A Novel Locus for Bakanae Disease Resistance, qBK4T, Identified in Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions of Mapping Population, and Bioassay
2.2. DNA Extraction and High-Throughput SNP Genotyping
2.3. QTL Mapping and Genome-Wide Association Study (GWAS)
3. Results
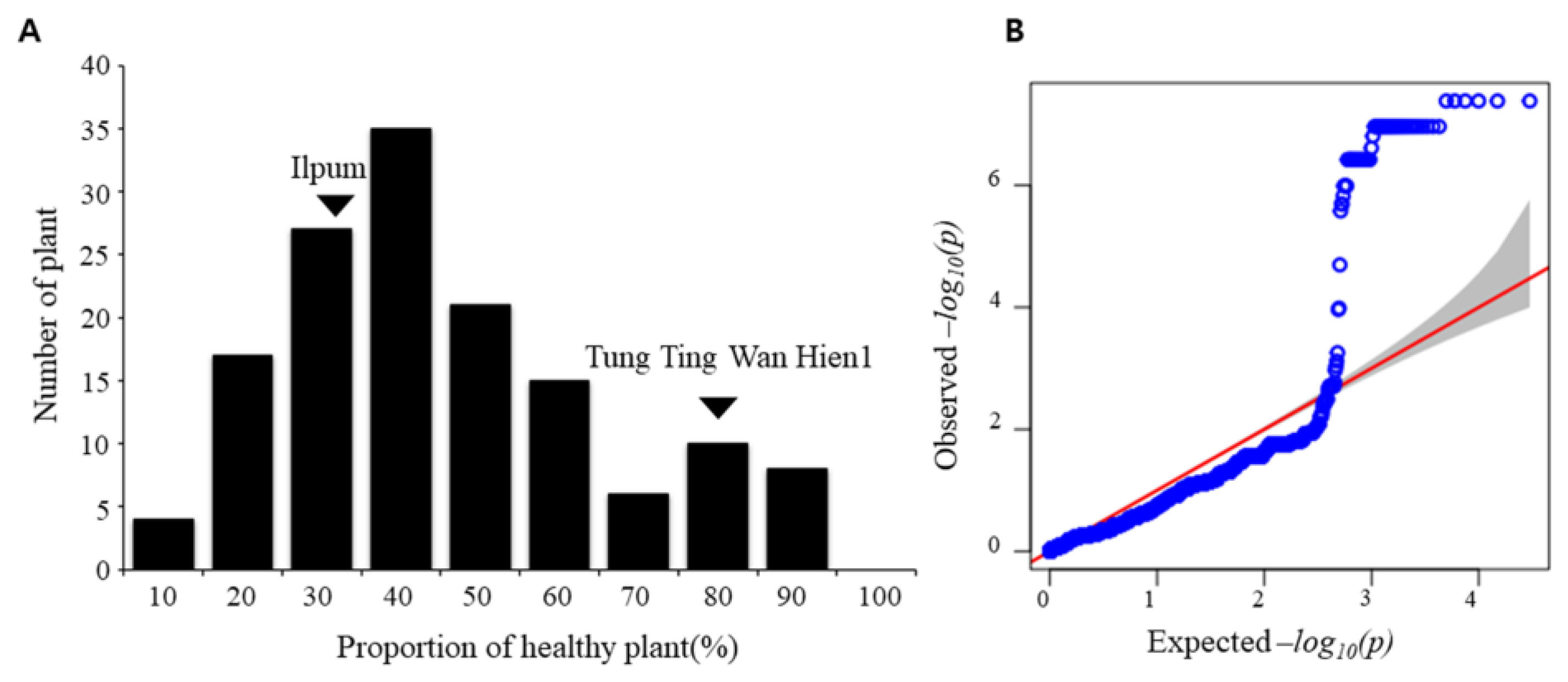
3.1. Phenotypic Response towards Bakanae Disease
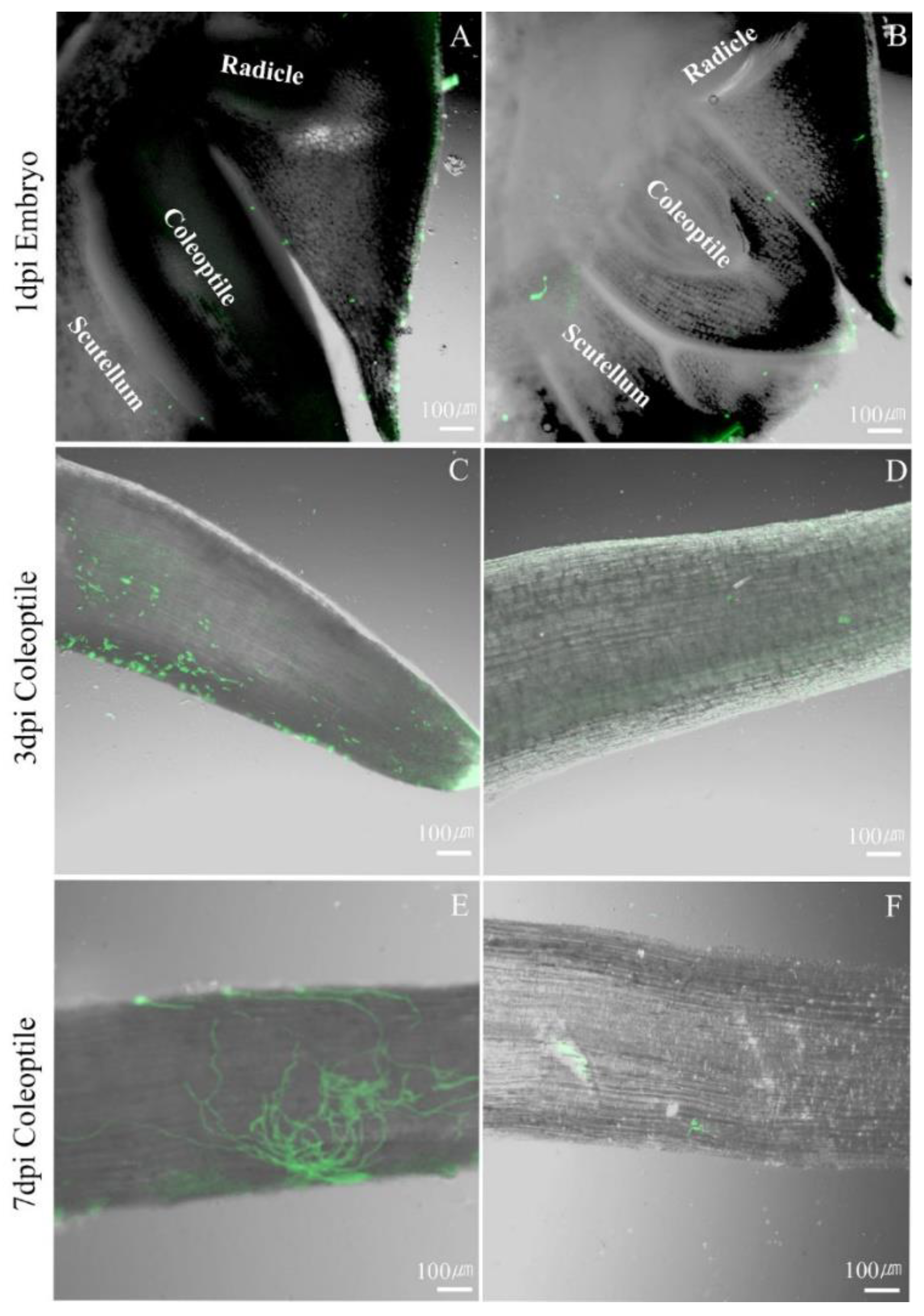
3.2. Differential Invasion Patterns of CF283-GFP Fusarium fujikuroi in the Early Stage of Infection
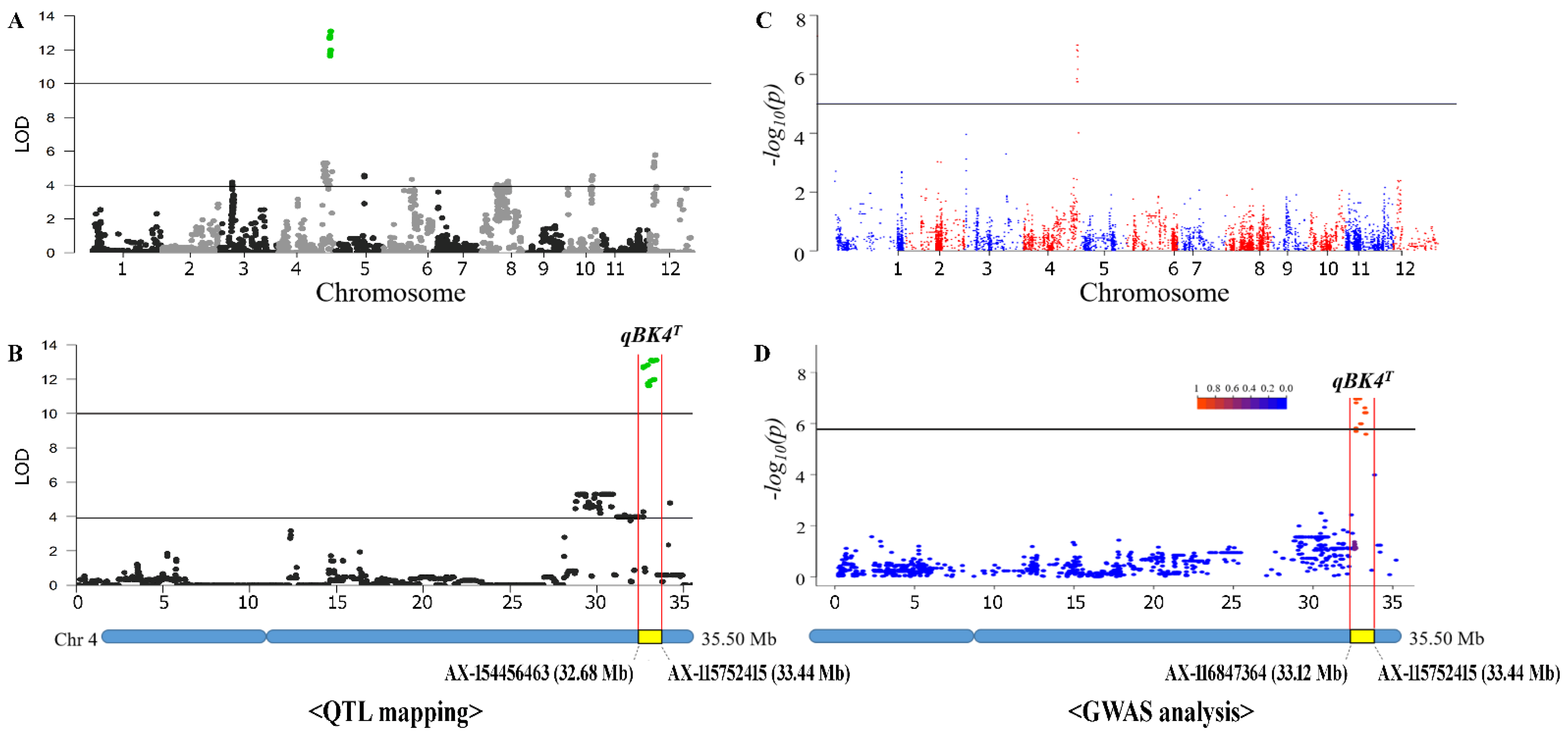
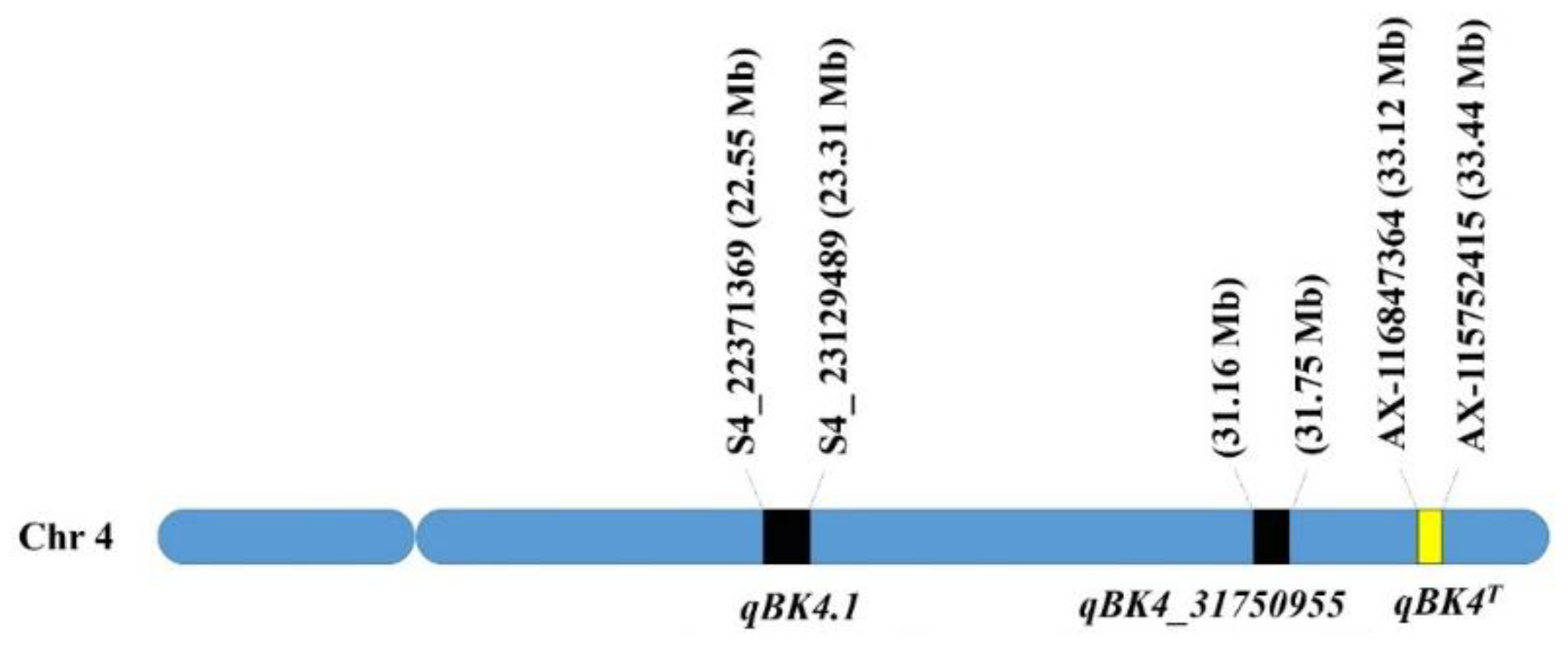
3.3. QTL Mapping and Genome-Wide Association Study (GWAS) Detected a Novel qBK4T Locus
4. Discussion
4.1. Differential Phenotypic Response of the Mapping Population towards Fusarium fujikuroi
4.2. GFP-Tagged Fusarium fujikuroi Localized in Embryo, and Stem of Susceptible Rice in the Early Stage of Infection
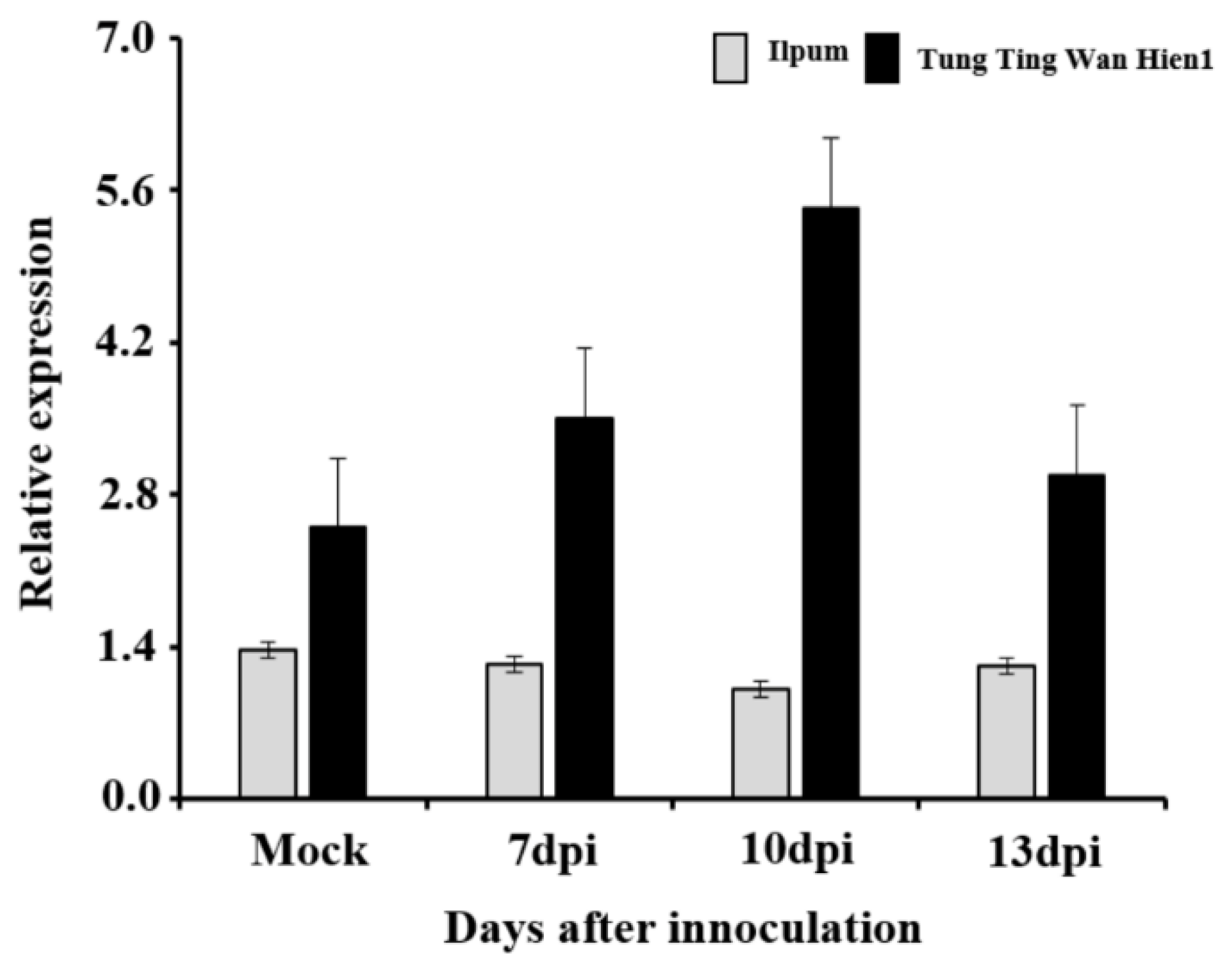
4.3. Novel qBK4T Harbors Genes Associated with Stress Signaling and Defense
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ito, S.; Kimura, J. Studies on the bakanae disease of the rice plant. Rep. Hokkaido Natl. Agric. Exp. Stn. 1931, 27, 1–95. [Google Scholar]
- Gupta, A.; Solanki, I.S.; Bashyal, B.; Singh, Y.; Srivastava, K. Bakanae of rice-an emerging disease in asia. J. Anim. Plant Sci. 2015, 25, 1499–1514. [Google Scholar]
- Wulff, E.G.; Sørensen, J.L.; Lübeck, M.; Nielsen, K.F.; Thrane, U.; Torp, J. Fusarium spp. associated with rice Bakanae: Ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 2010, 12, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Javed, N.; Sahi, S.T.; Cheema, N.M. Genetic management of bakanae disease of rice and evaluation of various fungicides against Fusarium moniliforme in vitro. Park. J. Phytopathol. 2011, 23, 103–107. [Google Scholar]
- Ou, S.H. Rice Diseases, 2nd ed.; Commonwealth Mycological Institute, Kew: Surrey, London, UK; Cabi Publishing: Wallingford, UK, 1985. [Google Scholar]
- Ora, N.; Faruq, A.; Islam, M.; Akhtar, N.; Rahman, M.M. Detection and identification of seed borne pathogens from some cultivated hybrid rice varieties in Bangladesh. Middel-East J. Sci. Res. 2011, 10, 482–488. [Google Scholar]
- Matic, S.; Gullino, M.L.; Spadaro, D. The puzzle of bakanae disease through interactions between Fusarium fujikuroi and rice. Front Biosci. 2017, 9, 333–344. [Google Scholar]
- Mew, T.W.; Gonzales, P.G. A Handbook of Rice Seedborne Fungi; International Rice Research Institute: Los Baňos, Philippines; Science Publishers, Inc., Enfield: New York, NY, USA, 2002. [Google Scholar]
- Singh, R.; Sunder, S. Foot rot and bakanae of rice: An overview. Rev. Plant Pathol. 2012, 5, 565–604. [Google Scholar]
- Desjardins, A.E.; Manandhar, H.K.; Plattner, R.D.; Manandhar, G.G.; Poling, S.M.; Maragos, C.M. Fusarium species from nepalese rice and production of mycotoxins and gibberellic acid by selected species. Appl. Environ. Microbiol. 2000, 66, 1020–1025. [Google Scholar] [CrossRef]
- Sunder, S.; Satyavir; Virk, K.S. Studies on correlation between bakanae incidence and yield loss in paddy. Indian Phytopathol. 1997, 50, 99–101. [Google Scholar]
- Singh, R.; Sunder, S. Foot rot and bakanae of rice: Retrospects and prospects. Int. J. Trop. Plant Dis. 1997, 15, 153–176. [Google Scholar]
- Yasin, S.; Khan, T.; Akhtar, K.; Anwer, M.; Ahmad, M. Economic evaluation of bakanae disease of rice. Mycopath 2003, 1, 115–117. [Google Scholar]
- Saremi, H.; Ammarellou, A.; Marefat, A.; Okhovvat, S. Binam a rice cultivar, resistant for root rot disease on rice caused by Fusarium moniliforme in northwest, Iran. Int. J. Bot. 2008, 4, 383–389. [Google Scholar] [CrossRef]
- Park, W.S.; Choi, H.W.; Han, S.S.; Shin, D.B.; Shim, H.K.; Jung, E.S.; Lee, S.W.; Lim, C.K.; Lee, Y.H. Control of bakanae disease of rice by seed soaking into the mixed solution of Procholraz and Fludioxnil. Res. Plant Dis. 2009, 15, 94–100. [Google Scholar] [CrossRef]
- Rosales, A.M.; Mew, T.W. Suppression of Fusarium moniliforme in rice by rice-associated antagonistic bacteria. Plant Dis. 1997, 81, 49–52. [Google Scholar] [CrossRef]
- Hayasaka, T.; Ishiguro, K.; Shibutani, K.; Namai, T. Seed disinfection using hot water immersion to control several seed-borne diseases of rice plants. Jpn. J. Phytopathol. 2001, 67, 26–32. [Google Scholar] [CrossRef]
- Lee, S.B.; Hur, Y.J.; Cho, J.H.; Lee, J.H.; Kim, T.H.; Cho, S.M.; Song, Y.C.; Seo, Y.S.; Lee, J.; Kim, T.S.; et al. Molecular mapping of qBK1WD, a major QTL for bakanae disease resistance in rice. Rice 2018, 11, 3. [Google Scholar] [CrossRef]
- Ogawa, K. Damage by “Bakanae” disease and its chemical control. Jpn. Pestic. Inf. 1988, 52, 13–15. [Google Scholar]
- Kim, J.M.; Hong, S.K.; Kim, W.G.; Lee, Y.K.; Yu, S.H.; Choi, H.W. Fungicide resistance of gibberella fujikuroi isolates causing rice bakanae disease and their progeny isolates. Korean J. Mycol. 2010, 38, 75–79. [Google Scholar] [CrossRef]
- Lee, Y.H.; Lee, M.J.; Choi, H.W.; Kim, S.T.; Park, J.W.; Myung, I.S.; Park, K.S.; Lee, S.W. Development of in vitro seedling screening method for selection of resistant rice against bakanae disease. Res. Plant Dis. 2011, 17, 288–294. [Google Scholar] [CrossRef]
- Yang, C.D.; Guo, L.B.; Li, X.M.; Ji, Z.J.; Ma, L.Y.; Qian, Q. Analysis of QTLs for resistance to rice bakanae disease. Chin. J. Rice Sci. 2006, 6, 657–659. [Google Scholar]
- Hur, Y.J.; Lee, S.B.; Kim, T.H.; Kwon, T.; Lee, J.H.; Shin, D.J.; Park, S.K.; Hwang, U.H.; Cho, J.H.; Yoon, Y.N.; et al. Mapping of qBk1, a major qtl for bakanae disease resistance in rice. Mol. Breed. 2015, 35, 78. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, N.; Hur, Y.J.; Cho, S.M.; Kim, T.H.; Lee, J.Y.; Cho, J.H.; Lee, J.H.; Song, Y.C.; Seo, Y.S.; et al. Fine mapping of qBK1, a major QTL for bakanae disease resistance in rice. Rice 2019, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Fiyaz, R.A.; Yadav, A.K.; Krishnan, S.G.; Ellur, R.K.; Bashyal, B.M.; Grover, N.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.; Singh, N.K. Mapping quantitative trait loci responsible for resistance to bakanae disease in rice. Rice 2016, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Kim, T.H.; Lee, G.S.; Kang, H.J.; Lee, S.B.; Suh, S.C.; Kim, S.L.; Choi, I.; Baek, J.; Kim, K.H. Mapping of a major quantitative trait locus for bakanae disease resistance in rice by genome resequencing. Mol. Genet. Genom. 2018, 293, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Volante, A.; Tondelli, A.; Aragona, M.; Valente, M.T.; Biselli, C.; Desiderio, F.; Bagnaresi, P.; Matic, S.; Gullino, M.L.; Infantino, A.; et al. Identification of bakanae disease resistance loci in japonica rice through genome wide association study. Rice 2017, 10, 29. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, N.; Jo, S.; Hur, Y.J.; Lee, J.Y.; Cho, J.H.; Lee, J.H.; Kang, J.W.; Song, Y.C.; Bombay, M.; et al. Mapping of a major QTL, qBK1z, for bakanae disease resistance in rice. Plants 2021, 10, 434. [Google Scholar] [CrossRef]
- Kang, D.Y.; Cheon, K.S.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Choi, I.; Baek, J.; Kim, K.; et al. Rice genome resequencing reveals a major quantitative trait locus for resistance to bakanae disease caused by Fusarium fujikuroi. Int. J. Mol. Sci. 2019, 20, 2598. [Google Scholar] [CrossRef]
- Kim, M.H.; Hur, Y.J.; Lee, S.B.; Kwon, T.; Hwang, U.H.; Park, S.K.; Yoon, Y.N.; Lee, J.H.; Cho, J.H.; Shin, D.J. Large-scale screening of rice accessions to evaluate resistance to bakanae disease. J. Gen. Plant Pathol. 2014, 80, 408–414. [Google Scholar] [CrossRef]
- Hur, Y.J.; Lee, S.B.; Shin, D.; Kim, T.H.; Cho, J.H.; Han, S.I.; Oh, S.H.; Lee, J.Y.; Son, Y.B.; Lee, J.H.; et al. Screening of rice germplasm for bakanae disease resistance in rice. Korean J. Breed. Sci. 2016, 48, 22–28. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Kim, K.W.; Nam, J.; Chu, S.H.; Phitaktansakul, R.; Yoo, J.M.; Kang, J.S.; Min, M.H.; Cheng, L.; Cao, Y.; Aung, K.M.; et al. Development of knu axiom oryza 580k genotyping array. In Proceedings of the KSBS & SABRAO International Conference on Plant Breeding for Sustainable Development, Gwangju, Korea, 2–5 July 2019; Volume 158, p. PCS02-58. [Google Scholar]
- Rana, A.; Sahgal, M.; Johri, B. Fusarium oxysporum: Genomics, diversity and plant–host interaction. In Developments in Fungal Biology and Applied Mycology; Satyanarayana, T., Deshmukh, S., Johri, B., Eds.; Springer: Singapore, 2017. [Google Scholar]
- Jansen, C.; Wettstein, D.V.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed]
- Elshafey, R.A.S.; Tahoon, A.M.; El-Emary, F.A. Analysis of varietal response to bakanae infection Fusarium fujikuroi and gibberellic acid through morphological, anatomical and hormonal changes in three rice varieties. J. Phytopathol. Pest Manag. 2018, 5, 63–87. [Google Scholar]
- Li, C.; Xu, M.; Cai, X.; Han, Z.; Si, J.; Chen, D. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 2022, 23, 3945. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [PubMed]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. Osjaz1 attenuates drought resistance by regulating ja and aba signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [PubMed]
- Grant, G.A. The act domain: A small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 2006, 281, 33825–33829. [Google Scholar] [CrossRef]
- Chen, C.Y.; Lin, P.H.; Chen, K.H.; Cheng, Y.S. Structural insights into arabidopsis ethylene response factor 96 with an extended n-terminal binding to gcc box. Plant Mol. Biol. 2020, 104, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene response factors in plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [PubMed]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by ap2/erf transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar]
- Mase, K.; Ishihama, N.; Mori, H.; Takahashi, H.; Kaminaka, H.; Kodama, M.; Yoshioka, H. Ethylene-responsive AP2/ERF transcription factor macd1 participates in phytotoxin-triggered programmed cell death. Mol. Plant Microbe Interact 2013, 26, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Lai, M.H.; Tung, C.W.; Wu, D.H.; Chang, F.Y.; Lin, T.C.; Chung, C.L. Genome-wide association mapping of gene loci affecting disease resistance in the rice-fusarium Fujikuroi pathosystem. Rice 2019, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.H.; Kabange, N.R.; Lee, J.Y.; Lee, S.M.; Cha, J.K.; Shin, D.J.; Cho, J.H.; Kang, J.W.; Ko, J.M.; Lee, J.H. Novel QTL associated with shoot branching identified in doubled haploid rice (Oryza sativa L.) under low nitrogen cultivation. Genes 2021, 12, 745. [Google Scholar] [CrossRef]
- Wang, P.; Li, Z.; Wei, J.; Zhao, Z.; Sun, D.; Cui, A. Na+/Ca2+ exchanger-like protein (AtNCLl) involved in salt stress in arabidopsis. J. Biol. Chem. 2012, 287, 44062–44070. [Google Scholar] [CrossRef]
- Schulze, D.H.; Muqhal, M.; Lederer, W.J.; Ruknudin, A.M. Sodium/Calcium exchanger (NCX1) macromolecular complex. J. Biol. Chem. 2003, 278, 28849–28855. [Google Scholar] [CrossRef]
- Chen, C.; Song, Y.; Zhuang, K.; Li, L.; Xia, Y.; Shen, Z. Proteomic analysis of copper-binding proteins in excess copper-stressed roots of two rice (Oryza sativa L.) varieties with different cu tolerances. PLoS ONE 2015, 10, e0125367. [Google Scholar]

| No. | MSU ID | Annotation |
|---|---|---|
| 1 | Os04g55640 | CCB4, protein of unknown function DUF579 |
| 2 | Os04g55650 | Oryzain alpha chain precursor (Copper-binding protein), hydrolase activity |
| 3 | Os04g55660 | GDSL-like lipase/acylhydrolase, hydrolase activity |
| 4 | Os04g55670 | Glycosyltransferase family 43 protein, transferase activity |
| 5 | Os04g55680 | Indole-3-acetate beta-glucosyltransferase |
| 6 | Os04g55690 | Cofactor assembly of complex C involved in photosystem II assembly |
| 7 | Os04g55700 | Exonuclease, nuclease activity |
| 8 | Os04g55720 | D-3-phosphoglycerate dehydrogenase, chloroplast precursor, nucleotide binding |
| 9 | Os04g55730 | Alpha-N-acetylglucosaminidase, hydrolase activity |
| 10 | Os04g55740 | Peroxidase precursor, catalytic activity, protein binding |
| 11 | Os04g55750 | OsWAK54—OsWAK short gene, kinase activity |
| 12 | Os04g55760 | OsWAK55, Wall-associated kinase (WAK), receptor-like protein kinase |
| 13 | Os04g55770 | GT1, Myeloblastosis (MYB)-like protein, SANT, one of the GT trihelix transcription |
| 14 | Os04g55780 | Arogenate dehydratase and ACT domains |
| 15 | Os04g55790 | GT1, myb-like, SANT family; GT-1, a myb-like protein, is one of the GT trihelix transcription |
| 16 | Os04g55800 | Sulfate transporter, transporter activity |
| 17 | Os04g55810 | Golgin subfamily A member 5; Members of this family of proteins are involved in maintaining Golgi structure. |
| 18 | Os04g55840 | Peptidase C65 Otubain domain containing protein encoding gene. This family of proteins conserved from plants to humans |
| 19 | Os04g55850 | Nuclease PA3 |
| 20 | Os04g55860 | Peptidyl-tRNA hydrolase |
| 21 | Os04g55920 | OsJAZ1, Zinc-finger protein |
| 22 | Os04g55940 | NCX1, sodium/calcium exchanger protein |
| 23 | Os04g55960 | NADPH reductase |
| 24 | Os04g55970 | AP2-like ethylene-responsive transcription factor AINTEGUMENTA |
| 25 | Os04g55980 | Glycine-rich RNA-binding, abscisic acid-inducible protein |
| 26 | Os04g56010 | Glycine-rich cell wall structural protein 1 precursor |
| 27 | Os04g56060 | Protein phosphorylation, protein serine/threonine, kinase activity |
| 28 | Os04g56070 | COP9 signalosome complex subunit 5b |
| 29 | Os04g56080 | S-locus-like receptor protein kinase, recognition of pollen, protein phosphorylation, protein serine/threonine kinase activity |
| 30 | Os04g56100 | One cut domain family member 3 |
| 31 | Os04g56110 | protein kinase, carbohydrate binding, kinase activity |
| 32 | Os04g56120 | protein kinase domain containing protein, kinase activity, carbohydrate binding |
| 33 | Os04g56130 | protein kinase domain containing protein, kinase activity, carbohydrate binding |
| 34 | Os04g56150 | AP2 domain containing protein, sequence-specific DNA binding transcription factor activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-B.; Lee, J.-Y.; Kang, J.-W.; Mang, H.; Kabange, N.R.; Seong, G.-U.; Kwon, Y.; Lee, S.-M.; Shin, D.; Lee, J.-H.; et al. A Novel Locus for Bakanae Disease Resistance, qBK4T, Identified in Rice. Agronomy 2022, 12, 2567. https://doi.org/10.3390/agronomy12102567
Lee S-B, Lee J-Y, Kang J-W, Mang H, Kabange NR, Seong G-U, Kwon Y, Lee S-M, Shin D, Lee J-H, et al. A Novel Locus for Bakanae Disease Resistance, qBK4T, Identified in Rice. Agronomy. 2022; 12(10):2567. https://doi.org/10.3390/agronomy12102567
Chicago/Turabian StyleLee, Sais-Beul, Ji-Yoon Lee, Ju-Won Kang, Hyunggon Mang, Nkulu Rolly Kabange, Gi-Un Seong, Youngho Kwon, So-Myeong Lee, Dongjin Shin, Jong-Hee Lee, and et al. 2022. "A Novel Locus for Bakanae Disease Resistance, qBK4T, Identified in Rice" Agronomy 12, no. 10: 2567. https://doi.org/10.3390/agronomy12102567
APA StyleLee, S.-B., Lee, J.-Y., Kang, J.-W., Mang, H., Kabange, N. R., Seong, G.-U., Kwon, Y., Lee, S.-M., Shin, D., Lee, J.-H., Cho, J.-H., Oh, K.-W., & Park, D.-S. (2022). A Novel Locus for Bakanae Disease Resistance, qBK4T, Identified in Rice. Agronomy, 12(10), 2567. https://doi.org/10.3390/agronomy12102567







