Remediation of Pasture Dieback Using Plant Growth Promotant
Abstract
:1. Introduction
2. Materials and Methods
2.1. Farm Description and Location
2.2. Experimental Plots
2.3. Sampling and Measurement
2.4. Soil Mineral Analysis
2.5. DNA Sequencing and Bioinformatics
3. Results
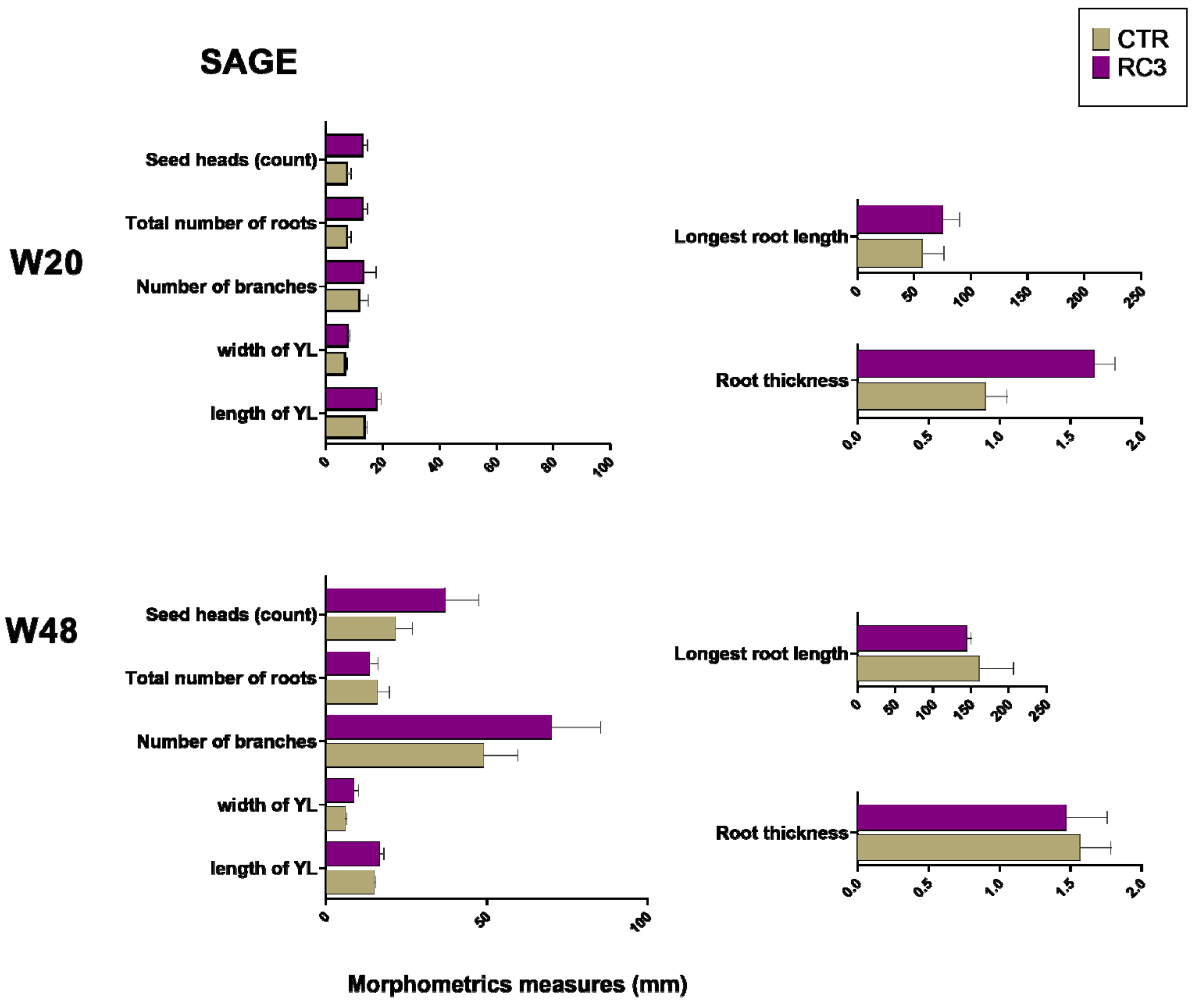
3.1. Plant Dry Matter, Morphometric Measurements and Soil Minerals
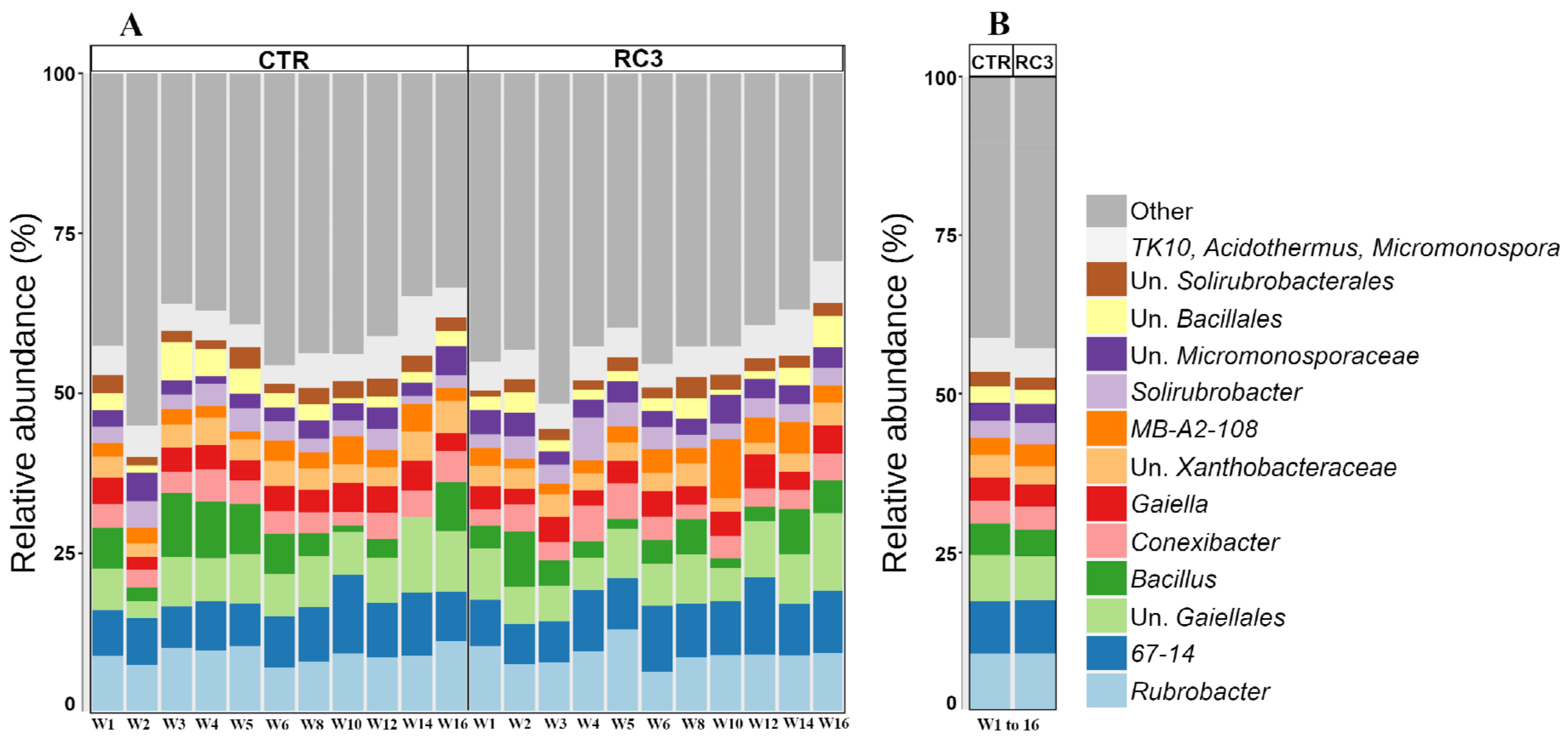
3.2. Soil Microbial Community Structure
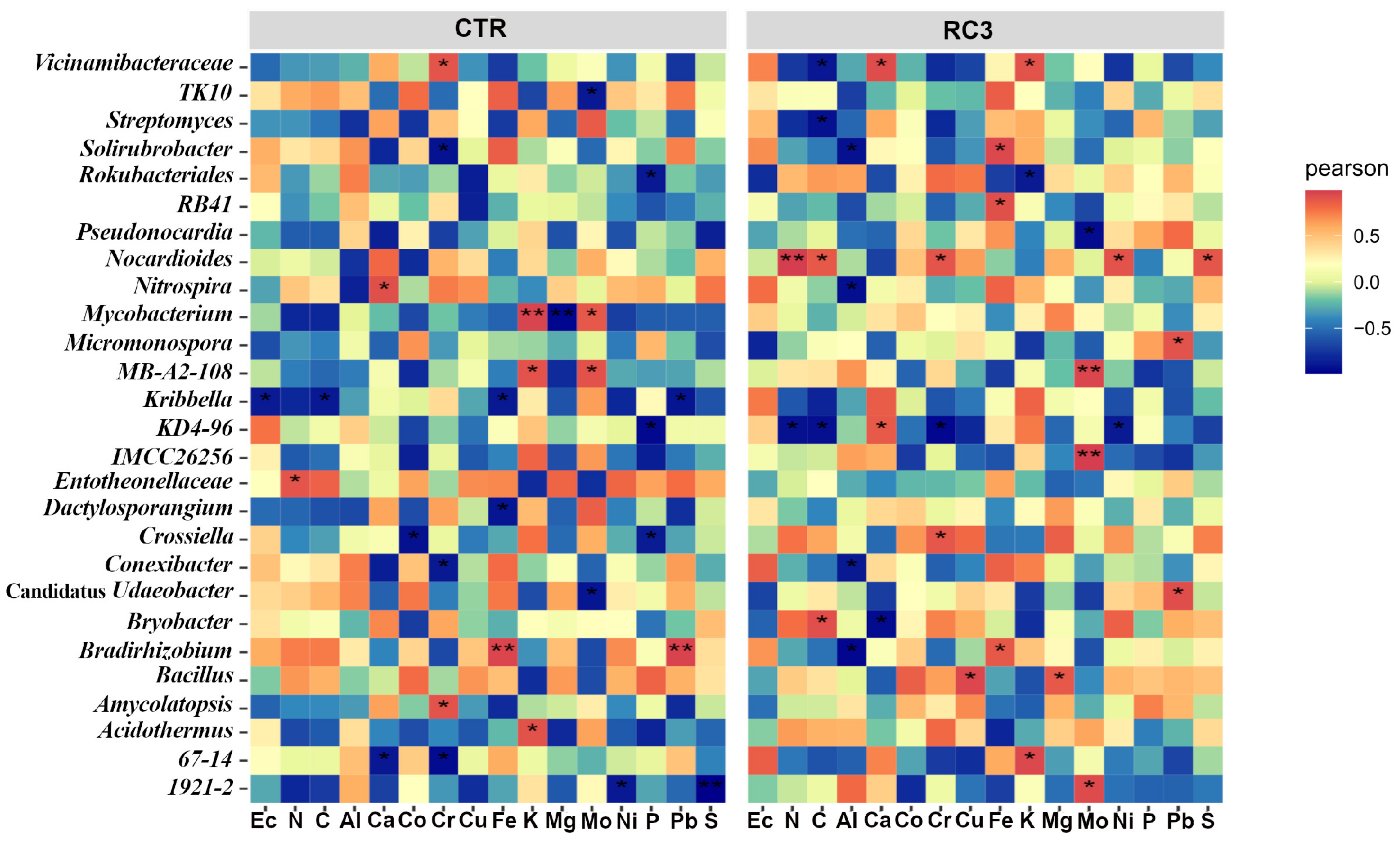
3.3. Correlation Analysis
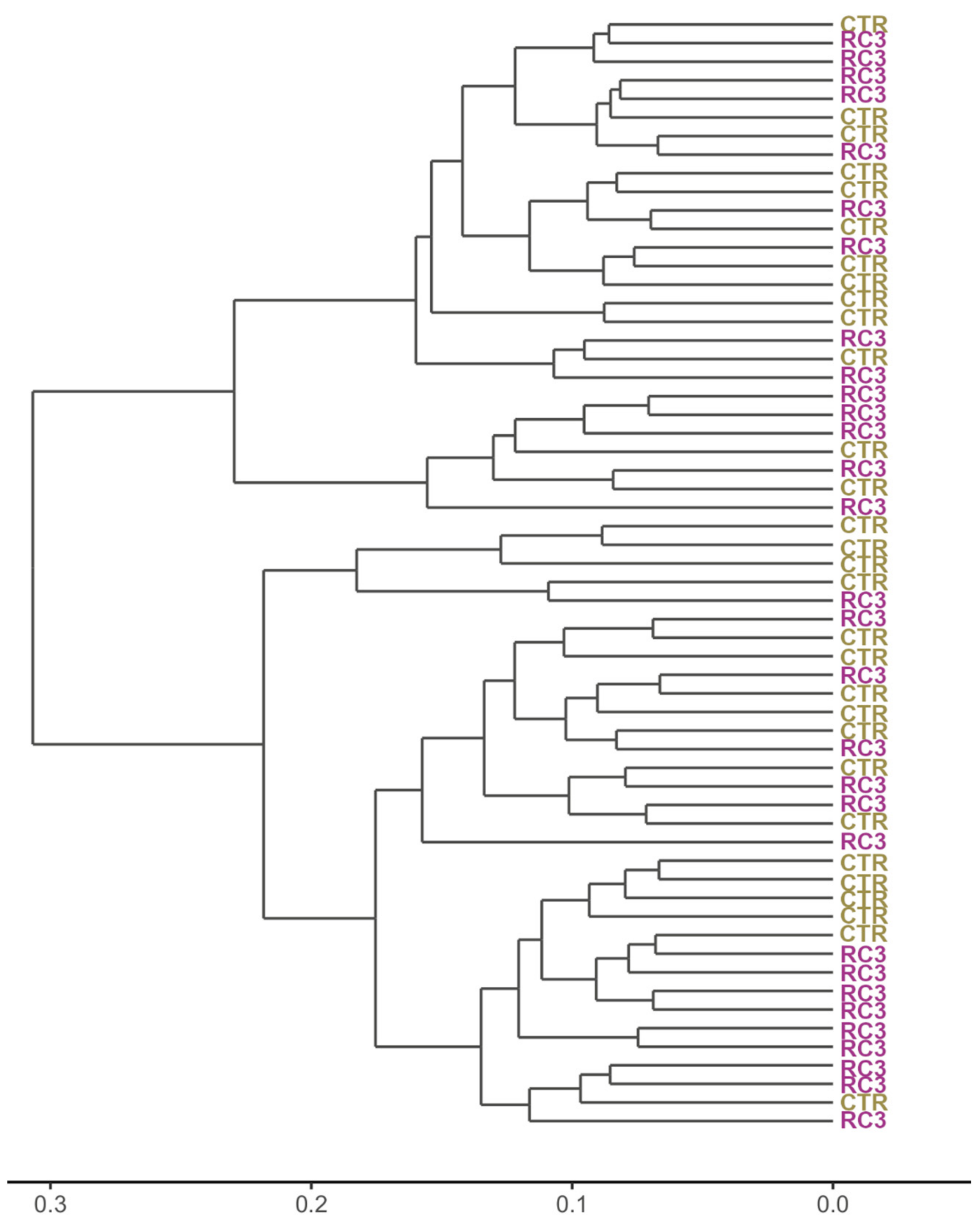
3.4. Beta Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, S.B.; Gotoh, T.; Greenwood, P.L. Current situation and future prospects for global beef production: Overview of special issue. Asian-Australas. J. Anim. Sci. 2018, 31, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, S.A.; Basarab, J.A.; Guan, L.L.; McAllister, T.A. Strategies to improve the efficiency of beef cattle production. Can. J. Anim. Sci. 2021, 101, 1–19. [Google Scholar] [CrossRef]
- Mottet, A.; Teillard, F.; Boettcher, P.; De’Besi, G.; Besbes, B. Domestic herbivores and food security: Current contribution, trends and challenges for a sustainable development. Animal 2018, 12, s188–s198. [Google Scholar] [CrossRef] [PubMed]
- Meat & Livestock Australia. State of the Industry Report-2020; MLA: North Sydney, Autralia, 2020. [Google Scholar]
- Hassan, F.-u.; Arshad, M.A.; Li, M.; Rehman, M.S.-u.; Loor, J.J.; Huang, J. Potential of Mulberry Leaf Biomass and Its Flavonoids to Improve Production and Health in Ruminants: Mechanistic Insights and Prospects. Animals 2020, 10, 2076. [Google Scholar] [CrossRef]
- Llonch, P.; Somarriba, M.; Duthie, C.A.; Troy, S.; Roehe, R.; Rooke, J.; Haskell, M.J.; Turner, S.P. Temperament and dominance relate to feeding behaviour and activity in beef cattle: Implications for performance and methane emissions. Animal 2018, 12, 2639–2648. [Google Scholar] [CrossRef] [Green Version]
- Insua, J.R.; Agnusdei, M.G.; Utsumi, S.A.; Berone, G.D. Morphological, environmental and management factors affecting nutritive value of tall fescue (Lolium arundinaceum). Crop Pasture Sci. 2018, 69, 1165–1172. [Google Scholar] [CrossRef]
- Tharmaraj, J.; Chapman, D.F.; Nie, Z.N.; Lane, A.P. Herbage accumulation, botanical composition, and nutritive value of five pasture types for dairy production in southern Australia. Aust. J. Agric. Res. 2008, 59, 127–138. [Google Scholar] [CrossRef]
- Cobon, D.H.; Stone, G.; Carter, J.; McKeon, G.; Zhang, B.S.; Heidemann, H. Native pastures and beef cattle show a spatially variable response to a changing climate in Queensland, Australia. Eur. J. Agron. 2020, 114. [Google Scholar] [CrossRef]
- Dineen, M.; McCarthy, B.; Ross, D.; Ortega, A.; Dillon, P.; Van Amburgh, M.E. Characterization of the nutritive value of perennial ryegrass (Lolium perenne L.) dominated pastures using updated chemical methods with application for the Cornell Net Carbohydrate and Protein System. Anim. Feed Sci. Technol. 2021, 272, 114752. [Google Scholar] [CrossRef]
- Buck, S. What Is Pasture Dieback? Queenland Government: Brisbane, Australia, 2021.
- Makiela, S.; Harrower, K.M. Overview of the current status of buffel grass dieback. Australas. Plant Dis. Notes 2008, 3, 12–16. [Google Scholar] [CrossRef]
- Hauxwell, C.; McNicholl, D. Mealybugs and Pasture Dieback; Meat and Livestock Australia: North Sydney, Australia, 2018. [Google Scholar]
- Thomson, M. Hidden Gems: An Epidemiological Investigation into the Association of Ground Pearls with Pasture Dieback; University of Queensland: Brisbane, Australia, 2019. [Google Scholar]
- Queensland Department of Environment and Sciences. Climate Change in Australia. Available online: https://www.climatechangeinaustralia.gov.au/ (accessed on 15 November 2022).
- Albiac, J.; Kahil, T.; Notivol, E.; Calvo, E. Agriculture and climate change: Potential for mitigation in Spain. Sci. Total Environ. 2017, 592, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Pathak, H. Impact, adaptation, and mitigation of climate change in Indian agriculture. Environ. Monit. Assess. 2022, 195, 52. [Google Scholar] [CrossRef]
- Zilli, M.; Scarabello, M.; Soterroni, A.C.; Valin, H.; Mosnier, A.; Leclere, D.; Havlik, P.; Kraxner, F.; Lopes, M.A.; Ramos, F.M. The impact of climate change on Brazil’s agriculture. Sci. Total Environ. 2020, 740, 139384. [Google Scholar] [CrossRef]
- Moore, A.D.; Ghahramani, A. Climate change and broadacre livestock production across southern Australia. 1. Impacts of climate change on pasture and livestock productivity, and on sustainable levels of profitability. Glob. Chang. Biol. 2013, 19, 1440–1455. [Google Scholar] [CrossRef]
- Whitton, M.M.; Ren, X.; Yu, S.J.; Irving, A.D.; Trotter, T.T.; Bajagai, Y.S.; Stanley, D. Sea minerals reduce dysbiosis, improve pasture productivity and plant morphometrics in pasture dieback affected soils. Sustainability 2022, 14, 14873. [Google Scholar] [CrossRef]
- Alder, R.; Tomlinson, D. Use of composition as growth promotant for plants. 2019361542, 16 October 2019. [Google Scholar]
- Yazici, E.; Ahlatci, F.; Yilmaz, E.; Celep, O.; Deveci, H. Precipitation of zinc from cyanide leach solutions using Trimercapto-s-triazine (TMT). Hydrometallurgy 2020, 191, 105206. [Google Scholar] [CrossRef]
- Ayoub, G.M.; Koopman, B.; Bitton, G.; Riedesel, K. Heavy metal detoxification by trimercapto-s-triazine (TMT) as evaluated by a bacterial enzyme assay. Environ. Toxicol. Chem. Int. J. 1995, 14, 193–196. [Google Scholar] [CrossRef]
- Wang, Q.; Zheng, C.; Zhang, J.; He, F.; Yao, Y.; Zhang, T.C.; He, C. Insights into the adsorption of Pb (II) over trimercapto-s-triazine trisodium salt-modified lignin in a wide pH range. Chem. Eng. J. Adv. 2020, 1, 100002. [Google Scholar] [CrossRef]
- Zhao, N.; Li, B.; Huang, H.; Lv, X.; Zhang, M.; Cao, L. Modification of kelp and sludge biochar by TMT-102 and NaOH for cadmium adsorption. J. Taiwan Inst. Chem. Eng. 2020, 116, 101–111. [Google Scholar] [CrossRef]
- Pohl, A. Removal of heavy metal ions from water and wastewaters by sulfur-containing precipitation agents. Water Air Soil Pollut. 2020, 231, 1–17. [Google Scholar] [CrossRef]
- Wang, D.-F.; Li, S.-H.; Wang, X.-Q.; Li, L.-X.; Zhang, X. Synergistic passivation of fly ash and TMT on heavy metals in sewage sludge. Sustainability 2018, 10, 4731. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Li, M.; Sun, Y.; Gao, H.; Mao, L.; Zhang, H.; Tao, H. Optimization of Solidification and Stabilization Efficiency of Heavy Metal Contaminated Sediment Based on Response Surface Methodology. Sustainability 2022, 14, 3306. [Google Scholar] [CrossRef]
- Ahlatci, F.; Yazici, E.; Yilmaz, E.; Celep, O.; Deveci, H. A novel method for the recovery of silver from thiosulphate leach solutions using trimercapto-s-triazine (TMT). Miner. Eng. 2022, 177, 107373. [Google Scholar] [CrossRef]
- Ihsan, M.Z.; Daur, I.; Alghabari, F.; Alzamanan, S.; Rizwan, S.; Ahmad, M.; Waqas, M.; Shafqat, W. Heat stress and plant development: Role of sulphur metabolites and management strategies. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2019, 69, 332–342. [Google Scholar] [CrossRef]
- Rency, A.S.; Pandian, S.; Ramesh, M. Influence of adenine sulphate on multiple shoot induction in Clitoria ternatea L. and analysis of phyto-compounds in in vitro grown plants. Biocatal. Agric. Biotechnol. 2018, 16, 181–191. [Google Scholar] [CrossRef]
- Tiku, A.R. Antimicrobial compounds and their role in plant defense. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Berlin/Heidelberg, Germany, 2018; pp. 283–307. [Google Scholar]
- Pope, J.; Eichenberg, R.; Birthisel, T. Use of humate dispersible granule technology as a soil amendment in turfgrass and horticultural soils. Appl. Turfgrass Sci. 2013, 10, 1. [Google Scholar] [CrossRef]
- Taha, S.S.; Osman, A.S. Influence of potassium humate on biochemical and agronomic attributes of bean plants grown on saline soil. J. Hortic. Sci. Biotechnol. 2018, 93, 545–554. [Google Scholar] [CrossRef]
- El-Naqma, K. The role of humate substances in controlling synergism and antagonism of nutrients uptake by potato plants. Environ. Biodivers. Soil Secur. 2020, 4, 149–165. [Google Scholar] [CrossRef]
- Jin, Q.; Zhang, Y.; Wang, Q.; Li, M.; Sun, H.; Liu, N.; Zhang, L.; Zhang, Y.; Liu, Z. Effects of potassium fulvic acid and potassium humate on microbial biodiversity in bulk soil and rhizosphere soil of Panax ginseng. Microbiol. Res. 2022, 254, 126914. [Google Scholar] [CrossRef]
- da Silva, M.S.R.D.; dos Santos, B.D.S.; da Silva, C.S.R.D.; da Silva, C.S.R.D.; Antunes, L.F.D.; dos Santos, R.M.; Santos, C.H.B.; Rigobelo, E.C. Humic Substances in Combination With Plant Growth-Promoting Bacteria as an Alternative for Sustainable Agriculture. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Tilman, D.; Furey, G.; Lehman, C. Soil carbon sequestration accelerated by restoration of grassland biodiversity. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, R.B.; Hergert, G.W.; Shapiro, C.A.; Wortmann, C.S. Guidelines for Soil Sampling; University of Nebraska–Lincoln: Lincoln, NE, USA, 2007. [Google Scholar]
- Wagi, S.; Ahmed, A. Bacillus spp.: Potent microfactories of bacterial IAA. PeerJ 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Idris, E.E.; Iglesias, D.J.; Talon, M.; Borriss, R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant-Microbe Interact. 2007, 20, 619–626. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.Z.; Zhang, Z.F.; Zhou, N.; Jiang, C.Y.; Wang, B.J.; Cai, L.; Liu, S.J. Diversity, Distribution and Co-occurrence Patterns of Bacterial Communities in a Karst Cave System. Front. Microbiol. 2019, 10, 1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, T.; Matsumoto, A.; Ōmura, S.; Takahashi, Y. Distribution and isolation of strains belonging to the order Solirubrobacterales. J. Antibiot. 2015, 68, 763–766. [Google Scholar] [CrossRef]
- Egas, C.; Barroso, C.; Froufe, H.J.C.; Pacheco, J.; Albuquerque, L.; da Costa, M.S. Complete genome sequence of the Radiation-Resistant bacterium Rubrobacter radiotolerans RSPS-4. Stand. Genomic. Sci. 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, A.; Adkins-Jablonsky, S.; Barreto Filho, M.M.; Schilling, M.; Dawson, A.; Heiser, S.; O’Connor, A.; Walker, M.; Roberts, Q.; Morris, J. Heavy metal pollution impacts soil bacterial community structure and antimicrobial resistance at the Birmingham 35th Avenue Superfund Site. bioRxiv 2022. [Google Scholar] [CrossRef]
- Peng, M.; Jia, H.; Wang, Q. The effect of land use on bacterial communities in saline–alkali soil. Curr. Microbiol. 2017, 74, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-W.; Asem, M.D.; Li, X.; Li, L.-Y.; Salam, N.; Alkhalifah, D.H.M.; Hozzein, W.N.; Nie, G.-X.; Li, W.-J. Blastococcus deserti sp. nov., isolated from a desert sample. Arch. Microbiol. 2019, 201, 193–198. [Google Scholar] [CrossRef]
- Castro, J.F.; Nouioui, I.; Sangal, V.; Choi, S.; Yang, S.-J.; Kim, B.-Y.; Trujillo, M.E.; Riesco, R.; del Carmen Montero-Calasanz, M.; Rahmani, T.P. Blastococcus atacamensis sp. nov., a novel strain adapted to life in the Yungay core region of the Atacama Desert. Int. J. Syst. Evol. Microbiol. 2018, 68, 2712–2721. [Google Scholar] [CrossRef] [PubMed]
- Foesel, B.U.; Rohde, M.; Overmann, J. Blastocatella fastidiosa gen. nov., sp. nov., isolated from semiarid savanna soil—The first described species of Acidobacteria subdivision 4. Syst. Appl. Microbiol. 2013, 36, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.W.; Li, J.H.; Koch, B.J.; Blazewicz, S.J.; Dijkstra, P.; Hayer, M.; Hofmockel, K.S.; Liu, X.J.A.; Mau, R.L.; Morrissey, E.M.; et al. Nutrients cause consolidation of soil carbon flux to small proportion of bacterial community. Nat. Commun. 2021, 12. [Google Scholar] [CrossRef]
- Thawai, C.; Thamsathit, W.; Kudo, T. Planosporangium thailandense sp. nov., isolated from soil from a Thai hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 1051–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaturro, M.; Chierico, F.D.; Motro, Y.; Chaldoupi, A.; Flountzi, A.; Moran-Gilad, J.; Girolamo, A.; Koutsiomani, T.; Krogulska, B.; Lindsay, D.; et al. Bacterial communities of premise plumbing systems in four European cities, and their association with culturable Legionella. bioRxiv 2008. Available online: https://www.biorxiv.org/content/10.1101/2022.08.12.503735v1 (accessed on 31 October 2022). [CrossRef]
- Finlay, B.J.; Black, H.I.; Brown, S.; Clarke, K.J.; Esteban, G.F.; Hindle, R.M.; Olmo, J.L.; Rollett, A.; Vickerman, K. Estimating the growth potential of the soil protozoan community. Protist 2000, 151, 69–80. [Google Scholar] [CrossRef]
- Rosenberg, K.; Bertaux, J.; Krome, K.; Hartmann, A.; Scheu, S.; Bonkowski, M. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J. 2009, 3, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhang, S.; Zhong, Q.; Gong, G.; Wang, G.; Guo, X.; Xu, X. Effects of soil chemical properties and fractions of Pb, Cd, and Zn on bacterial and fungal communities. Sci. Total Environ. 2020, 715, 136904. [Google Scholar] [CrossRef]
- Bill, M.; Chidamba, L.; Gokul, J.K.; Labuschagne, N.; Korsten, L. Bacterial community dynamics and functional profiling of soils from conventional and organic cropping systems. Appl. Soil. Ecol. 2021, 157. [Google Scholar] [CrossRef]
- Niemhom, N.; Thawai, C. Screening and Identification of Rare Actinomycetes Isolated from Soil in Pho Hin Dad Waterfall (Namtok Sam Lan National Park, Saraburi Province) for Antimicrobial Activities. In Proceedings of the RSU International Research Conference 2020, Pathum Thani, Thailand, 1 May 2020; pp. 654–664. [Google Scholar]
- Wang, Y.; Du, L.; Liu, H.; Long, D.; Huang, M.; Wang, Y.; Huang, S.; Jin, D. Halosulfuron methyl did not have a significant effect on diversity and community of sugarcane rhizosphere microflora. J. Hazard. Mater. 2020, 399, 123040. [Google Scholar] [CrossRef]
- Li, Q.; Wan, F.; Zhao, M. Distinct soil microbial communities under Ageratina adenophora invasions. Plant Biol. 2022, 24, 430–439. [Google Scholar] [CrossRef]
- Inaba, T.; Hori, T.; Navarro, R.R.; Ogata, A.; Hanajima, D.; Habe, H. Revealing sludge and biofilm microbiomes in membrane bioreactor treating piggery wastewater by non-destructive microscopy and 16S rRNA gene sequencing. Chem. Eng. J. 2018, 331, 75–83. [Google Scholar] [CrossRef]
- Canisares, L.P.; Poffenbarger, H.; Brodie, E.L.; Sorensen, P.O.; Karaoz, U.; Villegas, D.M.; Arango, J.; Momesso, L.; Crusciol, C.A.C.; Cantarella, H. Legacy effects of intercropping and nitrogen fertilization on soil N cycling, nitrous oxide emissions, and the soil microbial community in tropical maize production. Front. Soil Sci. 2021, 17. [Google Scholar] [CrossRef]
- Gschwend, F.; Hartmann, M.; Mayerhofer, J.; Hug, A.S.; Enkerli, J.; Gubler, A.; Meuli, R.G.; Frey, B.; Widmer, F. Site and land-use associations of soil bacteria and fungi define core and indicative taxa. FEMS Microbiol. Ecol. 2021, 97, fiab165. [Google Scholar] [CrossRef] [PubMed]
- Yajun, W.; Chongchong, G.; Tianjing, C.; Jinshou, L.; Yan, X.; Dafang, F. Adaptability of enhanced bioretention cell for nitrogen and phosphorus removal under two antibiotics stress. Ecotoxicol. Environ. Saf. 2022, 230, 113114. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Putri, A.L.; Setiawan, R. Isolation and screening of actinomycetes producing cellulase and xylanase from Mamasa soil, West Sulawesi. In Proceedings of the IOP Conference Series: Earth and Environmental Science; IOP Publishing Ltd.: Bristol, UK, 2019; p. 012035. [Google Scholar]
- Zhang, Y.; Wu, X.-h.; LIi, X.-g.; Duan, T.-t.; Xu, J.; Dong, F.-s.; Liu, X.-g.; Zheng, Y.-q. The Response of Soil Microbial Community to Repeated Application Clomazone. Biotechnol. Bull. 2020, 36, 64. [Google Scholar]
- Zhang, K.; Li, K.; Tong, M.; Xia, Y.; Cui, Y.; Liu, Z.; Chen, Q.; Li, Q.; Hu, F.; Yang, F. Distribution Pattern and Influencing Factors of Heavy Metal Resistance Genes in the Yellow River Sediments of Henan Section. Int. J. Environ. Res. Public Health 2022, 19, 10724. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.K.; Bitla, U.M.; Sorty, A.M.; Singh, D.P.; Gupta, V.K.; Wakchaure, G.; Kumar, S. Mitigation of salinity stress in wheat seedlings due to the application of phytohormone-rich culture filtrate extract of methylotrophic actinobacterium Nocardioides sp. NIMMe6. Front. Microbiol. 2020, 11, 2091. [Google Scholar] [CrossRef] [PubMed]
- Girardot, F.; Allégra, S.; Pfendler, S.; Conord, C.; Rey, C.; Gillet, B.; Hughes, S.; Bouchardon, A.E.; Hua, A.; Paran, F. Bacterial diversity on an abandoned, industrial wasteland contaminated by polychlorinated biphenyls, dioxins, furans and trace metals. Sci. Total Environ. 2020, 748, 141242. [Google Scholar] [CrossRef]



| CTR | RC3 | |||||
|---|---|---|---|---|---|---|
| Genus | Rho | p | Signif. | Rho | p | Signif. |
| Actinomadura | −0.356 | 0.049 | * | 0.038 | 0.843 | |
| Actinoplanes | −0.490 | 0.005 | ** | −0.239 | 0.213 | |
| Asanoa | −0.454 | 0.010 | * | −0.193 | 0.316 | |
| BIrii41 | −0.480 | 0.006 | ** | −0.295 | 0.1206 | |
| Blastococcus | −0.404 | 0.024 | * | −0.421 | 0.023 | * |
| Conexibacter | 0.282 | 0.125 | 0.408 | 0.028 | * | |
| Craurococcus-Caldovatus | 0.020 | 0.913 | −0.401 | 0.031 | * | |
| Cryptosporangium | −0.395 | 0.028 | * | −0.042 | 0.827 | |
| Dactylosporangium | −0.434 | 0.015 | * | 0.086 | 0.658 | |
| Dongia | −0.406 | 0.023 | * | 0.139 | 0.471 | |
| Ellin6067 | −0.474 | 0.007 | ** | −0.049 | 0.797 | |
| MND1 | 0.358 | 0.048 | * | −0.019 | 0.922 | |
| Marmoricola | −0.695 | 0.000014 | *** | 0.061 | 0.752 | |
| Planosporangium | 0.225 | 0.224 | 0.387 | 0.038 | * | |
| Pyxidicoccus | −0.369 | 0.041 | * | −0.045 | 0.815 | |
| RB41 | −0.112 | 0.547 | 0.433 | 0.019 | * | |
| Reyranella | −0.473 | 0.007 | ** | −0.204 | 0.287 | |
| Saccharopolyspora | −0.359 | 0.047 | * | −0.010 | 0.957 | |
| Skermanella | −0.404 | 0.024 | * | −0.025 | 0.898 | |
| Subgroup 10 | −0.379 | 0.036 | * | 0.134 | 0.488 | |
| Vicinamibacteraceae | −0.363 | 0.045 | * | −0.207 | 0.282 | |
| mle1-7 | −0.472 | 0.007 | ** | 0.193 | 0.315 | |
| Distance | Effect of | R2 | p | Significance |
|---|---|---|---|---|
| Unweighted Unifrac | Treatment | 0.017 | 0.298 | |
| Time (weeks) | 0.238 | 0.001 | *** | |
| Weighted Unifrac | Treatment | 0.012 | 0.548 | |
| Time (weeks) | 0.248 | 0.019 | ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whitton, M.M.; Ren, X.; Yu, S.J.; Trotter, T.; Stanley, D.; Bajagai, Y.S. Remediation of Pasture Dieback Using Plant Growth Promotant. Agronomy 2022, 12, 3153. https://doi.org/10.3390/agronomy12123153
Whitton MM, Ren X, Yu SJ, Trotter T, Stanley D, Bajagai YS. Remediation of Pasture Dieback Using Plant Growth Promotant. Agronomy. 2022; 12(12):3153. https://doi.org/10.3390/agronomy12123153
Chicago/Turabian StyleWhitton, Maria M., Xipeng Ren, Sung J. Yu, Tieneke Trotter, Dragana Stanley, and Yadav S. Bajagai. 2022. "Remediation of Pasture Dieback Using Plant Growth Promotant" Agronomy 12, no. 12: 3153. https://doi.org/10.3390/agronomy12123153
APA StyleWhitton, M. M., Ren, X., Yu, S. J., Trotter, T., Stanley, D., & Bajagai, Y. S. (2022). Remediation of Pasture Dieback Using Plant Growth Promotant. Agronomy, 12(12), 3153. https://doi.org/10.3390/agronomy12123153









