Potential of Omics to Control Diseases and Pests in the Coconut Tree
Abstract
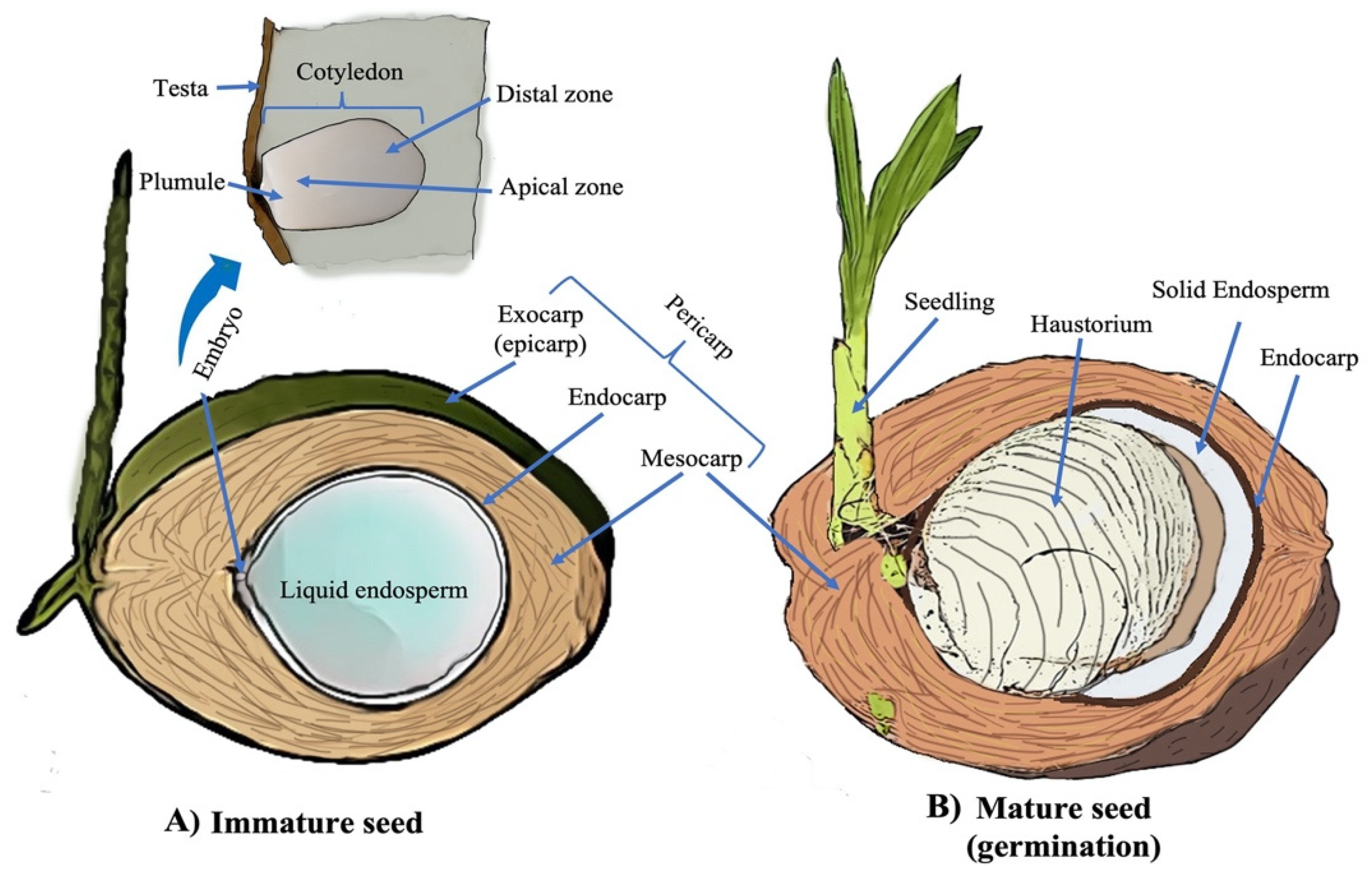
:1. Introduction
2. Coconut Pests and Diseases
3. Control Methods of Pests and Diseases in Coconut Crop
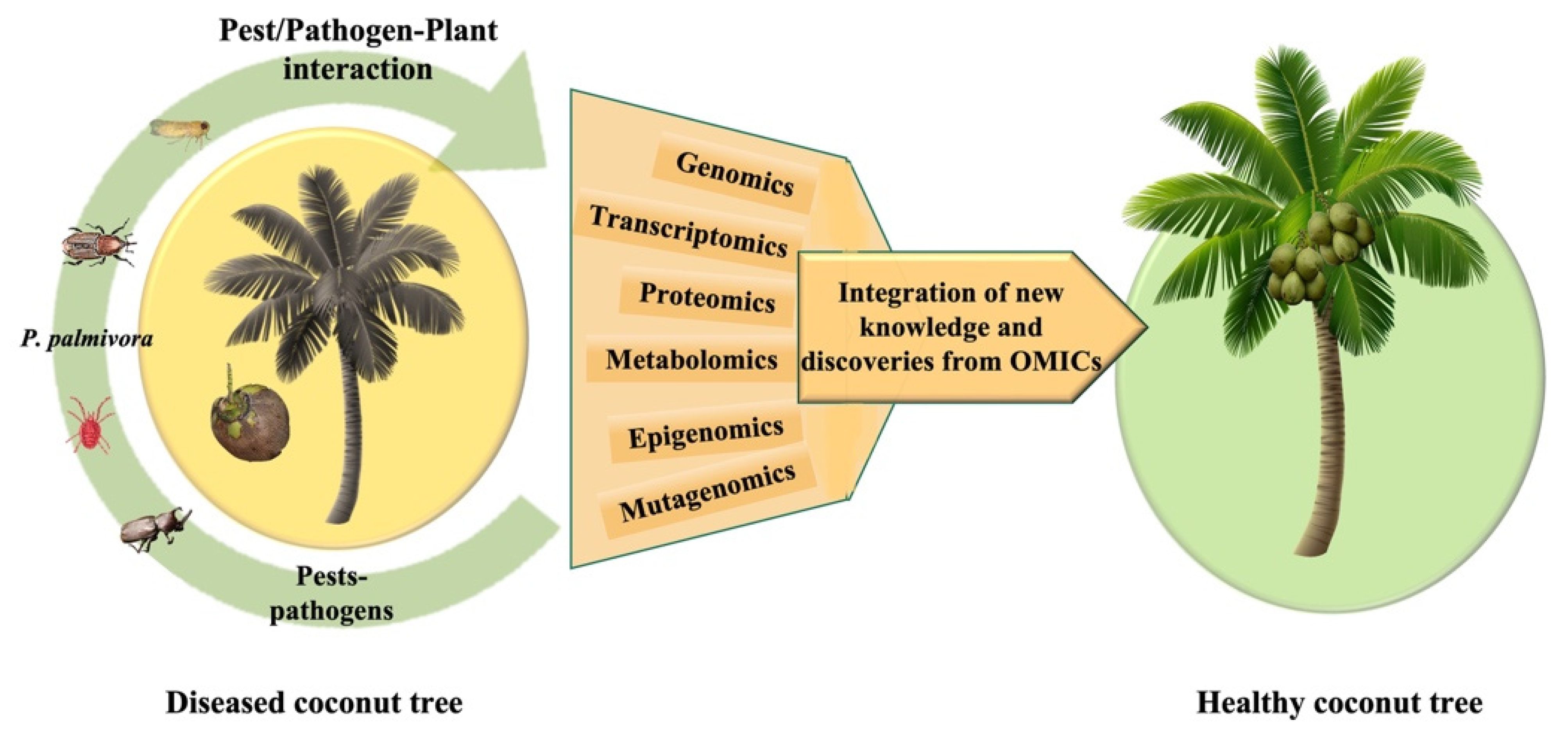
4. Omics and Its Application to the Study of the Coconut Palm
4.1. Genomics
4.2. Transcriptomics
4.3. Proteomics
4.4. Metabolomics
5. Opportunities and Challenges Going forward: From Omics-Based Results to Solutions for Phytosanitary Problems in the Coconut Palm
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manikantan, M.R.; Pandiselvam, R.; Beegum, S.; Mathew, A.C. Harvest and Postharvest Technology. In The Coconut Palm (Cocos nucifera L.)—Research and Development Perspectives; Krishnakumar, V., Thampan, P.K., Nair, M.A., Eds.; Springer: Singapore, 2018; pp. 635–722. ISBN 9789811327544. [Google Scholar]
- Niral, V.; Jerard, B.A. Botany, Origin and Genetic Resources of Coconut. In The Coconut Palm (Cocos nucifera L.)—Research and Development Perspectives; Krishnakumar, V., Thampan, P.K., Nair, M.A., Eds.; Springer: Singapore, 2018; pp. 57–111. ISBN 9789811327544. [Google Scholar]
- Zhang, L.; Tu, L.; Liang, Y.; Chen, Q.; Li, Z.; Li, C.; Wang, Z.; Li, W. Coconut-Based Activated Carbon Fibers for Efficient Adsorption of Various Organic Dyes. RSC Adv. 2018, 8, 42280–42291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zehr, R. Hydrating with Coconut Water, Water or Gatorade® Results in Similar Basketball Fitness & Skill Performance During a Simulated Basketball Game. Electron. Thesis Diss. Repos. 2019, 56. Available online: https://ir.lib.uwo.ca/etd/6497 (accessed on 30 October 2022).
- Mardesci, H.; Santosa; Nazir, N.; Hadiguna, R.A. Determination of Value-Added and Contributing Organization in the Development of Coconut Water-Based Agro Industry. IOP Conf. Ser. Earth Environ. Sci. 2021, 709, 012062. [Google Scholar] [CrossRef]
- Kaur, K.; Chhikara, N.; Sharma, P.; Garg, M.K.; Panghal, A. Coconut Meal: Nutraceutical Importance and Food Industry Application. Foods Raw Mater. 2019, 7, 419–427. [Google Scholar] [CrossRef]
- Islas-Flores, I.; Tzec-Simá, M. Research Opportunities on the Coconut (Cocos nucifera L.) Using New Technologies. South Afr. J. Bot. 2021, 141, 414–420. [Google Scholar] [CrossRef]
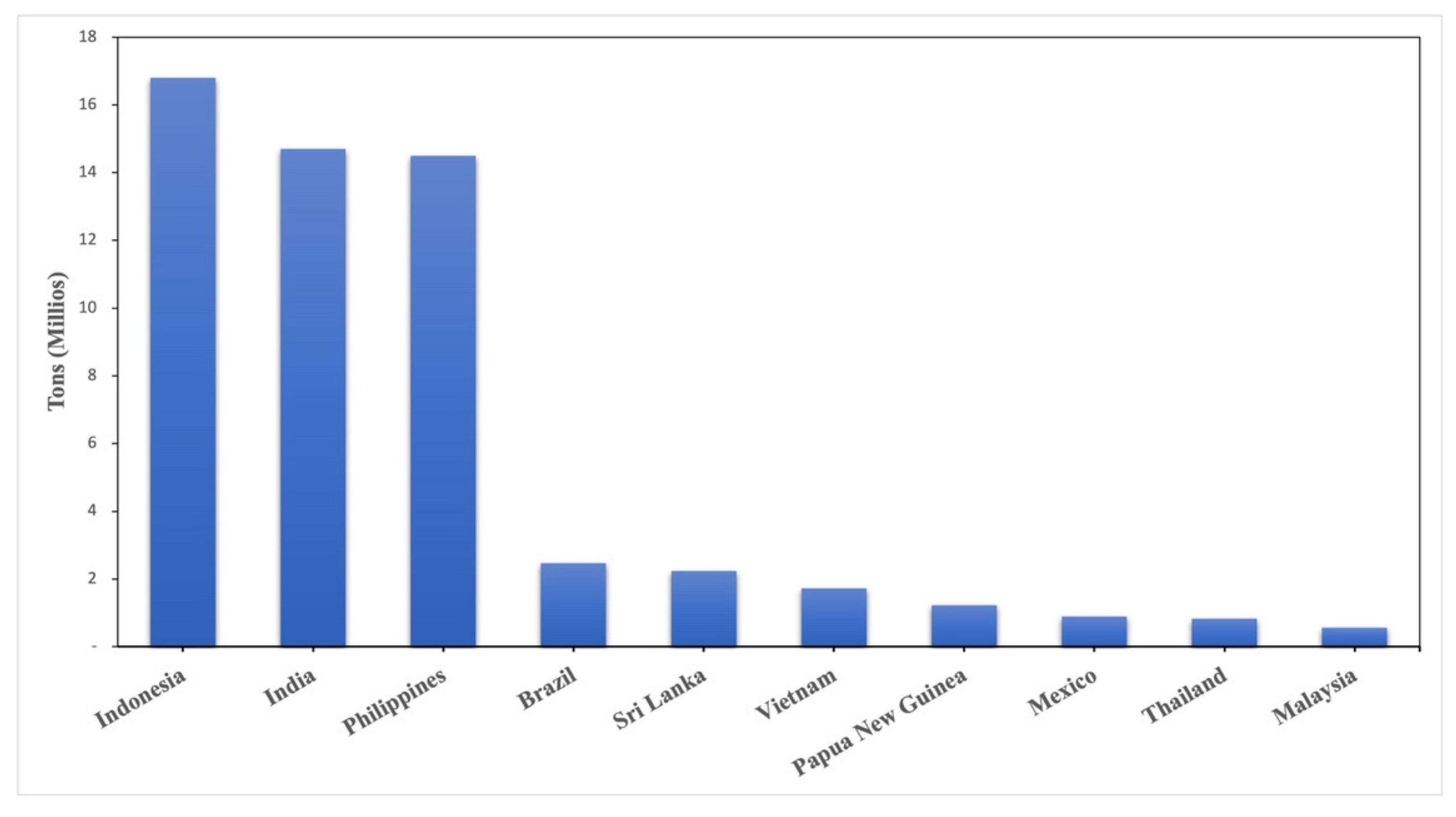
- FAO (Food Agriculture Organization) Production of Coconuts: Top 10 Producers. Available online: https://www.fao.org/faostat/es/#rankings/countries_by_commodity (accessed on 28 October 2022).
- Statista 2020 Coconut Production Worldwide by Leading Country 2020. Available online: https://www.statista.com/statistics/1040499/world-coconut-production-by-leading-producers/ (accessed on 28 October 2022).
- Kumara, A.D.N.T.; Chandrashekharaiah, M.; Kandakoor, S.B.; Chakravarthy, A.K. Status and Management of Three Major Insect Pests of Coconut in the Tropics and Subtropics. In New Horizons in Insect Science: Towards Sustainable Pest Management; Chakravarthy, A.K., Ed.; Springer India: New Delhi, India, 2015; pp. 359–381. ISBN 978-81-322-2089-3. [Google Scholar]
- Peña, J.E.; Mannion, C.M.; Howard, F.W.; Hoy, M.A. Raoiella indica (Prostigmata: Tenuipalpidae): The Red Palm Mite: A Potential Invasive Pest of Palms and Bananas and Other Tropical Crops of Florida: ENY-837/IN681, 11/2006. EDIS 2006, 27, 1–8. [Google Scholar] [CrossRef]
- Viteri Jumbo, L.O.; Teodoro, A.V.; Rêgo, A.S.; Haddi, K.; Galvão, A.S.; de Oliveira, E.E. The Lacewing Ceraeochrysa caligata as a Potential Biological Agent for Controlling the Red Palm Mite Raoiella Indica. PeerJ 2019, 7, e7123. [Google Scholar] [CrossRef] [Green Version]
- Dalbon, V.A.; Acevedo, J.P.M.; Ribeiro Junior, K.A.L.; Ribeiro, T.F.L.; da Silva, J.M.; Fonseca, H.G.; Santana, A.E.G.; Porcelli, F. Perspectives for Synergic Blends of Attractive Sources in South American Palm Weevil Mass Trapping: Waiting for the Red Palm Weevil Brazil Invasion. Insects 2021, 12, 828. [Google Scholar] [CrossRef]
- Almarinez, B.J.M.; Barrion, A.T.; Navasero, M.V.; Navasero, M.M.; Cayabyab, B.F.; Carandang, J.S.R.; Legaspi, J.C.; Watanabe, K.; Amalin, D.M. Biological Control: A Major Component of the Pest Management Program for the Invasive Coconut Scale Insect, Aspidiotus Rigidus Reyne, in the Philippines. Insects 2020, 11, 745. [Google Scholar] [CrossRef]
- Shelomi, M.; Lin, S.-S.; Liu, L.-Y. Transcriptome and Microbiome of Coconut rhinoceros Beetle (Oryctes Rhinoceros) Larvae. BMC Genom. 2019, 20, 957. [Google Scholar] [CrossRef] [Green Version]
- Izaitul Aida, I.; Mohd Rasdi, Z.; Ismail, R.; Ismeazilla, M.B.; Mohd Faizol, K.; Muhammad Shakir, Z.; Nur Fakriyah, A.; Noor Shuhaina, S.M. Susceptibility and Resistant of Different Host Varieties of Oil Palm and Coconut Palm towards Pest, Rhinoceros Beetle (Oryctes rhinoceros). Asian J. Agric. Rural Dev. 2020, 10, 56–67. [Google Scholar] [CrossRef]
- Chandrashekharaiah; Chakravarthy, A.K. Pheromone Mass Trapping Technology for Management of Coconut Black-Headed Caterpillar, Opisina Arenosella Walker (Lepidoptera: Oecophoridae) in Southern Karnataka; University of Agricultural Sciences, GKVK: Bangalore, India, 2013. [Google Scholar]
- Gangaraj, K.P.; Rajesh, M.K. Dataset of Dual RNA-Sequencing of Phytophthora palmivora Infecting Coconut (Cocos Nucifera L.). Data Brief 2020, 30, 105455. [Google Scholar] [CrossRef] [PubMed]
- Avinash, B.; Kotresh, K.; Neelagund, S. Coconut’s Bud Rot by Phytophthora palmivora: A Destructive Disease. J. Mycol. Mycol. Sci. 2022, 5, 6. [Google Scholar] [CrossRef]
- Gurr, G.M.; Johnson, A.C.; Ash, G.J.; Wilson, B.A.L.; Ero, M.M.; Pilotti, C.A.; Dewhurst, C.F.; You, M.S. Coconut Lethal Yellowing Diseases: A Phytoplasma Threat to Palms of Global Economic and Social Significance. Front. Plant Sci. 2016, 7, 1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemmati, C.; Nikooei, M.; Al-Sadi, A.M. Four Decades of Research on Phytoplasma Diseases of Palms: A Review. Int. J. Agric. Biol. 2020, 24, 14. [Google Scholar]
- Oropeza-Salín, C.; Sáenz, L.; Narvaez, M.; Nic-Matos, G.; Córdova, I.; Myrie, W.; Ortíz, C.F.; Ramos, E. Dealing with Lethal Yellowing and Related Diseases in Coconut. In Coconut Biotechnology: Towards the Sustainability of the ‘Tree of Life’; Adkins, S., Foale, M., Bourdeix, R., Nguyen, Q., Biddle, J., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 169–197. ISBN 978-3-030-44987-2. [Google Scholar]
- Huda-Shakirah, A.R.; Mohamed Nor, N.M.I.; Zakaria, L.; Leong, Y.-H.; Mohd, M.H. Lasiodiplodia Theobromae as a Causal Pathogen of Leaf Blight, Stem Canker, and Pod Rot of Theobroma cacao in Malaysia. Sci. Rep. 2022, 12, 8966. [Google Scholar] [CrossRef]
- Dheepa, R.; Goplakrishnan, C.; Kamalakannan, A.; Nakkeeran, S.; Mahalingam, C.A.; Suresh, J. Coconut Nut Rot Disease in India: Prevalence, Characterization of Pathogen and Standardization of Inoculation Techniques. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 2046–2057. [Google Scholar] [CrossRef]
- Rajeswari, E.; Ramjegathesh, R.; Sivakumar, V.; Praneetha, S.; Pugalendhi, L.; Maheswarandhippa, H.P. Occurrence and Distribution of Coconut Diseases in Tamil Nadu, India. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 3859–3869. [Google Scholar] [CrossRef]
- Ushamalini, C.; Kumar, P.A.; Parthasarathy, S. Efficacy of Fungicides for the Management of Coconut Leaf Blight. Plant Dis. Res. 2019, 34, 29. [Google Scholar] [CrossRef]
- Ma, Z.; Yoshimura, M.A.; Michailides, T.J. Identification and Characterization of Benzimidazole Resistance in Monilinia fructicola from Stone Fruit Orchards in California. Appl. Environ. Microbiol. 2003, 69, 7145–7152. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Wang, J.; Wang, H.; Bao, Y.; Li, Y.; Govindaraju, M.; Yao, W.; Chen, B.; Zhang, M. Molecular Characterization of Carbendazim Resistance of Fusarium Species Complex That Causes Sugarcane Pokkah Boeng Disease. BMC Genom. 2019, 20, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Brankovics, B.; van der Lee, T.A.J.; Waalwijk, C.; van Diepeningen, A.A.D.; Xu, J.; Xu, J.; Chen, W.; Feng, J. A Single-Nucleotide-Polymorphism-Based Genotyping Assay for Simultaneous Detection of Different Carbendazim-Resistant Genotypes in the Fusarium graminearum Species Complex. PeerJ 2016, 4, e2609. [Google Scholar] [CrossRef] [PubMed]
- McCoy, R.E. Petiole injection of coconut palm, a method to prevent permanent trunk injury during antibiotic treatment for lethal yellowing. Proc Fla State Hort Soc 1977, 90, 114–117. [Google Scholar]
- Gopal, M.; Gupta, A.; Thomas, G.V. Prospects of Using Metarhizium anisopliae to Check the Breeding of Insect Pest, Oryctes rhinoceros L. in Coconut Leaf Vermicomposting Sites. Bioresour. Technol. 2006, 97, 1801–1806. [Google Scholar] [CrossRef]
- Huger, A.M. The Oryctes Virus: Its Detection, Identification, and Implementation in Biological Control of the Coconut Palm Rhinoceros Beetle, Oryctes rhinoceros (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 2005, 89, 78–84. [Google Scholar] [CrossRef]
- Paudel, S.; Mansfield, S.; Villamizar, L.F.; Jackson, T.A.; Marshall, S.D.G. Can Biological Control Overcome the Threat from Newly Invasive Coconut Rhinoceros Beetle Populations (Coleoptera: Scarabaeidae)? A Review. Ann. Entomol. Soc. Am. 2021, 114, 247–256. [Google Scholar] [CrossRef]
- Marshall, S.D.G.; Moore, A.; Vaqalo, M.; Noble, A.; Jackson, T.A. A New Haplotype of the Coconut rhinoceros Beetle, Oryctes rhinoceros, Has Escaped Biological Control by Oryctes Rhinoceros Nudivirus and Is Invading Pacific Islands. J. Invertebr. Pathol. 2017, 149, 127–134. [Google Scholar] [CrossRef]
- Basheer, P.P.M.; Thomas, S.K. Indian Treepie Dendrocitta vagabunda parvula (Latham, 1790) (Passeriformes: Corvidae) as a Natural Enemy of the Pests of Coconut and Areca Palm Plantations. J. Biopestic. 2012, 5, 205. [Google Scholar]
- Rehman, G.; Mamoon-ur-Rashid, M. Evaluation of Entomopathogenic Nematodes against Red Palm Weevil, Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Insects 2022, 13, 733. [Google Scholar] [CrossRef]
- Almarinez, B.J.M.; Amalin, D.M.; Carandang VI, J.S.R.; Navasero, M.V.; Navasero, M.M. First Philippine Record of the Parasitoid, Comperiella Sp. (Hymenoptera: Encyrtidae): A Potential Biocontrol Agent against Aspidiotus rigidus (Hemiptera: Diaspididae). J. Appl. Entomol. 2015, 139, 237–240. [Google Scholar] [CrossRef]
- Abhishek, T.; Dwivedi, S. Review on Integrated Pest Management of Coconut Crop. Int. J. Entomol. Res. 2021, 6, 115–120. [Google Scholar]
- Serrana, J.M.; Ishitani, N.; Carvajal, T.M.; Almarinez, B.J.M.; Barrion, A.T.; Amalin, D.M.; Watanabe, K. Unraveling the Genetic Structure of the Coconut Scale Insect Pest (Aspidiotus rigidus Reyne) Outbreak Populations in the Philippines. Insects 2019, 10, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lantican, D.V.; Strickler, S.R.; Canama, A.O.; Gardoce, R.R.; Mueller, L.A.; Galvez, H.F. De Novo Genome Sequence Assembly of Dwarf Coconut (Cocos nucifera L. ‘Catigan Green Dwarf’) Provides Insights into Genomic Variation Between Coconut Types and Related Palm Species. G3 GenesGenomesGenetics 2019, 9, 2377–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Bocs, S.; Fan, H.; Armero, A.; Baudouin, L.; Xu, P.; Xu, J.; This, D.; Hamelin, C.; Iqbal, A.; et al. Coconut Genome Assembly Enables Evolutionary Analysis of Palms and Highlights Signaling Pathways Involved in Salt Tolerance. Commun. Biol. 2021, 4, 105. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Jasrotia, R.S.; Iquebal, M.A.; Jaiswal, S.; Angadi, U.B.; Rai, A.; Kumar, D. Deciphering Genes Associated with Root Wilt Disease of Coconut and Development of Its Transcriptomic Database (CnTDB). Physiol. Mol. Plant Pathol. 2017, 100, 255–263. [Google Scholar] [CrossRef]
- Xu, D.; Yang, H.; Zhuo, Z.; Lu, B.; Hu, J.; Yang, F. Characterization and Analysis of the Transcriptome in Opisina arenosella from Different Developmental Stages Using Single-Molecule Real-Time Transcript Sequencing and RNA-Seq. Int. J. Biol. Macromol. 2021, 169, 216–227. [Google Scholar] [CrossRef]
- Christiaens, O.; Niu, J.; Nji Tizi Taning, C. RNAi in Insects: A Revolution in Fundamental Research and Pest Control Applications. Insects 2020, 11, 415. [Google Scholar] [CrossRef]
- Lü, J.; Liu, Z.; Guo, W.; Guo, M.; Chen, S.; Li, H.; Yang, C.; Zhang, Y.; Pan, H. Feeding Delivery of DsHvSnf7 Is a Promising Method for Management of the Pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). Insects 2019, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Saand, M.A.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Fan, H.; Wang, F. ITRAQ-Based Comparative Proteomic Analysis of Two Coconut Varieties Reveals Aromatic Coconut Cold-Sensitive in Response to Low Temperature. J. Proteom. 2020, 220, 103766. [Google Scholar] [CrossRef]
- D’Amato, A.; Fasoli, E.; Righetti, P.G. Harry Belafonte and the Secret Proteome of Coconut Milk. J. Proteom. 2012, 75, 914–920. [Google Scholar] [CrossRef]
- Saha, B.; Sircar, G.; Pandey, N.; Gupta Bhattacharya, S. Mining Novel Allergens from Coconut Pollen Employing Manual De Novo Sequencing and Homology-Driven Proteomics. J. Proteome Res. 2015, 14, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, X.; Lan, Q.; Lai, X.; Luo, Z.; Yang, G. Proteomic Profile of Coconuts. Eur. Food Res. Technol. 2016, 242, 449–455. [Google Scholar] [CrossRef]
- Lakshmi, J.; Bhavyashree, U.; Karun, A. ComparatIve Proteome Analysis of Zygotic and Somatic Embryogenesis in Coconut. In Proceedings of the 3rd International Symposium on Coconut Research and Development, Karasagod, India, 10–12 December 2016. ICAR-CPCRI. [Google Scholar]
- Lin, Y.; Wang, Y.; Ji, Z.; Le, X. Isolation, Purification, and Identification of Coconut Protein through SDS-PAGE, HPLC, and MALDI-TOF/TOF-MS. Food Anal. Methods 2020, 13, 1246–1254. [Google Scholar] [CrossRef]
- Kotecka-Majchrzak, K.; Sumara, A.; Fornal, E.; Montowska, M. Proteomic Analysis of Oilseed Cake: A Comparative Study of Species-specific Proteins and Peptides Extracted from Ten Seed Species. J. Sci. Food Agric. 2021, 101, 297–306. [Google Scholar] [CrossRef]
- Wang, J.; Yao, L.; Li, B.; Meng, Y.; Ma, X.; Lai, Y.; Si, E.; Ren, P.; Yang, K.; Shang, X.; et al. Comparative Proteomic Analysis of Cultured Suspension Cells of the Halophyte Halogeton glomeratus by ITRAQ Provides Insights into Response Mechanisms to Salt Stress. Front. Plant Sci. 2016, 7, 110. [Google Scholar] [CrossRef]
- Rasool, K.G.; Khan, M.A.; Tufail, M.; Husain, M.; Mehmood, K.; Mukhtar, M.; Takeda, M.; Aldawood, A.S. Differential Proteomic Analysis of Date Palm Leaves Infested with the Red Palm Weevil (Coleoptera: Curculionidae). Fla. Entomol. 2018, 101, 290–298. [Google Scholar] [CrossRef] [Green Version]
- Abdel Farag El-Shafie, H.; Romeno Faleiro, J. Red Palm Weevil Rhynchophorus ferrugineus (Coleoptera: Curculionidae): Global Invasion, Current Management Options, Challenges and Future Prospects. In Invasive Species—Introduction Pathways, Economic Impact, and Possible Management Options; El-Shafie, H., Ed.; IntechOpen: London, UK, 2020; ISBN 978-1-78985-849-5. [Google Scholar]
- Yang, H.; Xu, D.; Zhuo, Z.; Hu, J.; Lu, B. Transcriptome and Gene Expression Analysis of Rhynchophorus Ferrugineus (Coleoptera: Curculionidae) during Developmental Stages. PeerJ 2020, 8, e10223. [Google Scholar] [CrossRef]
- Hall, D.R.; Harte, S.J.; Farman, D.I.; Ero, M.; Pokana, A. Identification of Components of the Aggregation Pheromone of the Guam Strain of Coconut Rhinoceros Beetle, Oryctes Rhinoceros, and Determination of Stereochemistry. J. Chem. Ecol. 2022, 48, 289–301. [Google Scholar] [CrossRef]
- Ding, B.; Wang, H.; Al-Saleh, M.A.; Löfstedt, C.; Antony, B. Bioproduction of (Z, E)-9,12-tetradecadienyl Acetate (ZETA), the Major Pheromone Component of Plodia, Ephestia, and Spodoptera Species in Yeast. Pest Manag. Sci. 2022, 78, 1048–1059. [Google Scholar] [CrossRef]
- Lara-Pérez, L.A.; Oros-Ortega, I.; Córdova-Lara, I.; Estrada-Medina, H.; O’Connor-Sánchez, A.; Góngora-Castillo, E.; Sáenz-Carbonell, L. Seasonal Shifts of Arbuscular Mycorrhizal Fungi in Cocos nucifera Roots in Yucatan, Mexico. Mycorrhiza 2020, 30, 269–283. [Google Scholar] [CrossRef]
- Andrade-Torres, A.; Oropeza, C.; Sáenz, L.; González-Estrada, T.; Ramírez-Benítez, J.E.; Becerril, K.; Chan, J.L.; Rodríguez-Zapata, L.C. Transient Genetic Transformation of Embryogenic Callus of Cocos Nucifera. Biologia 2011, 66, 790–800. [Google Scholar] [CrossRef] [Green Version]
- Keijzer, P.; van Bueren, E.T.L.; Engelen, C.J.M.; Hutten, R.C.B. Breeding Late Blight Resistant Potatoes for Organic Farming—A Collaborative Model of Participatory Plant Breeding: The Bioimpuls Project. Potato Res. 2022, 65, 349–377. [Google Scholar] [CrossRef]
- Baudouin, L.; Lebrun, P.; Rognon, F.; Ritter, E. Use of Molecular Markers for Coconut Improvement: Status and Prospects. Available online: https://agritrop.cirad.fr/535232/ (accessed on 4 December 2022).
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A Perspective on RNAi-Based Biopesticides. Front. Plant Sci. 2020, 11, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, A.; Höfle, L.; Werner, B.T.; Imani, J.; Schmidt, A.; Jelonek, L.; Kogel, K. SIGS vs. HIGS: A Study on the Efficacy of Two DsRNA Delivery Strategies to Silence Fusarium FgCYP51 Genes in Infected Host and Non-host Plants. Mol. Plant Pathol. 2019, 20, 1636–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, D.; Hamby, R.; Sanchez, J.N.; Cai, Q.; Yan, Q.; Jin, H. RNAs—A New Frontier in Crop Protection. Curr. Opin. Biotechnol. 2021, 70, 204–212. [Google Scholar] [CrossRef]
- Gong, L.; Chen, Y.; Hu, Z.; Hu, M. Testing Insecticidal Activity of Novel Chemically Synthesized SiRNA against Plutella xylostella under Laboratory and Field Conditions. PLoS ONE 2013, 8, e62990. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Guan, R.; Miao, X. Lepidopteran Insect Species-Specific, Broad-Spectrum, and Systemic RNA Interference by Spraying DsRNA on Larvae. Entomol. Exp. Appl. 2015, 155, 218–228. [Google Scholar] [CrossRef]
- Killiny, N.; Hajeri, S.; Tiwari, S.; Gowda, S.; Stelinski, L.L. Double-Stranded RNA Uptake through Topical Application, Mediates Silencing of Five CYP4 Genes and Suppresses Insecticide Resistance in Diaphorina citri. PLoS ONE 2014, 9, e110536. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Ren, B.; Zeng, B.; Shen, J. Improving RNAi Efficiency for Pest Control in Crop Species. BioTechniques 2020, 68, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Hamby, R.; Wang, M.; Qiao, L.; Jin, H. Synthesizing Fluorescently Labeled DsRNAs and SRNAs to Visualize Fungal RNA Uptake. In RNA Tagging; Heinlein, M., Ed.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2020; Volume 2166, pp. 215–225. ISBN 978-1-07-160711-4. [Google Scholar]
- Machado, A.K.; Brown, N.A.; Urban, M.; Kanyuka, K.; Hammond-Kosack, K.E. RNAi as an Emerging Approach to Control Fusarium Head Blight Disease and Mycotoxin Contamination in Cereals: RNAi-Mediated Control of Plant Pathogenic Fungi. Pest Manag. Sci. 2018, 74, 790–799. [Google Scholar] [CrossRef] [Green Version]
- Šečić, E.; Kogel, K.-H. Requirements for Fungal Uptake of DsRNA and Gene Silencing in RNAi-Based Crop Protection Strategies. Curr. Opin. Biotechnol. 2021, 70, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Dreischhoff, S.; Das, I.S.; Jakobi, M.; Kasper, K.; Polle, A. Local Responses and Systemic Induced Resistance Mediated by Ectomycorrhizal Fungi. Front. Plant Sci. 2020, 11, 590063. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Kusajima, M.; Fukagawa, M.; Okumura, Y.; Nakajima, M.; Akiyama, K.; Asami, T.; Yoneyama, K.; Kato, H.; Nakashita, H. Response of Tomatoes Primed by Mycorrhizal Colonization to Virulent and Avirulent Bacterial Pathogens. Sci. Rep. 2022, 12, 4686. [Google Scholar] [CrossRef]
- Department of Environment Government Tonga the Fight against the Coconut Beetles. RIDGE REEF APPROACH Work. COMMUNITIES. 2019. Available online: https://www.environment.gov.to/2019/09/10/the-fight-against-the-coconut-beetles/ (accessed on 12 December 2022).
- Agriculture and Agriproducts Coconut Products Market Size, Share & Trends | Forecast, 2026. Available online: https://www.alliedmarketresearch.com/coconut-products-market (accessed on 29 October 2022).
- ENY998/IN1240: Oxytetracycline Hydrochloride (OTC-HCl) Application for Control of Palm Phytoplasmas. Available online: https://edis.ifas.ufl.edu/publication/IN1240 (accessed on 4 December 2022).
- Soto, N.; Humphries, A.R.; Mou, D.; Helmick, E.E.; Glover, J.P.; Bahder, B.W. Effect of Oxytetracycline-Hydrochloride on Phytoplasma Titer and Symptom Progression of the 16SrIV-D Phytoplasma in Cabbage Palms from Florida. Plant Dis. 2020, 104, 2330–2337. [Google Scholar] [CrossRef] [PubMed]
- Hollomon, D. Does Agricultural Use of Azole Fungicides Contribute to Resistance in the Human Pathogen Aspergillus fumigatus?: Agricultural Azole Fungicides and A. Fumigatus Resistance. Pest Manag. Sci. 2017, 73, 1987–1993. [Google Scholar] [CrossRef] [PubMed]
- Hegde, K.; Mathew, N.; Shivashankara, A.R.; Prabhu, A.N.; Baliga, M.S. Hepatoprotective Effects of Picroliv. In Bioactive Food as Dietary Interventions for Liver and Gastrointestinal Disease; Elsevier: Amsterdam, The Netherlands, 2013; pp. 685–695. ISBN 978-0-12-397154-8. [Google Scholar]
- Rajesh, M.K.; Ramesh, S.V.; Perera, L.; Kole, C. (Eds.) The Coconut Genome; Springer: Cham, Switzerland, 2021; ISBN 978-3-030-76648-1. [Google Scholar]
- Syazliana, S.N.; Khairuddin, F. Risk Assessment on Factors Affecting Coconut Field Operation Among Smallholders to Enhance Coconut Production. Int. J. Acad. Res. Bus. Soc. Sci. 2022, 1, 13. [Google Scholar]
- Singh, P.; Kaur, A. Chapter 2—A Systematic Review of Artificial Intelligence in Agriculture. In Deep Learning for Sustainable Agriculture; Cognitive Data Science in Sustainable Computing; Poonia, R.C., Singh, V., Nayak, S.R., Eds.; Academic Press: London, UK, 2022; pp. 57–80. ISBN 978-0-323-85214-2. [Google Scholar]
- Alsanea, M.; Habib, S.; Khan, N.F.; Alsharekh, M.F.; Islam, M.; Khan, S. A Deep-Learning Model for Real-Time Red Palm Weevil Detection and Localization. J. Imaging 2022, 8, 170. [Google Scholar] [CrossRef]
| Study | Organism | Targetable Genes | Functions | Observations | Reference |
|---|---|---|---|---|---|
| Genomics of coconut | Coconut | Resistance gene analogs (RGA) | Resistance to pests and diseases | The identification of particular RGAs for resistance to specific pathogens and pests is necessary | [40] |
| Transcriptomics of root wilt disease of coconut (causal agent phytoplasma) | Coconut | NBS-LRR domain proteins | Signal transduction to induce the plant defense against pathogen attack | Up-regulated | [42] |
| Transcriptomics of root wilt disease of coconut (causal agent phytoplasma) | Coconut | PR1, PR4 | pathogenesis-related proteins | Supports the defense mechanism against RWD | [42] |
| Transcriptomics of root wilt disease of coconut (causal agent phytoplasma) | Coconut | PTI5-like gene | pathogenesis-related genes transcriptional activator | Highly up-regulated | [42] |
| Transcriptomics of root wilt disease of coconut (causal agent phytoplasma) | Coconut | HSP70 | A chaperone involved in hypersensitive response in plant defense mechanism | Highly up-regulated | [42] |
| Transcriptomics of C. nucifera and Phytophthora palmivora interaction | Coconut | 64,639 coconut transcripts | -- | Draft of dataset. Needs to be characterized to identify targetable genes | [18] |
| Transcriptomics of C. nucifera and Phytophthora palmivora interaction | P. palmivora | 9168 P. palmivora transcripts | -- | Draft of dataset. Needs to be characterized to identify targetable genes | [18] |
| Transcriptome of the beetle Oryctes rhinoceros | O. rhinoceros | β-1,4-endoglucanases and cellobiases | Degradation of the coconut cell wall. Feeding | Up-regulated in the intestine of the beetle larvae | [15] |
| Transcriptome of the moth, Euphestia (Cadra) cautella | The genes codifying 2-carbon fatty acid desaturase, fatty acid reductase and acetyl transferase | Involved in the biosynthesis of the pheromone (Z,E)-9,12-tetradecadienyl acetate | Power pheromone that induces aggregation in this moth. Its production is by heterologous expression of the three enzymes. | [58] | |
| Transcriptome of Rhynchophorus ferrugineus | R. ferrugineus (red palm weevil) | zf-C2H2, ZBTB, TF-bZIP transcription factors | Regulate growth, development and immunity in insects | Up-regulated in larvae and pupa stages | [56] |
| Transcriptome of R. ferrugineus | R. ferrugineus (red palm weevil) | serine hydrolases and peptidases | Proteases with role in feeding | Up-regulated in larvae stage | [56] |
| Proteomics of interaction of date palm leaves (close coconut relative) and R. ferrugineus | Date palm | homologs to the late blight resistant protein R1B-8 and late blight resistant protein RIA-10 | Up-regulated in date palm infested with this red palm weevil | Identification of orthologs of these RGAs is necessary in coconut as well as the elucidation of its role in red palm weevil infestation | [54] |
| Metabolomics studies of the beetle Oryctes rhinoceros, haplotype G | O. rhinoceros, | The pheromone ethyl (R)-4-methyloctanoate | The “R” enantiomer is a strong attractant for males and females of rhinoceros beetles | Genes involved in R)-4-methyloctanoate biosynthesis need to be identified for further production of this pheromone by genetic engineering | [57] |
| Metagenomics identification of mycorrhizal fungi associated with coconut roots | Coconut | Species of Glomus, Sclerocystis, Rhizophagus, Redeckera, and Diversispora genera | Mycorrhizal fungi improve nutrient uptake from the soil and modulate plant defenses against insects and pathogens. | These were the most abundant species associated with coconut roots | [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tzec-Simá, M.; Félix, J.W.; Granados-Alegría, M.; Aparicio-Ortiz, M.; Juárez-Monroy, D.; Mayo-Ruiz, D.; Vivas-López, S.; Gómez-Tah, R.; Canto-Canché, B.; Berezovski, M.V.; et al. Potential of Omics to Control Diseases and Pests in the Coconut Tree. Agronomy 2022, 12, 3164. https://doi.org/10.3390/agronomy12123164
Tzec-Simá M, Félix JW, Granados-Alegría M, Aparicio-Ortiz M, Juárez-Monroy D, Mayo-Ruiz D, Vivas-López S, Gómez-Tah R, Canto-Canché B, Berezovski MV, et al. Potential of Omics to Control Diseases and Pests in the Coconut Tree. Agronomy. 2022; 12(12):3164. https://doi.org/10.3390/agronomy12123164
Chicago/Turabian StyleTzec-Simá, Miguel, Jean Wildort Félix, María Granados-Alegría, Mónica Aparicio-Ortiz, Dilery Juárez-Monroy, Damian Mayo-Ruiz, Saraí Vivas-López, Rufino Gómez-Tah, Blondy Canto-Canché, Maxim V. Berezovski, and et al. 2022. "Potential of Omics to Control Diseases and Pests in the Coconut Tree" Agronomy 12, no. 12: 3164. https://doi.org/10.3390/agronomy12123164







