Tomato Seedlings after Rhizospheric Inoculation with Azospirillum baldaniorum or Paraburkholderia graminis. Agronomy. 2022; 12(10):2537. https://doi.org/10.3390/agronomy12102537
Chicago/Turabian StyleCaradonia, Federica, Matteo Buti, Alessia Flore, Roberto Gatti, Caterina Morcia, Valeria Terzi, Domenico Ronga, Lionel Moulin, Enrico Francia, and Justyna Anna Milc. 2022. "Characterization of Leaf Transcriptome of Grafted Tomato Seedlings after Rhizospheric Inoculation with Azospirillum baldaniorum or Paraburkholderia graminis" Agronomy 12, no. 10: 2537. https://doi.org/10.3390/agronomy12102537
APA StyleCaradonia, F., Buti, M., Flore, A., Gatti, R., Morcia, C., Terzi, V., Ronga, D., Moulin, L., Francia, E., & Milc, J. A. (2022). Characterization of Leaf Transcriptome of Grafted Tomato Seedlings after Rhizospheric Inoculation with Azospirillum baldaniorum or Paraburkholderia graminis. Agronomy, 12(10), 2537. https://doi.org/10.3390/agronomy12102537









