Genetic Resistance of Switchgrass to Rust Evaluated in a Composite Upland × Lowland Population in Lab and Field Settings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Design
2.2. Statistical Analysis
2.3. Responses to Selection
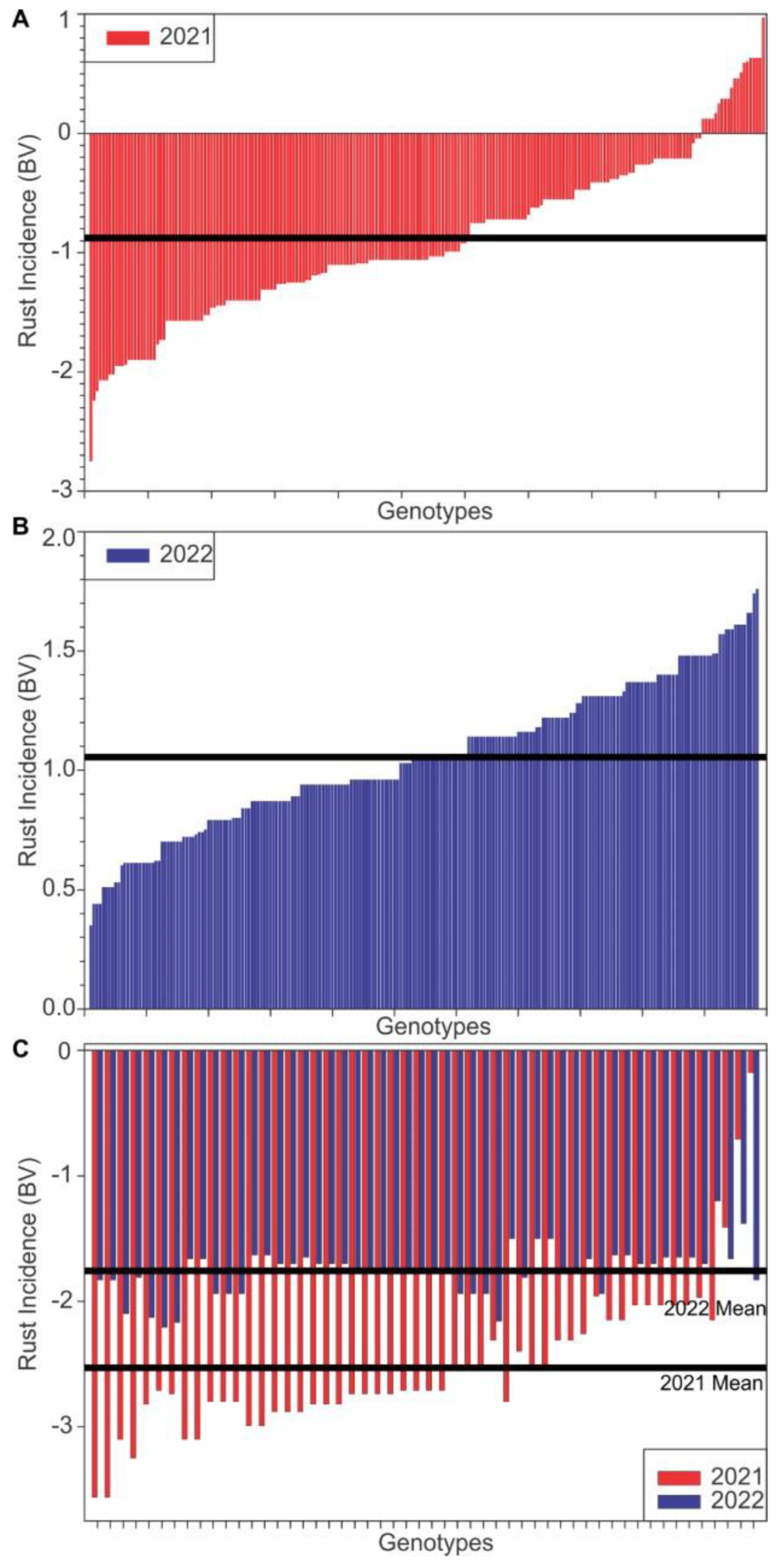
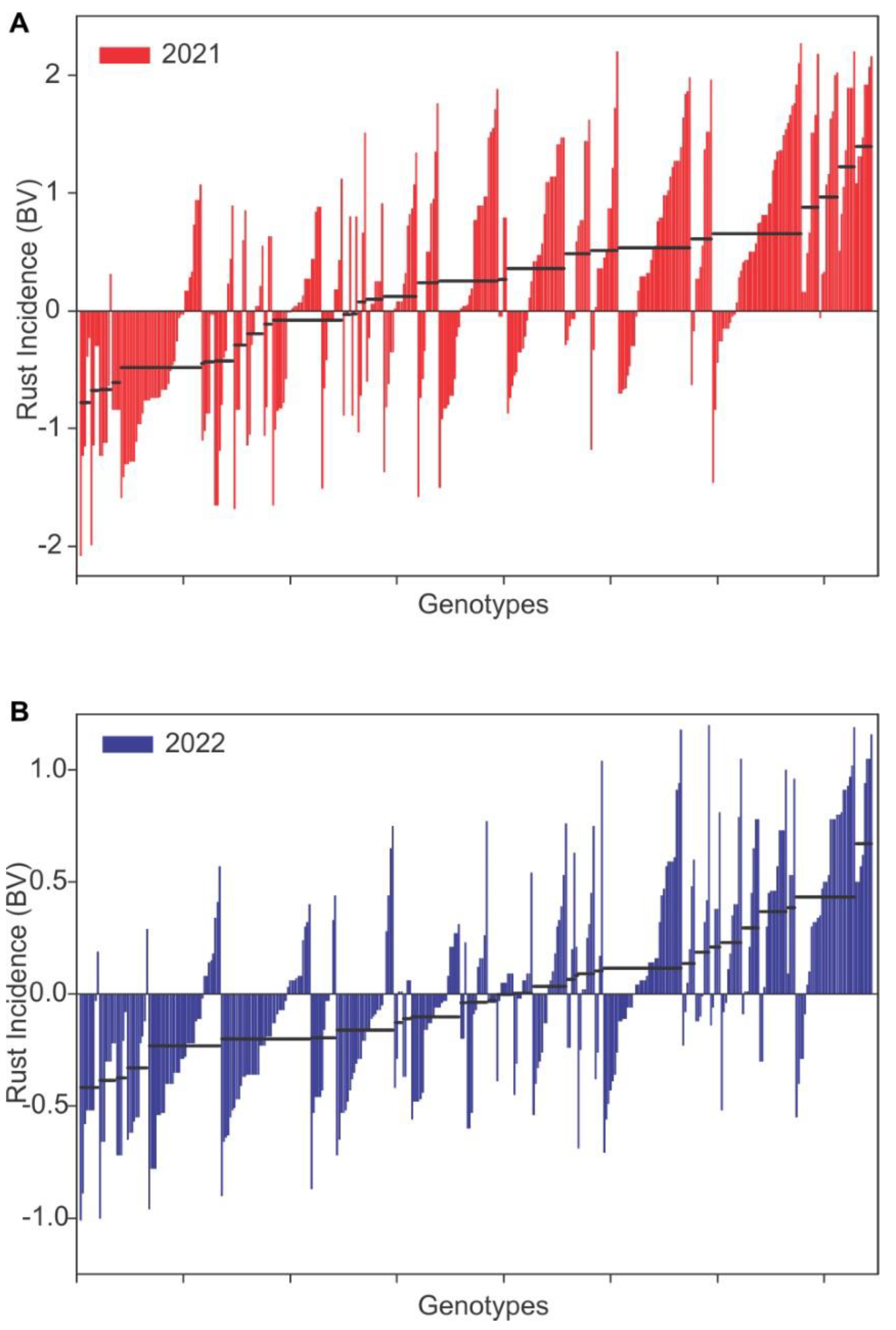
3. Results
3.1. Genetic Parameters
3.2. Predicted and Realized Responses to Selection
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vogel, K.P. Switchgrass. In Warm-Season (C4) Grasses; Moser, L.E., Burson, B.L., Sollenberger, L.E., Eds.; ASA, CSSA, and SSSA: Madison, WI, USA, 2004; pp. 561–588. [Google Scholar]
- Mitchell, R.; Vogel, K.P.; Sarath, G. Managing and enhancing switchgrass as a bioenergy feedstock. Biofuels Bioprod. Biorefining 2008, 2, 530–539. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, S.B.; Kszos, L.A. Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 2005, 28, 515–535. [Google Scholar] [CrossRef]
- Edmé, S.; Mitchell, R.; Sarath, G. Genetic parameters and prediction of breeding values in switchgrass bred for bioenergy. Crop Sci. 2017, 57, 1464–1474. [Google Scholar] [CrossRef]
- Nayak, S.; Bhandari, H.; Sams, C.; Sykes, V.; Hilafu, H.; Dalid, C.; Senseman, S.; Pantalone, V. Genetic variation for biomass yield and predicted genetic gain in lowland switchgrass “Kanlow”. Agronomy 2020, 10, 1845. [Google Scholar] [CrossRef]
- Casler, M.D.; Tobias, C.M.; Kaeppler, S.M.; Buell, C.R.; Wang, Z.-Y.; Cao, P.; Schmutz, J.; Ronald, P. The switchgrass genome: Tools and strategies. Plant Genome 2011, 4, 273–282. [Google Scholar] [CrossRef]
- Edmé, S.J.; Sarath, G.; Palmer, N.; Yuen, G.; Muhle, A.; Mitchell, R.; Tatineni, S.; Tobias, C. Genetic (co)variation and accuracy of selection for resistance to viral mosaic disease and production traits in an inter-ecotypic switchgrass breeding population. Crop Sci. 2021, 61, 1652–1665. [Google Scholar] [CrossRef]
- Vogel, K.P.; Mitchell, R.B.; Casler, M.D.; Sarath, G. Registration of ‘Liberty’ Switchgrass. J. Plant Regist. 2014, 8, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Tilhou, N.W.; Casler, M.D. Biomass yield improvement in switchgrass through genomic prediction of flowering time. GCB Bioenergy 2022, 14, 1023–1034. [Google Scholar] [CrossRef]
- Ma, Y. Rust Diseases on Switchgrass (Panicum virgatum). Master’s Thesis, University of Nebraska-Lincoln, Lincoln, NE, USA, 2015. [Google Scholar]
- Sykes, V.; Allen, F.L.; Mielenz, J.R.; Stewart, C.N.; Windham, M.T.; Hamilton, C.Y.; Rodriguez, M., Jr.; Yee, K.L. Reduction of ethanol yield from switchgrass infected with rust caused by Puccinia emaculata. BioEnergy Res. 2015, 9, 239–247. [Google Scholar] [CrossRef]
- Demers, J.E.; Liu, M.; Hambleton, S.; Castlebury, L.A. Rust fungi on Panicum. Mycologia 2017, 109, 1–17. [Google Scholar] [CrossRef]
- Cheng, Q.; Windham, A.S.; Lamour, K.H.; Saxton, A.M.; Windham, M.T. Evaluation of variation in switchgrass (Panicum virgatum L.) cultivars for rust (Puccinia emaculata) resistance. J. Environ. Hortic. 2019, 37, 127–135. [Google Scholar] [CrossRef]
- Van Wallendael, A.; Bonnette, J.; Juenger, T.E.; Fritschi, F.B.; Fay, P.A.; Mitchell, R.B.; Lloyd-Reilley, J.; Rouquette, F.M.; Bergstrom, G.C.; Lowry, D.B. Geographic variation in the genetic basis of resistance to leaf rust between locally adapted ecotypes of the biofuel crop switchgrass (Panicum virgatum). New Phytol. 2020, 227, 1696–1708. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Serba, D.D.; Ishiga, Y.; Szabo, L.; Mittal, S.; Bhandari, H.; Bouton, J.; Mysore, K.; Saha, M. Characterization of the rust fungus, Puccinia emaculata, and evaluation of genetic variability for rust resistance in switchgrass populations. Bioenergy Res. 2013, 6, 458–468. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, D.M.; Boe, A.; Jin, Y. Genetic variation for Puccinia emaculata infection in switchgrass. Crop Sci. 2003, 43, 755–759. [Google Scholar] [CrossRef]
- Palmer, N.; Alvarez, S.; Naldrett, M.J.; Muhle, A.; Sarath, G.; Edmé, S.J.; Tatineni, S.; Mitchell, R.B.; Yuen, G. Dynamic reconfiguration of switchgrass proteomes in response to rust (Puccinia novopanici) infection. Sci. Rep. 2022; submitted. [Google Scholar]
- SAS Institute Inc. SAS/STAT® 14.3 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2017. [Google Scholar]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates, Inc.: Sunderland, UK, 1998. [Google Scholar]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Addison Wesley Longman: Harlow, UK, 1996. [Google Scholar]
- Eberhart, S.A.; Newell, L.C. Variation in domestic collections of switchgrass, Panicum virgatum L. Agronomy J. 1959, 51, 613–616. [Google Scholar] [CrossRef]
- Newell, L.C.; Eberhart, S.A. Clone and progeny evaluation in the improvement of switchgrass, Panicum virgatum L. Crop Sci. 1961, 1, 117–121. [Google Scholar] [CrossRef] [Green Version]
- Milano, E.R.; Lowry, D.B.; Juenger, T.E. The genetic basis of upland/lowland ecotype divergence in switchgrass (Panicum virgatum). G3 Genes|Genomes|Genet. 2016, 6, 3561–3570. [Google Scholar] [CrossRef] [Green Version]
- Harvell, C.; Mitchell, C.; Ward, J.; Altizer, S.; Dobson, A.; Ostfeld, R.S.; Samuel, M.D. Climate warming and disease risks for terrestrial and marine biota. Science 2002, 296, 2158–2163. [Google Scholar] [CrossRef] [Green Version]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Silva, P.H.M.; Miranda, A.C.; Moraes, M.L.T.; Furtado, E.L.; Stape, J.L.; Alvares, C.A.; Sentelhas, P.C.; Mori, E.S.; Sebbenn, A.M. Selecting for rust (Puccinia psidii) resistance in Eucalyptus grandis in São Paulo State, Brazil. For. Ecol. Manag. 2013, 303, 91–97. [Google Scholar] [CrossRef]
- Gravert, C.E.; Munkvold, G.P. Fungi and diseases associated with cultivated switchgrass in Iowa. J. Iowa Acad. Sci. 2002, 109, 30–34. [Google Scholar]
- Robinson, G.K. That BLUP is a good thing: The estimation of random effects. Stat. Sci. 1991, 6, 15–32. [Google Scholar]
- Piepho, H.P.; Möhring, J.; Melchinger, A.E.; Büchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 2008, 161, 209–228. [Google Scholar] [CrossRef]
- Vogel, K.P.; Jung, H.-J.G. Genetic modification of herbaceous plants for feed and fuel. Crit. Rev. Plant Sci. 2001, 20, 15–49. [Google Scholar] [CrossRef]
- Serba, D.D.; Uppalapati, S.R.; Mukherjee, S.; Krom, N.; Tang, Y.; Mysore, K.S.; Saha, M.C. Transcriptome profiling of rust resistance in switchgrass using RNA-Seq analysis. Plant Genome 2015, 8, plantgenome2014-10. [Google Scholar] [CrossRef]
| Parameters | Estimate | Standard Error | Z-Value | p-Value |
|---|---|---|---|---|
| Family | 0.244 | 0.102 | 2.39 | 0.008 |
| Block (Rep) | 0.083 | 0.066 | 1.25 | 0.105 |
| Family × Rep | 0.238 | 0.084 | 2.82 | 0.002 |
| Residual | 0.868 | 0.057 | 15.22 | <0.0001 |
| h2f | 0.73 | 0.093 | ||
| h2i | 0.68 | 0.25 |
| Parameters | Estimate | Standard Error | z-Ratio | p-Value |
|---|---|---|---|---|
| Family | 0.0956 | 0.03614 | 2.65 | 0.0041 |
| Block (Rep) | 0.1252 | 0.053 | 2.36 | 0.0091 |
| Family × Rep | 0.03521 | 0.03749 | 0.94 | 0.1738 |
| Residual | 0.6603 | 0.04287 | 15.4 | <0.0001 |
| h2f | 0.63 | 0.09 | ||
| h2i | 0.42 | 0.14 |
| Parameters | Estimate | Standard Error | z-Ratio | p-Value |
|---|---|---|---|---|
| Family | 0.0844 | 0.09714 | 0.87 | 0.1924 |
| Block (Rep) | 0 | - | - | - |
| Family × Rep | 0.2944 | 0.1482 | 1.99 | 0.0235 |
| Residual | 2.2638 | 0.1749 | 12.94 | <0.0001 |
| Populations | BV a (Mean) | Gains_Pred b | Top 10% c | Gains_Pred |
|---|---|---|---|---|
| Δfam-2021 | 3.06 | |||
| Δfam-2022 | 2.12 | |||
| Δi-2021 | 1.43 | |||
| Δi-2022 | 0.70 | |||
| Kanlow | −2.452 | |||
| Gen 2_family | 0.21 d | −1.08 | −0.68 | −0.72 |
| Gen 2_individual | −1.25 | −0.49 | ||
| Gen 3_ family | −0.889 | −0.64 | −0.889 | −0.64 |
| Gen 3_ individual | −1.99 | −0.19 | ||
| Gains_realized (%) | ||||
| Gen 3/2_ family | 31 | |||
| Gen 3/2_ individual | 59 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edmé, S.J.; Palmer, N.A.; Sarath, G.; Muhle, A.A.; Mitchell, R.; Yuen, G. Genetic Resistance of Switchgrass to Rust Evaluated in a Composite Upland × Lowland Population in Lab and Field Settings. Agronomy 2022, 12, 3137. https://doi.org/10.3390/agronomy12123137
Edmé SJ, Palmer NA, Sarath G, Muhle AA, Mitchell R, Yuen G. Genetic Resistance of Switchgrass to Rust Evaluated in a Composite Upland × Lowland Population in Lab and Field Settings. Agronomy. 2022; 12(12):3137. https://doi.org/10.3390/agronomy12123137
Chicago/Turabian StyleEdmé, Serge J., Nathan A. Palmer, Gautam Sarath, Anthony A. Muhle, Rob Mitchell, and Gary Yuen. 2022. "Genetic Resistance of Switchgrass to Rust Evaluated in a Composite Upland × Lowland Population in Lab and Field Settings" Agronomy 12, no. 12: 3137. https://doi.org/10.3390/agronomy12123137






