Assessing the Suitability of Selection Approaches and Genetic Diversity Analysis for Early Detection of Salt Tolerance of Barley Genotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Germination Experiment and Their Traits Measurements
2.3. Seedling Experiment and Their Traits Measurements
2.4. Stress Tolerance Index
2.5. DNA Extraction and PCR Amplification
2.6. Statistical Analysis
3. Results
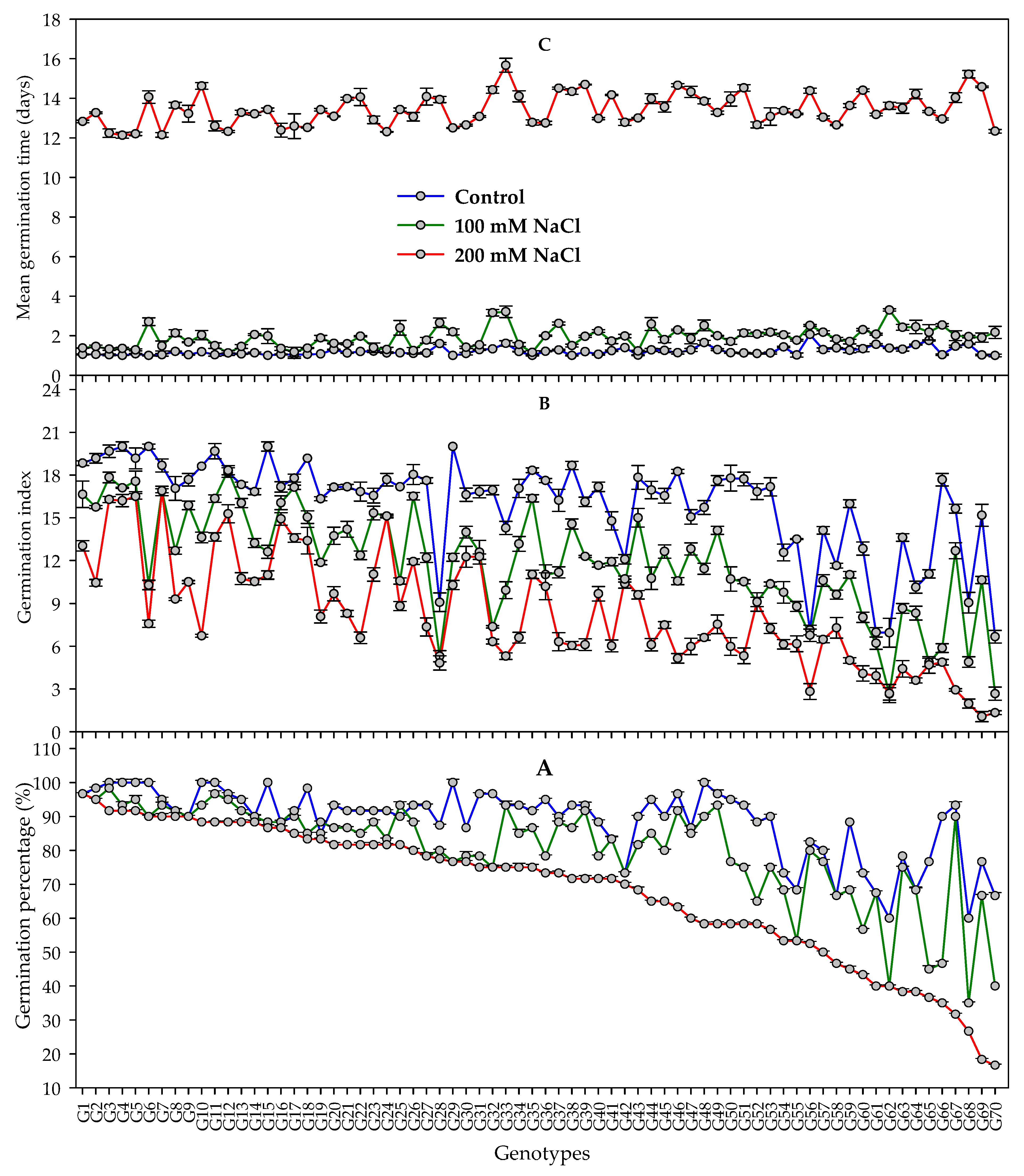
3.1. Germination Traits
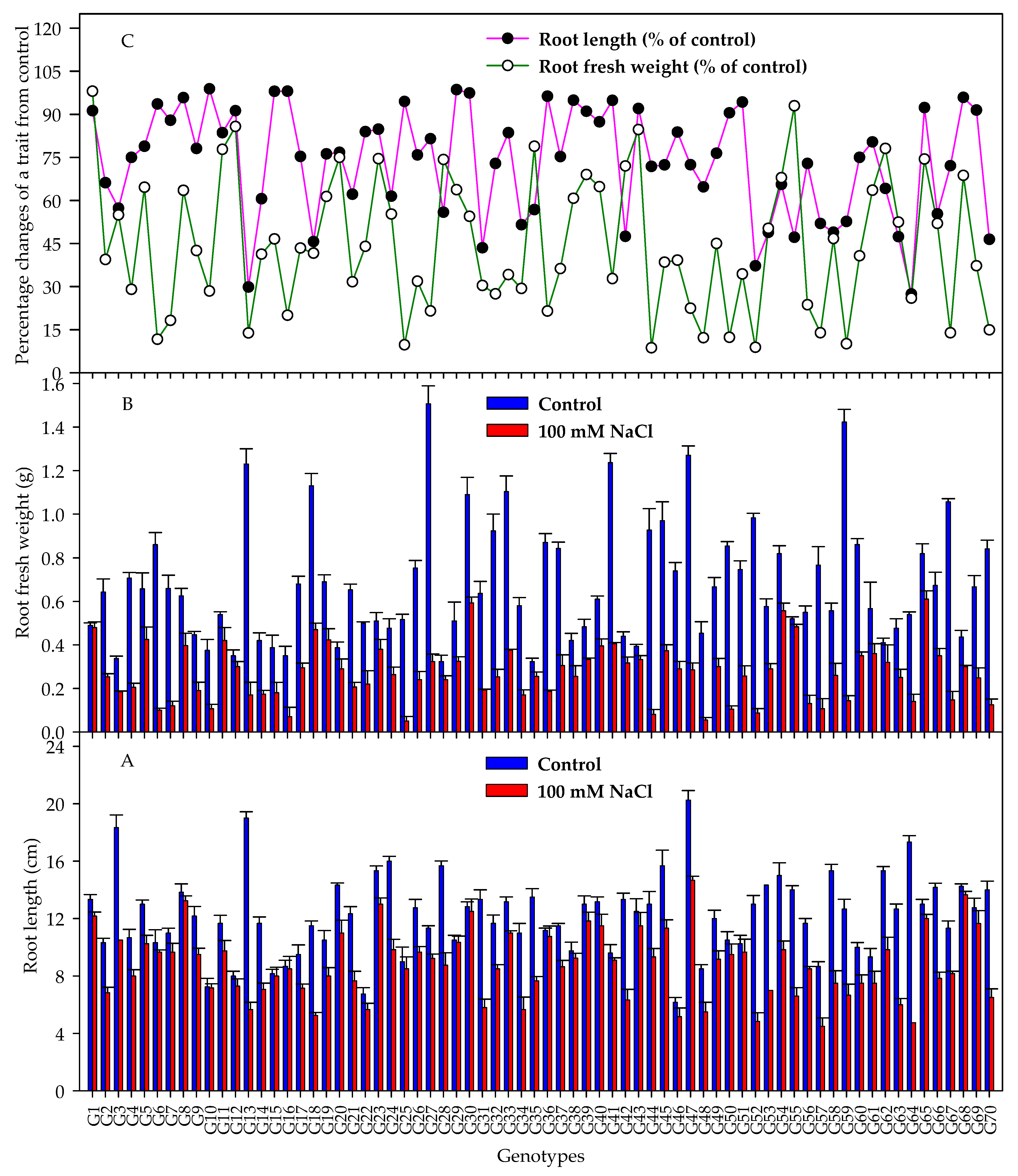
3.2. Seedling Traits
3.3. Correlation Analysis
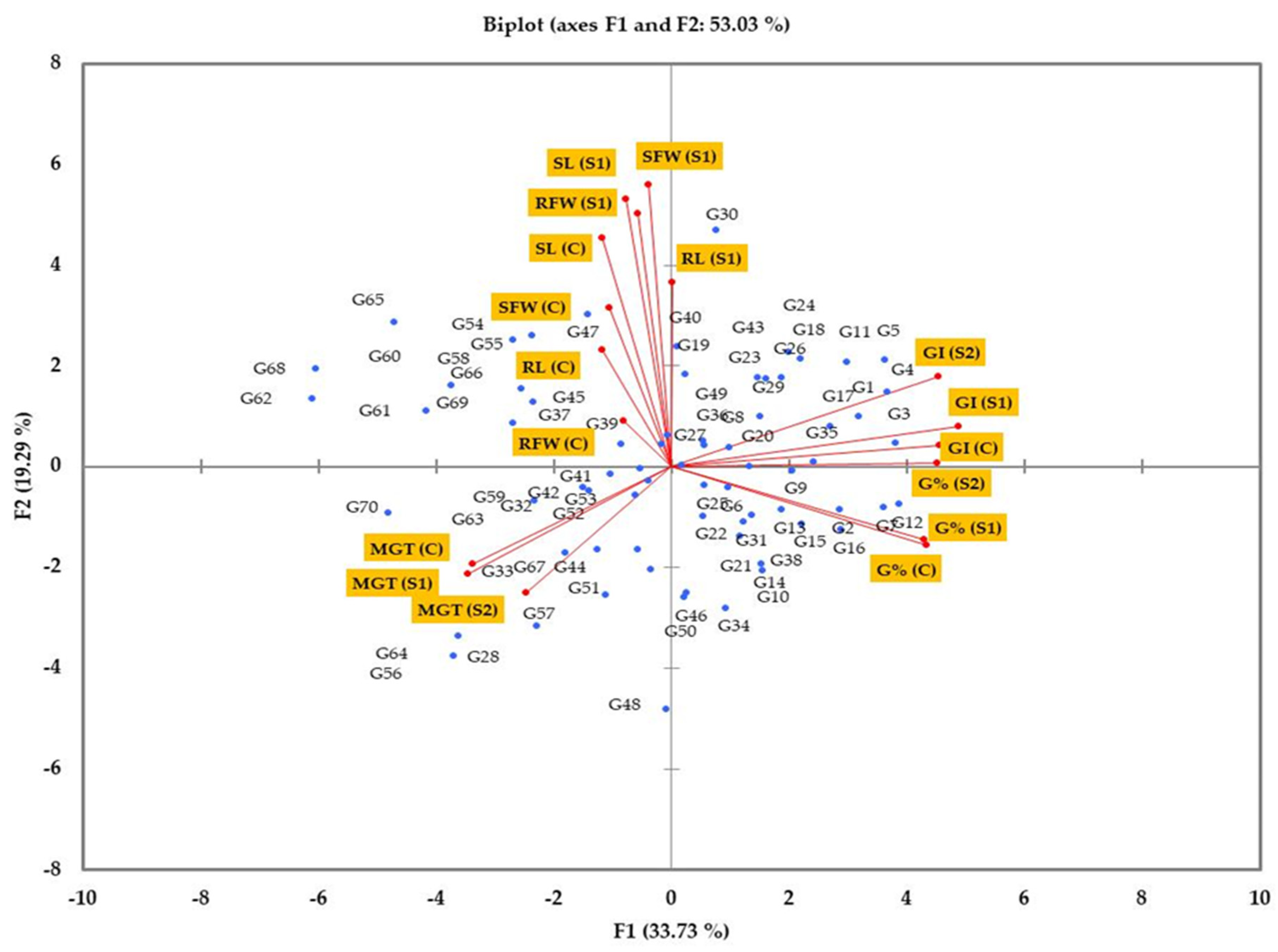
3.4. Principal Component Analysis
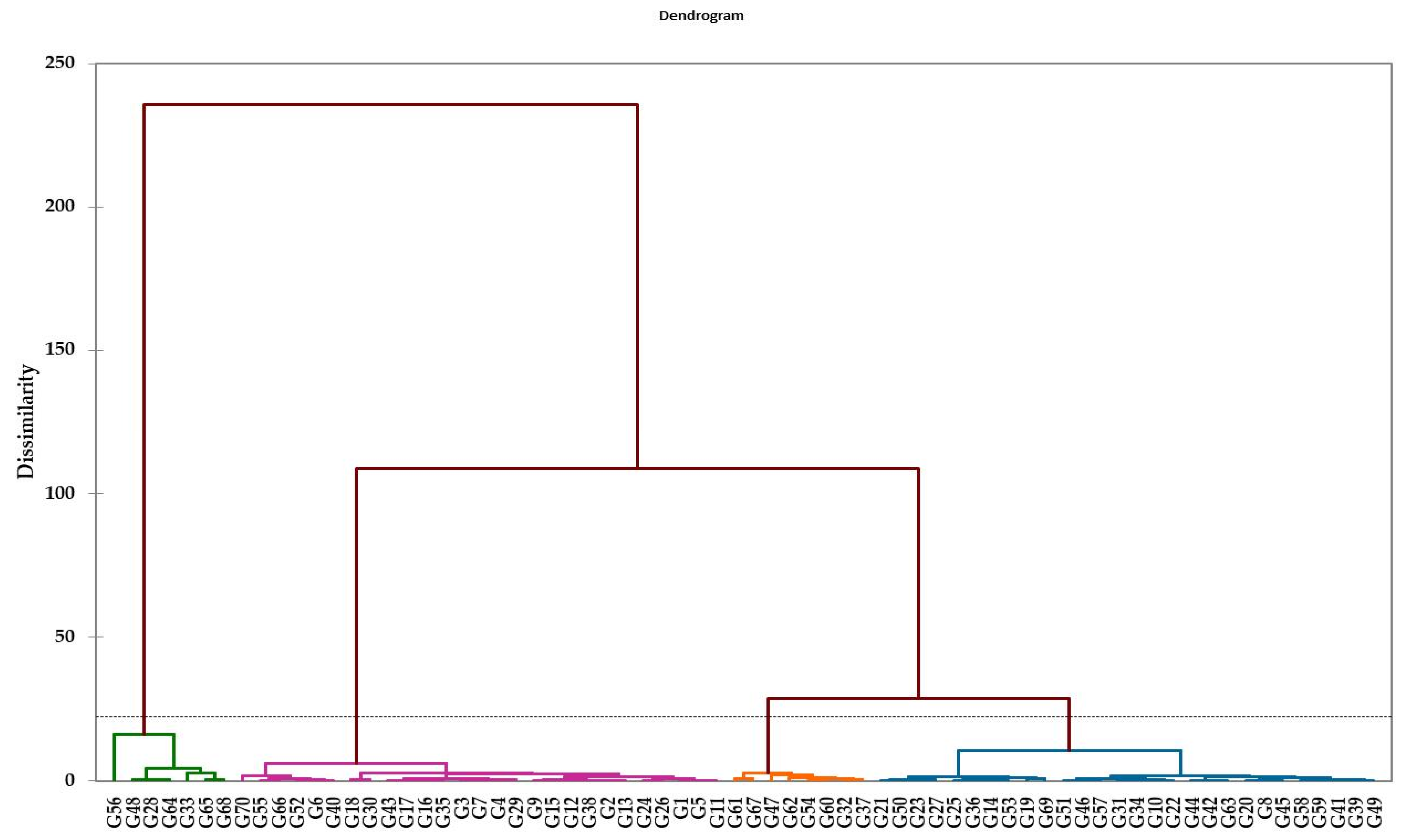
3.5. Stress Tolerance Index and Cluster Analysis
3.6. Ranking of Genotypes for their Relative Salt Tolerance at Two Growth Stages
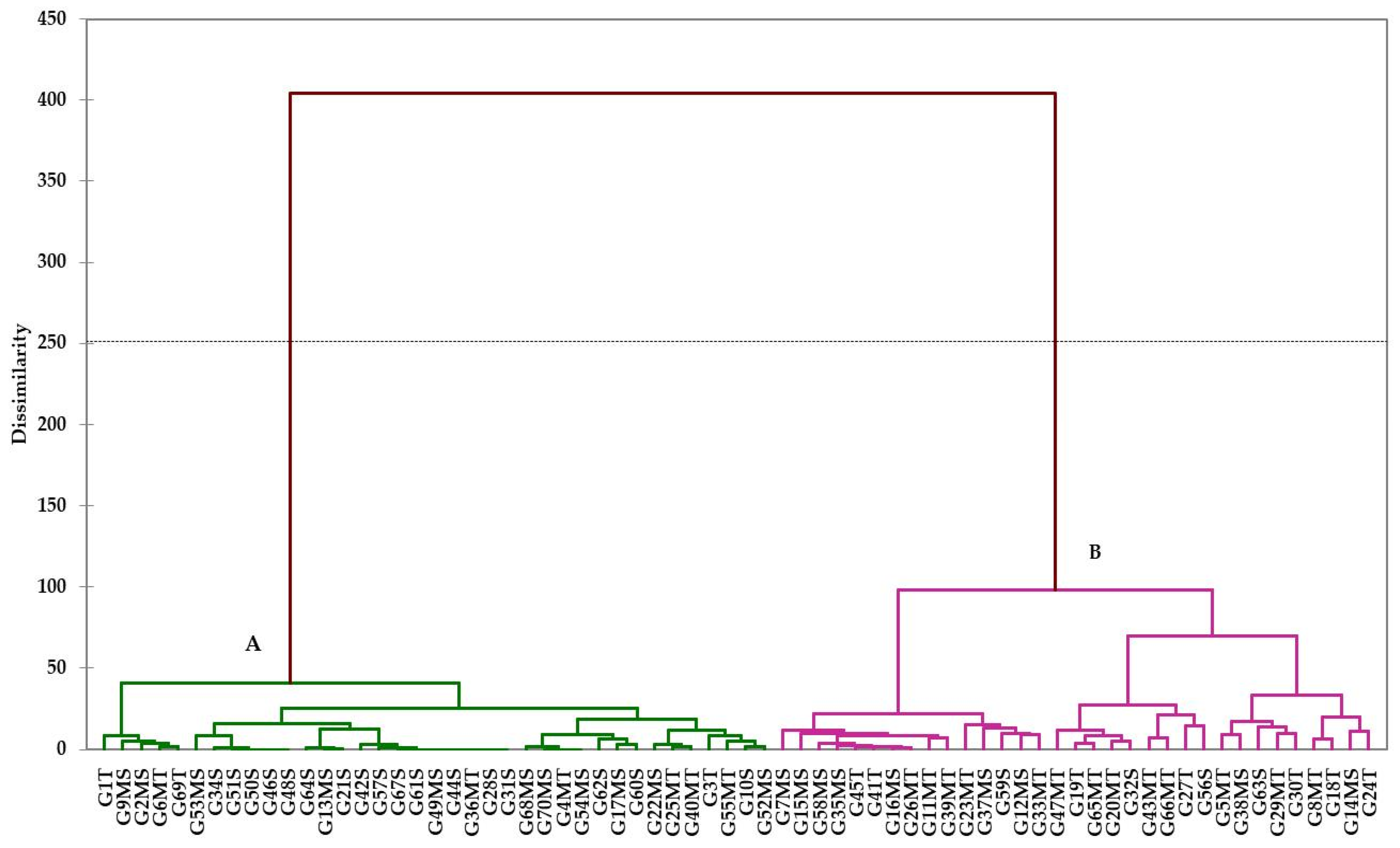
3.7. SRAP Analysis
4. Discussion
4.1. Ability of Evaluating Salt Tolerance of Barley Genotypes at Early Stage Based on Germination Indices
4.2. Ability of Evaluating Salt Tolerance of Barley Genotypes at Early Stage Based on Seedling Traits
4.3. Relationship between the Salt Tolerance during Germination and Seedling Stages
4.4. Detecting the Salt Tolerance of Genotypes Using Multivariate Analysis
4.5. Detecting the Salt Tolerance of Genotypes Based on SRAP
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Butcher, K.; Wick, A.F.; DeSutter, T.; Chatterjee, A.; Harmon, J. Soil salinity: A threat to global food security. Agron. J. 2016, 108, 2189–2200. [Google Scholar] [CrossRef]
- Mansour, E.; Moustafa, E.S.; Desoky, E.-S.M.; Ali, M.M.; Yasin, M.A.; Attia, A.; Alsuhaibani, N.; Tahir, M.U.; El-Hendawy, S. Multidimensional evaluation for detecting salt tolerance of bread wheat genotypes under actual saline field growing conditions. Plants 2020, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Lei, Y.; Liu, Q.; Hettenhausen, C.; Cao, G.; Tan, Q.; Zhao, W.; Lin, H.; Wu, J. Salt-tolerant and-sensitive alfalfa (Medicago sativa) cultivars have large variations in defense responses to the lepidopteran insect Spodoptera litura under normal and salt stress condition. PLoS ONE 2017, 12, e0181589. [Google Scholar] [CrossRef] [PubMed]
- Tigabu, E.; Tesfaye, K. Response of sorghum (Sorghum bicolor (L.) Moench) genotypes to NaCl levels at early growth stages. Afr. J. Agric. Res. 2012, 7, 5711–5718. [Google Scholar]
- UÇARLI, C. Physiological and molecular effects of exogenous gibberellin (GA3) treatment on germination of barley seeds under salt stress. Adıyaman Univ. J. Sci. 2021, 11, 227–243. [Google Scholar] [CrossRef]
- El-Hendawy, S.; Elshafei, A.; Al-Suhaibani, N.; Alotabi, M.; Hassan, W.; Dewir, Y.H.; Abdella, K. Assessment of the salt tolerance of wheat genotypes during the germination stage based on germination ability parameters and associated SSR markers. J. Plant Interact. 2019, 14, 151–163. [Google Scholar] [CrossRef]
- Hakim, M.; Juraimi, A.S.; Hanafi, M.; Selamat, A.; Ismail, M.R.; Karim, S.R. Studies on seed germination and growth in weed species of rice field under salinity stress. J. Environ. Biol. 2011, 32, 529. [Google Scholar]
- Dehnavi, A.R.; Zahedi, M.; Ludwiczak, A.; Perez, S.C.; Piernik, A. Effect of salinity on seed germination and seedling development of sorghum (Sorghum bicolor (L.) Moench) genotypes. Agronomy 2020, 10, 859. [Google Scholar] [CrossRef]
- Biabani, A.; Heidari, H.; Vafaie-Tabar, M. Salinity effect of stress on germination of wheat cultivars. Int. J. Agric. Food Sci. Sci. Technol. Technol. 2013, 4, 263–268. [Google Scholar]
- Kano, M.; Inukai, Y.; Kitano, H.; Yamauchi, A. Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant Soil 2011, 342, 117–128. [Google Scholar] [CrossRef]
- EL Hendawy, S.; Hu, Y.; Sakagami, J.-I.; Schmidhalter, U. Screening Egyptian wheat genotypes for salt tolerance at early growth stages. Int. J. Plant Prod. 2011, 5, 283–298. [Google Scholar]
- Colmer, T.; Munns, R.; Flowers, T. Improving salt tolerance of wheat and barley: Future prospects. Aust. J. Exp. Agric. 2005, 45, 1425–1443. [Google Scholar] [CrossRef]
- Chen, Z.; Pottosin, I.I.; Cuin, T.A.; Fuglsang, A.T.; Tester, M.; Jha, D.; Zepeda-Jazo, I.; Zhou, M.; Palmgren, M.G.; Newman, I.A.; et al. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 2007, 145, 1714–1725. [Google Scholar] [CrossRef]
- Wu, D.; Qiu, L.; Xu, L.; Ye, L.; Chen, M.; Sun, D.; Chen, Z.; Zhang, H.; Jin, X.; Dai, F. Genetic variation of HvCBF genes and their association with salinity tolerance in Tibetan annual wild barley. PLoS ONE 2011, 6, e22938. [Google Scholar] [CrossRef][Green Version]
- Witzel, K.; Weidner, A.; Surabhi, G.K.; Börner, A.; Mock, H.P. Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J. Exp. Bot. 2009, 60, 3545–3557. [Google Scholar] [CrossRef]
- Mano, Y.; Nakazumi, H.; Takeda, K. Varietal variation in and effects of some major genes on salt tolerance at the germination stage in barley. Jpn. J. Breed. 1996, 46, 227–233. [Google Scholar] [CrossRef]
- Mano, Y.; Takeda, K. Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 1997, 94, 263–272. [Google Scholar] [CrossRef]
- Xue, D.; Huang, Y.; Zhang, X.; Wei, K.; Westcott, S.; Li, C.; Chen, M.; Zhang, G.; Lance, R. Identification of QTLs associated with salinity tolerance at late growth stage in barley. Euphytica 2009, 169, 187–196. [Google Scholar] [CrossRef]
- Shelden, M.C.; Roessner, U.; Sharp, R.E.; Tester, M.; Bacic, A. Genetic variation in the root growth response of barley genotypes to salinity stress. Funct. Plant Biol. 2013, 40, 516–530. [Google Scholar] [CrossRef]
- Gupta, S.; Rupasinghe, T.; Callahan, D.L.; Natera, S.H.; Smith, P.; Hill, C.B.; Roessner, U.; Boughton, B.A. Spatio-temporal metabolite and elemental profiling of salt stressed barley seeds during initial stages of germination by MALDI-MSI and µ-XRF spectrometry. Front. Plant Sci. 2019, 10, 1139. [Google Scholar] [CrossRef]
- Shelden, M.C.; Dias, D.A.; Jayasinghe, N.S.; Bacic, A.; Roessner, U. Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J. Exp. Bot. 2016, 67, 3731–3745. [Google Scholar] [CrossRef] [PubMed]
- Ghoulam, C.; Fares, K. Effect of salinity on seed germination and early seedling growth of sugar beet (Beta vulgaris L.). Seed Sci. Technol. 2001, 29, 357–364. [Google Scholar]
- Punia, H.; Tokas, J.; Malik, A.; Singh, S.; Phogat, D.S.; Bhuker, A.; Mor, V.S.; Rani, A.; Sheokand, R.N. Discerning morpho-physiological and quality traits contributing to salinity tolerance acquisition in sorghum [Sorghum bicolor (L.) Moench]. S. Afr. J. Bot. 2021, 140, 409–418. [Google Scholar] [CrossRef]
- El-Hamamsy, S.; Behairy, R. Effect of salinity stress on seedling vigor and biochemical characters of Egyptian barley landraces (Hordeum vulgare L.). Midle East J. Appl. 2015, 5, 786–796. [Google Scholar]
- Garland, S.H.; Lewin, L.; Abedinia, M.; Henry, R.; Blakeney, A.J.E. The use of microsatellite polymorphisms for the identification of Australian breeding lines of rice (Oryza sativa L.). Euphytica 1999, 108, 53–63. [Google Scholar] [CrossRef]
- Abdel-Hamid, A.M. Physiological and molecular markers for salt tolerance in four barley cultivars. Eur. Sci. J. 2014, 10, 252–272. [Google Scholar]
- Yang, P.; Liu, X.; Yang, W.; Feng, Z. Diversity analysis of the developed qingke (hulless barley) cultivars representing different growing regions of the Qinghai-Tibet Plateau in China using sequencerelated amplified polymorphism (SRAP) markers. Afr. J. Biotechnol. 2010, 9, 8530–8538. [Google Scholar]
- Mariey, S. Genetic diversity study of Egyptian barley cultivars using sequence-related amplified polymorphism (SRAP) analysis for water stress tolerance. J. Sustain. Agric. Sci. 2018, 44, 21–37. [Google Scholar] [CrossRef]
- Al-Mudaris, M. Notes on various parameters recording the speed of seed germination. Der Trop.-J. Agric. Trop. Subtrop. 1998, 99, 147–154. [Google Scholar]
- Fernandez, G.C. Effective selection criteria for assessing plant stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and Other Food Crops in Temperature and Water Stress, Shanhua, Taiwan, 13–16 August 1992; pp. 257–270. [Google Scholar]
- Doyle, J. DNA protocols for plants. In Molecular Techniques in Taxonomy; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- Naceur, A.B.; Chaabane, R.; El-Faleh, M.; Abdelly, C.; Ramla, D.; Nada, A.; Sakr, M. Genetic diversity analysis of North Africa’s barley using SSR markers. J. Genet. Eng. Biotechnol. 2012, 10, 13–21. [Google Scholar] [CrossRef]
- El-Hendawy, S.E.; Hu, Y.; Yakout, G.M.; Awad, A.M.; Hafiz, S.E.; Schmidhalter, U. Evaluating salt tolerance of wheat genotypes using multiple parameters. Eur. J. Agron. 2005, 22, 243–253. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Team, R. RStudio: Integrated Development for R. RStudio. Available online: http://www.rstudio.com/ (accessed on 22 January 2021).
- Galili, T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Beattie, K.A. Worldwide Bayesian Causal Impact Analysis of Vaccine Administration on Deaths and Cases Associated with COVID-19: A BigData Analysis of 145 Countries. ResearchGate 2021. Available online: https://www.datascienceassn.org/content/worldwide-bayesian-causal-impact-analysis-vaccine-administration-deaths-and-cases-associated (accessed on 13 December 2022).
- Peterson, B.G.; Carl, P.; Boudt, K.; Bennett, R.; Ulrich, J.; Zivot, E.; Lestel, M.; Balkissoon, K.; Wuertz, D.; Peterson, M.B.G. Performance Analytics: Econometric Tools for Performance and Risk Analysis. R Package Version 1. Available online: https://cran.r-project.org/web/packages/PerformanceAnalytics/index.html (accessed on 15 August 2021).
- Wickham, H.; Hester, J.; Chang, W. Devtools: Tools to Make Developing R Packages Easier. R Package Version 2.3.1. 2020. Available online: https://CRAN.R-project.org/package=devtools (accessed on 13 December 2020).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. R package Version 3.3.2. 2020. Available online: https://CRAN.R-project.org/package=ggplot2 (accessed on 13 December 2021).
- Kassambara, A.; Kassambara, M.A. Package ‘ggpubr’. R Package Version 0.3.5; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Qiu, L.; Wu, D.; Ali, S.; Cai, S.; Dai, F.; Jin, X.; Wu, F.; Zhang, G. Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor. Appl. Genet. 2011, 122, 695–703. [Google Scholar] [CrossRef]
- Gul, B.; Ansari, R.; Flowers, T.J.; Khan, M.A. Germination strategies of halophyte seeds under salinity. Environ. Exp. Bot. 2013, 92, 4–18. [Google Scholar] [CrossRef]
- Bai, Q.; Wang, X.; Chen, X.; Shi, G.; Liu, Z.; Guo, C.; Xiao, K. Wheat miRNA TaemiR408 acts as an essential mediator in plant tolerance to Pi deprivation and salt stress via modulating stress-associated physiological processes. Front. Plant Sci. 2018, 9, 499. [Google Scholar] [CrossRef]
- Mwando, E.; Angessa, T.T.; Han, Y.; Li, C. Salinity tolerance in barley during germination—Homologs and potential genes. J. Zhejiang Univ.-Sci. B. 2020, 21, 93–121. [Google Scholar] [CrossRef]
- Wahid, A.; Perveen, M.; Gelani, S.; Basra, S.M. Pretreatment of seed with H2O2 improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. J. Plant Physiol. 2007, 164, 283–294. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.; Shen, J.; Baum, M.; Ogbonnaya, F.; Léon, J.; Ballvora, A. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Tao, R.; Ding, J.; Li, C.; Zhu, X.; Guo, W.; Zhu, M. Evaluating and screening of agro-physiological indices for salinity stress tolerance in wheat at the seedling stage. Front. Plant Sci. 2021, 12, 646175. [Google Scholar] [CrossRef]
- Almodares, A.; Hadi, M.; Dosti, B. Effects of salt stress on germination percentage and seedling growth in sweet sorghum cultivars. J. Biol. Sci. 2007, 7, 1492–1495. [Google Scholar] [CrossRef]
- Kinfemichael, G.A.; Melkamu, G.W. Response of some lowland growing sorghum (Sorghum bicolor L. Moench) accessions to salt stress during germination and seedling growth. Afr. J. Agric. Res. 2008, 3, 044–048. [Google Scholar]
- Jamil, M.; Lee, D.B.; Jung, K.Y.; Ashraf, M.; Lee, S.C.; Rha, E.S. Effect of salt (NaCl) stress on germination and early seedling growth of four vegetable species. J. Cent. Eur. Agric. 2006, 7, 273–282. [Google Scholar]
- Khan, M.A.; Khan, M.A.; Weber, D.J. Ecophysiology of High Salinity Tolerant Plants; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006; Volume 40. [Google Scholar]
- Gomes-Filho, E.; Lima, C.R.F.M.; Costa, J.H.; da Silva, A.C.M.; da Guia Silva Lima, M.; de Lacerda, C.F.; Prisco, J.T. Cowpea ribonuclease: Properties and effect of NaCl-salinity on its activation during seed germination and seedling establishment. Plant Cell Rep. 2008, 27, 147–157. [Google Scholar] [CrossRef]
- Kazemi, K.; Eskandari, H. Effects of salt stress on germination and early seedling growth of rice (Oryza sativa) cultivars in Iran. Afr. J. Biotechnol. 2011, 10, 17789–17792. [Google Scholar] [CrossRef]
- Malik, J.A.; AlQarawi, A.A.; AlZain, M.N.; Dar, B.A.; Habib, M.M.; Ibrahim, S.N.S. Effect of salinity and temperature on the seed germination and seedling growth of desert forage grass Lasiurus scindicus Henr. Sustainability 2022, 14, 8387. [Google Scholar] [CrossRef]
- Ayaz, F.; Kadioglu, A.; Turgut, R. Water stress effects on the content of low molecular weight carbohydrates and phenolic acids in Ctenanthe setosa (Rosc.) Eichler. Can. J. Plant Sci. 2000, 80, 373–378. [Google Scholar] [CrossRef]
- Promila, K.; Kumar, S. Vigna radiata seed germination under salinity. Biol. Plant. 2000, 43, 423–426. [Google Scholar] [CrossRef]
- Othman, Y.; Al-Karaki, G.; Al-Tawaha, A.; Al-Horani, A. Variation in germination and ion uptake in barley genotypes under salinity conditions. World J. Agric. Sci. 2006, 2, 11–15. [Google Scholar]
- Aflaki, F.; Sedghi, M.; Pazuki, A.; Pessarakli, M. Investigation of seed germination indices for early selection of salinity tolerant genotypes: A case study in wheat. Emir. J. Food Agric. 2017, 29, 222–226. [Google Scholar] [CrossRef]
- Khan, W.D.; Tanveer, M.; Shaukat, R.; Ali, M.; Pirdad, F. An overview of salinity tolerance mechanism in plants. In Salt and Drought Stress Tolerance in Plants; Tanveer, M., Hasanuzzaman, M., Eds.; Springer: Cham, Switzerland, 2020; pp. 1–16. [Google Scholar]
- Shah, A.N.; Tanveer, M.; Abbas, A.; Fahad, S.; Baloch, M.S.; Ahmad, M.I.; Saud, S.; Song, Y. Targeting salt stress coping mechanisms for stress tolerance in Brassica: A research perspective. Plant Physiol. Biochem. 2021, 158, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Uzair, M.; Ali, M.; Fiaz, S.; Attia, K.; Khan, N.; Al-Doss, A.A.; Khan, M.R.; Ali, Z. The characterization of wheat genotypes for salinity tolerance using morpho-physiological indices under hydroponic conditions. Saudi J. Biol. Sci. 2022, 29, 103299. [Google Scholar] [CrossRef] [PubMed]
- Shuba, A.; Patil, S.S.; Rajeshwari, M. Seed priming with endophytes: A novel approach for future prospects. J. Pharmacogn. Phytochem. 2019, 8, 1442–1446. [Google Scholar]
- El-Hendawy, S.; Ruan, Y.; Hu, Y.; Schmidhalter, U. A comparison of screening criteria for salt tolerance in wheat under field and controlled environmental conditions. J. Agron. Crop Sci. 2009, 195, 356–367. [Google Scholar] [CrossRef]
- Bai, J.; Yan, W.; Wang, Y.; Yin, Q.; Liu, J.; Wight, C.; Ma, B. Screening oat genotypes for tolerance to salinity and alkalinity. Front. Plant Sci. 2018, 9, 1302. [Google Scholar] [CrossRef]
- Rasel, M.; Tahjib-Ul-Arif, M.; Hossain, M.; Sayed, M.; Hassan, L. Discerning of rice landraces (Oryza sativa L.) for morpho-physiological, antioxidant enzyme activity, and molecular markers’ responses to induced salt stress at the seedling stage. J. Plant Growth Regul. 2020, 39, 41–59. [Google Scholar] [CrossRef]
- El-Bastawisy, Z.M.; El-Katony, T.M.; Abd El-Fatah, S.N. Genotypic variability in salt tolerance of Vicia faba during germination and early seedling growth. J. King Saud Univ. Sci. 2018, 30, 270–277. [Google Scholar] [CrossRef]
- Foti, C.; Khah, E.; Pavli, O. Germination profiling of lentil genotypes subjected to salinity stress. Plant Biol. 2019, 21, 480–486. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Ashraf, M. Salt-induced variation in some potential physiochemical attributes of two genetically diverse spring wheat (Triticum aestivum L.) cultivars: Photosynthesis and photosystem II efficiency. Pak. J. Bot. 2012, 44, 53–64. [Google Scholar]
- Radi, A.A.; Farghaly, F.A.; Hamada, A.M. Physiological and biochemical responses of salt-tolerant and salt-sensitive wheat and bean cultivars to salinity. J. Biol. Earth Sci. 2013, 3, 72–88. [Google Scholar]
- Taïbi, K.; Taïbi, F.; Abderrahim, L.A.; Ennajah, A.; Belkhodja, M.; Mulet, J.M. Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defence systems in Phaseolus vulgaris L. S. Afr. J. Bot. 2016, 105, 306–312. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Shoukat, E.; Ahmed, M.Z.; Abideen, Z.; Azeem, M.; Ibrahim, M.; Gul, B.; Khan, M.A. Short and long term salinity induced differences in growth and tissue specific ion regulation of Phragmites karka. Flora 2020, 263, 151550. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Sanjani, S.; Nikkhah-Chamanabad, H.; Mehrvar, M.R.; Asadi, A.; Amini, A. Identification of salt-tolerant barley genotypes using multiple-traits index and yield performance at the early growth and maturity stages. Bull. Natl. Res. Cent. 2021, 45, 117. [Google Scholar] [CrossRef]
- Jadidi, O.; Etminan, A.; Azizi-Nezhad, R.; Ebrahimi, A.; Pour-Aboughadareh, A. Physiological and molecular responses of barley genotypes to salinity stress. Genes 2022, 13, 2040. [Google Scholar] [CrossRef] [PubMed]
- Al-Dakheel, A.J.; Hussain, M.I. Genotypic variation for salinity tolerance in Cenchrus ciliaris L. Front. Plant Sci. 2016, 7, 1090. [Google Scholar] [CrossRef]
- Aslam, M.; Maqbool, M.A.; Zaman, Q.U.; Shahid, M.; Akhtar, M.A.; Rana, A.S. Comparison of different tolerance indices and PCA biplot analysis for assessment of salinity tolerance in lentil (Lens culinaris) genotypes. Int. J. Agric. Biol. 2017, 19, 470–478. [Google Scholar] [CrossRef]
- Magudeeswari, P.; Sastry, E.V.; Devi, T.R. Principal component (PCA) and cluster analyses for plant nutrient traits in baby corn (Zea mays L.). Indian J. Agric. Res. 2019, 3, 353–357. [Google Scholar] [CrossRef]
- Alam, M.S.; Hossain, S.; Ali, M.A.; Hossain, M.G.; Islam, M.F. Assessment of genetic divergence in tomato (Solanum lycopersicum L.) through clustering and principal component analysis. J. Agric. Sci. Eng. Innov. 2020, 1, 10–14. [Google Scholar]
- Ahmadi, J.; Pour-Aboughadareh, A.; Fabriki-Ourang, S.; Mehrabi, A.A.; Siddique, K.H.M. Screening wild progenitors of wheat for salinity stress at early stages of plant growth: Insight into potential sources of variability for salinity adaptation in wheat. Crop Pasture Sci. 2018, 69, 649–658. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Poczai, P. A dataset on multi-trait selection approaches for screening desirable wild relatives of wheat. Data Brief 2021, 39, 107541. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sengar, R.S.; Kulshreshtha, N.; Datta, D.; Tomar, R.S.; Rao, V.P.; Garg, D.; Ojha, A. Assessment of multiple tolerance indices for salinity stress in bread wheat (Triticum aestivum L.). J. Agric. Sci. Eng. Innov. 2015, 7, 49–57. [Google Scholar] [CrossRef]
- El-Hendawy, S.; Al-Suhaibani, N.; Mubushar, M.; Tahir, M.U.; Refay, Y.; Tola, E. Potential Use of hyperspectral reflectance as a high-throughput nondestructive phenotyping tool for assessing salt tolerance in advanced spring wheat lines under field conditions. Plants 2021, 10, 2512. [Google Scholar] [CrossRef]
- Ahmadi, J.; Pour-Aboughadareh, A.; Fabriki Ourang, S.; Khalili, P.; Poczai, P. Unraveling salinity stress responses in ancestral and neglected wheat species at early growth stage: A baseline for utilization in future wheat improvement programs. Physiol. Mol. Biol. Plants 2020, 26, 537–549. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Ahmadi, J.; Mehrabi, A.A.; Etminan, A.; Moghaddam, M.; Siddique, K.H. Physiological responses to drought stress in wild relatives of wheat: Implications for wheat improvement. Acta Physiol. Plant. 2017, 39, 106. [Google Scholar] [CrossRef]
- Alam, M.S.; Tester, M.; Fiene, G.; Mousa, M.A.A. Early growth stage characterization and the biochemical responses for salinity stress in tomato. Plants 2021, 10, 712. [Google Scholar] [CrossRef]
- Mohi-Ud-Din, M.; Hossain, M.A.; Rohman, M.M.; Uddin, M.N.; Haque, M.S.; Ahmed, J.U.; Hossain, A.; Hassan, M.M.; Mostofa, M.G. Multivariate analysis of morpho-physiological traits reveals differential drought tolerance potential of bread wheat genotypes at the seedling stage. Plants 2021, 10, 879. [Google Scholar] [CrossRef]
- Zaefizadeh, M.; Goliev, R. Diversity and relationships among durum wheat landraces (subconvars) by SRAP and phenotypic marker polymorphism. Res. J. Biol. Sci. 2009, 4, 960–966. [Google Scholar]
- El-Rawy, M.A.E.; Youssef, M. Evaluation of drought and heat tolerance in wheat based on seedling traits and molecular analysis. J. Crop Sci. Biotechnol. 2014, 17, 183–189. [Google Scholar] [CrossRef]
- Abulela, H.A.; El Shafee, E.; Farag, H.M.; Yacoub, I.H.; Elarabi, N.I. Evaluation of the morpho-physiological traits and the genetic diversity of some Egyptian bread wheat cultivars under salt stress conditions. Cereal Res. Commun. 2022, 50, 733–753. [Google Scholar] [CrossRef]

| Genotype Name | Abb. | Source | Genotype Name | Abb. | Source |
|---|---|---|---|---|---|
| Giza121 | G1 | Egypt | Giza121/Local/12-1 | G36 | Saudi Arabia |
| Beecher | G2 | ICARDA | Giza123/Local/2 | G37 | Saudi Arabia |
| Lignee/Local/174-29-8-3 | G3 | Saudi Arabia | ER/Apm | G38 | ICARDA |
| Begona | G4 | ICARDA | Giza121/Local/1-10-5-5 | G39 | Saudi Arabia |
| Carbo | G5 | ICARDA | Arar/h.spont.19-12 | G40 | ICARDA |
| Lignee | G6 | CIMMYT | C.C.89/Giza124 | G41 | Saudi Arabia |
| Giza123/Local/1 | G7 | Saudi Arabia | Gustoe/Local/4 | G42 | Saudi Arabia |
| Wl2291 | G8 | ICARDA | Sahrawy/Local/4 | G43 | Saudi Arabia |
| Giza134 | G9 | Egypt | Giza121/Local/4 | G44 | Saudi Arabia |
| Tadmor | G10 | ICARDA | Gustoe | G45 | Commercial |
| Dd-21 | G11 | ICARDA | Giza126 | G46 | Egypt |
| Lignee/Local/4 | G12 | Saudi Arabia | Justo/Local/5 | G47 | Saudi Arabia |
| Sahrawy | G13 | Egypt | Giza121/Justo-3 | G48 | Saudi Arabia |
| Justo/Local/88-29-10 | G14 | Saudi Arabia | Gustoe/Local/2 | G49 | Saudi Arabia |
| Barley Gp | G15 | Pakistan | C.C.89/Giza123 | G50 | Saudi Arabia |
| Sahrawy/Local/1 | G16 | Saudi Arabia | Giza123/Justo-4 | G51 | Saudi Arabia |
| Giza123/Local/4 | G17 | Saudi Arabia | Giza121/Local/5 | G52 | Saudi Arabia |
| Waveney | G18 | ICARDA | Rehane | G53 | CIMMYT |
| Giza127 | G19 | ARC-Egypt | Lignee/Local/182-30-9-4 | G54 | Saudi Arabia |
| Granado | G20 | ICARDA | Giza123/Local/24-8-1 | G55 | Saudi Arabia |
| Local (Assir) | G21 | Saudi Arabia | Giza123/Local/40-18-4 | G56 | Saudi Arabia |
| Carina/moroc9-75 | G22 | ICARDA | Gustoe/Local/1 | G57 | Saudi Arabia |
| Justo/Local/59-13 | G23 | Saudi Arabia | Giza123/Local/30-11-3 | G58 | Saudi Arabia |
| Giza121/Local/3 | G24 | Saudi Arabia | Rihana/Lignee | G59 | ICARDA |
| 316-80 | G25 | ICARDA | Lignee/Local/5 | G60 | Saudi Arabia |
| Justo/Local/80-28-9 | G26 | Saudi Arabia | Sahrawy/Local/5 | G61 | Saudi Arabia |
| Giza123 | G27 | Egypt | Giza123/Local/3 | G62 | Saudi Arabia |
| Assala-04 | G28 | ICARDA | Gp No. 5 | G63 | Pakistan |
| Giza 133 | G29 | Egypt | Giza123/Local/30-7 | G64 | Saudi Arabia |
| Giza123/Local/15-14-7-7 | G30 | Saudi Arabia | Justo/Local/56-12 | G65 | Saudi Arabia |
| Lignee/Local/3 | G31 | Saudi Arabia | ILBA | G66 | CIMMYT |
| Armelle | G32 | ICARDA | Gustoe/Local/3 | G67 | Saudi Arabia |
| Giza124 | G33 | Egypt | Lignee/Local/1 | G68 | Saudi Arabia |
| Giza126-1 | G34 | Egypt | Giza2000 | G69 | Egypt |
| Giza121/Local/1 | G35 | Saudi Arabia | SLb42-046 | G70 | ICARDA |
| Combination | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| 1 | TGAGTCCAAACCGGATA | GACTGCGTACGAATTAAT |
| 2 | TGAGTCCAAACCGGATA | GACTGCGTACGAATTTGC |
| 3 | TGAGTCCAAACCGGATA | GACTGCGTACGAATTGAC |
| 4 | TGAGTCCAAACCGGAGC | GACTGCGTACGAATTAAT |
| 5 | TGAGTCCAAACCGGAGC | GACTGCGTACGAATTTGC |
| 6 | TGAGTCCAAACCGGAGC | GACTGCGTACGAATTGAC |
| 7 | GAGTCCAAACCGGAAG | GACTGCGTACGAATTAAT |
| 8 | GAGTCCAAACCGGAAG | GACTGCGTACGAATTTGC |
| 9 | GAGTCCAAACCGGAAG | GACTGCGTACGAATTGAC |
| Traits | Source | F Value | p Value |
|---|---|---|---|
| SL | Genotypes (G) | 19.48 | 0.000 *** |
| Salinity (S) | 2395.83 | 0.000 *** | |
| G×S | 8.26 | 0.000 *** | |
| RL | Genotypes (G) | 5.68 | 0.000 *** |
| Salinity (S) | 143.45 | 0.000 *** | |
| G×S | 5.56 | 0.000 *** | |
| SFW | Genotypes (G) | 4.97 | 0.000 *** |
| Salinity (S) | 476.52 | 0.000 *** | |
| G×S | 4.17 | 0.000 *** | |
| RFW | Genotypes (G) | 2.66 | 0.000 *** |
| Salinity (S) | 322.76 | 0.000 *** | |
| G×S | 3.07 | 0.000 *** |
| Traits | G% (C) | G% (S1) | G% (S2) | GI (C) | GI (S1) | GI (S2) | MGT (C) | MGT (S1) | MGT (S2) | SL (C) | SL (S1) | RL (C) | RL (S1) | SFW (C) | SFW (S1) | RFW (C) | RFW (S1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G% | *** | *** | *** | *** | *** | ** | ** | ns | ** | ** | ** | ns | ns | ns | ns | ns | |
| G% (S1) | 0.79 | *** | *** | *** | *** | ** | ** | ns | ** | ** | ** | ns | ns | ns | ns | ns | |
| G% (S2) | 0.72 | 0.79 | *** | *** | *** | ** | ** | ** | ** | ns | ns | ns | ns | ns | ns | ns | |
| GI (C) | 0.86 | 0.68 | 0.69 | *** | *** | *** | *** | ** | ns | ns | ns | ns | ns | ns | ns | ns | |
| GI (S1) | 0.68 | 0.81 | 0.75 | 0.78 | *** | *** | *** | ** | ns | ns | ns | ns | ns | ns | ns | ns | |
| GI (S2) | 0.56 | 0.59 | 0.79 | 0.65 | 0.81 | *** | *** | *** | ns | ns | ns | ns | ns | ns | ns | ns | |
| MGT(C) | −0.41 | −0.29 | −0.44 | −0.71 | −0.55 | −0.52 | *** | ** | ns | ns | ns | ns | ns | ns | ns | ns | |
| MGT(S1) | −0.28 | −0.36 | −0.41 | −0.46 | −0.77 | −0.66 | 0.50 | *** | ns | ns | ns | ns | ns | ns | ns | ns | |
| MGT(S2) | −0.13 | −0.10 | −0.29 | −0.25 | −0.41 | −0.72 | 0.44 | 0.55 | ns | ns | ns | ns | ns | ns | ns | ns | |
| SL (C) | −0.25 | −0.34 | −0.26 | −0.03 | −0.13 | −0.05 | −0.13 | −0.06 | −0.12 | ** | ns | ns | *** | ** | ** | ** | |
| SL (S1) | −0.26 | −0.27 | −0.10 | −0.07 | −0.10 | 0.07 | −0.11 | 0.02 | −0.21 | 0.44 | ns | ** | ns | *** | ns | *** | |
| RL (C) | −0.28 | −0.24 | −0.17 | −0.16 | −0.11 | −0.02 | 0.12 | −0.02 | −0.11 | 0.22 | 0.12 | ** | ns | ns | ns | ns | |
| RL (S1) | 0.02 | 0.02 | 0.08 | 0.05 | 0.09 | 0.06 | 0.01 | −0.07 | 0.10 | 0.18 | 0.42 | 0.35 | ns | ** | ns | *** | |
| SFW (C) | −0.23 | −0.18 | −0.18 | −0.08 | −0.08 | −0.09 | −0.04 | −0.06 | 0.00 | 0.57 | 0.16 | 0.10 | −0.02 | ns | *** | ** | |
| SFW (S1) | −0.18 | −0.18 | 0.04 | 0.01 | −0.01 | 0.08 | −0.13 | −0.05 | −0.08 | 0.37 | 0.75 | 0.06 | 0.44 | 0.22 | ns | *** | |
| RFW (C) | 0.06 | −0.08 | −0.15 | 0.04 | −0.09 | −0.19 | 0.03 | 0.09 | 0.21 | 0.36 | −0.16 | 0.12 | 0.02 | 0.57 | −0.05 | ns | |
| RFW (S1) | −0.19 | −0.18 | 0.00 | −0.07 | −0.01 | 0.06 | 0.02 | −0.13 | −0.09 | 0.27 | 0.45 | 0.20 | 0.42 | 0.26 | 0.69 | 0.11 |
| Traits | PC1 | PC2 | PC3 | PC4 | PC5 |
|---|---|---|---|---|---|
| G% (C) | 0.817 | −0.210 | 0.145 | 0.352 | −0.030 |
| G% (S1) | 0.825 | −0.225 | 0.038 | 0.319 | 0.086 |
| G% (S2) | 0.862 | 0.010 | −0.094 | 0.215 | 0.033 |
| GI (C) | 0.870 | 0.062 | 0.203 | 0.199 | −0.167 |
| GI (S1) | 0.930 | 0.114 | 0.057 | 0.002 | 0.133 |
| GI (S2) | 0.865 | 0.257 | −0.102 | −0.225 | 0.100 |
| MGT (C) | −0.642 | −0.278 | −0.162 | 0.207 | 0.267 |
| MGT (S1) | −0.659 | −0.306 | −0.056 | 0.366 | −0.189 |
| MGT (S2) | −0.471 | −0.360 | 0.134 | 0.677 | −0.040 |
| SL (C) | −0.224 | 0.654 | 0.447 | −0.129 | −0.102 |
| SL (S1) | −0.147 | 0.766 | −0.324 | 0.059 | −0.292 |
| RL (C) | −0.226 | 0.334 | −0.035 | −0.133 | 0.797 |
| RL (S1) | 0.003 | 0.527 | −0.262 | 0.521 | 0.387 |
| SFW (C) | −0.203 | 0.453 | 0.712 | −0.052 | −0.062 |
| SFW (S1) | −0.074 | 0.807 | −0.247 | 0.271 | −0.289 |
| RFW (C) | −0.156 | 0.131 | 0.836 | 0.242 | 0.168 |
| RFW (S1) | −0.107 | 0.724 | −0.131 | 0.255 | 0.060 |
| Eigenvalue | 5.73 | 3.28 | 1.79 | 1.51 | 1.17 |
| Variability (%) | 33.73 | 19.29 | 10.54 | 8.86 | 6.91 |
| Cumulative % | 33.73 | 53.03 | 63.57 | 72.43 | 79.33 |
| Gen. | G% (S1) | G% (S2) | GI (S1) | GI (S2) | MGT (S1) | MGT (S2) | SL (S1) | RL (S1) | SFW (S1) | RFW (S1) | Gen. | G% (S1) | G% (S2) | GI (S1) | GI (S2) | MGT (S1) | MGT (S2) | SL (S1) | RL (S1) | SFW (S1) | RFW (S1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 1.19 | 1.19 | 1.23 | 0.96 | 0.98 | 9.17 | 0.44 | 1.08 | 0.46 | 0.50 | G36 | 0.95 | 0.89 | 0.76 | 0.70 | 1.64 | 10.49 | 0.72 | 0.80 | 0.48 | 0.35 |
| G2 | 1.19 | 1.19 | 1.18 | 0.78 | 1.04 | 9.48 | 0.42 | 0.47 | 0.45 | 0.35 | G37 | 1.01 | 0.84 | 0.71 | 0.40 | 2.27 | 12.59 | 0.80 | 0.66 | 0.84 | 0.55 |
| G3 | 1.25 | 1.17 | 1.37 | 1.25 | 0.93 | 8.60 | 0.62 | 1.28 | 0.18 | 0.13 | G38 | 1.03 | 0.85 | 1.06 | 0.44 | 1.02 | 9.75 | 0.47 | 0.60 | 0.37 | 0.23 |
| G4 | 1.19 | 1.17 | 1.34 | 1.27 | 0.92 | 8.24 | 0.89 | 0.57 | 0.68 | 0.31 | G39 | 1.09 | 0.85 | 0.77 | 0.38 | 1.59 | 11.88 | 0.62 | 1.02 | 0.76 | 0.34 |
| G5 | 1.21 | 1.17 | 1.32 | 1.24 | 0.94 | 8.98 | 0.70 | 0.89 | 0.68 | 0.60 | G40 | 0.88 | 0.81 | 0.78 | 0.65 | 1.61 | 9.32 | 1.13 | 1.01 | 0.57 | 0.52 |
| G6 | 1.15 | 1.15 | 0.80 | 0.59 | 1.84 | 9.56 | 0.82 | 0.67 | 0.49 | 0.18 | G41 | 0.88 | 0.76 | 0.69 | 0.35 | 1.46 | 11.95 | 0.48 | 0.58 | 0.57 | 1.07 |
| G7 | 1.13 | 1.09 | 1.23 | 1.23 | 1.04 | 8.54 | 0.38 | 0.71 | 0.23 | 0.17 | G42 | 0.68 | 0.65 | 0.49 | 0.51 | 1.88 | 12.11 | 0.56 | 0.56 | 0.45 | 0.30 |
| G8 | 1.07 | 1.05 | 0.85 | 0.62 | 1.74 | 11.15 | 0.53 | 1.22 | 0.37 | 0.53 | G43 | 0.94 | 0.78 | 1.05 | 0.67 | 0.85 | 8.98 | 0.85 | 0.96 | 0.49 | 0.28 |
| G9 | 1.03 | 1.03 | 1.10 | 0.73 | 1.18 | 9.30 | 0.65 | 0.77 | 0.34 | 0.18 | G44 | 1.03 | 0.79 | 0.71 | 0.40 | 2.25 | 12.16 | 0.52 | 0.81 | 0.17 | 0.16 |
| G10 | 1.19 | 1.12 | 0.99 | 0.49 | 1.63 | 11.76 | 0.58 | 0.35 | 0.33 | 0.09 | G45 | 0.92 | 0.74 | 0.82 | 0.48 | 1.52 | 11.52 | 0.52 | 1.18 | 0.33 | 0.78 |
| G11 | 1.23 | 1.12 | 1.26 | 1.05 | 1.05 | 8.85 | 0.84 | 0.76 | 0.79 | 0.49 | G46 | 1.13 | 0.78 | 0.75 | 0.37 | 1.77 | 11.32 | 0.30 | 0.21 | 0.37 | 0.46 |
| G12 | 1.17 | 1.09 | 1.30 | 1.09 | 0.86 | 9.40 | 0.44 | 0.39 | 0.23 | 0.22 | G47 | 0.94 | 0.66 | 0.75 | 0.35 | 1.61 | 12.46 | 1.01 | 1.98 | 0.98 | 0.78 |
| G13 | 1.11 | 1.07 | 1.09 | 0.73 | 1.06 | 9.70 | 0.29 | 0.72 | 0.25 | 0.45 | G48 | 1.15 | 0.74 | 0.70 | 0.41 | 2.82 | 15.54 | 0.19 | 0.31 | 0.08 | 0.05 |
| G14 | 1.03 | 1.01 | 0.87 | 0.69 | 1.58 | 10.14 | 0.34 | 0.55 | 0.25 | 0.16 | G49 | 1.15 | 0.72 | 0.97 | 0.52 | 1.76 | 11.69 | 0.70 | 0.73 | 0.71 | 0.43 |
| G15 | 1.12 | 1.10 | 0.99 | 0.86 | 1.34 | 9.12 | 0.67 | 0.44 | 0.18 | 0.15 | G50 | 0.93 | 0.71 | 0.74 | 0.42 | 1.32 | 10.81 | 0.24 | 0.66 | 0.16 | 0.19 |
| G16 | 0.99 | 0.97 | 1.08 | 1.00 | 0.97 | 8.90 | 0.51 | 0.49 | 0.12 | 0.05 | G51 | 0.89 | 0.69 | 0.73 | 0.37 | 1.63 | 11.11 | 0.33 | 0.66 | 0.16 | 0.41 |
| G17 | 1.05 | 0.97 | 1.19 | 0.94 | 0.83 | 8.90 | 0.82 | 0.45 | 0.39 | 0.43 | G52 | 0.73 | 0.66 | 0.60 | 0.60 | 1.54 | 9.41 | 0.62 | 0.42 | 0.28 | 0.18 |
| G18 | 1.06 | 1.04 | 1.13 | 1.00 | 0.98 | 8.94 | 0.67 | 0.40 | 1.12 | 1.14 | G53 | 0.86 | 0.65 | 0.70 | 0.49 | 1.66 | 9.98 | 0.63 | 0.67 | 0.32 | 0.36 |
| G19 | 0.96 | 0.90 | 0.76 | 0.52 | 1.38 | 9.84 | 0.69 | 0.56 | 1.36 | 0.63 | G54 | 0.64 | 0.50 | 0.48 | 0.30 | 1.99 | 12.98 | 0.82 | 0.98 | 0.92 | 0.98 |
| G20 | 1.03 | 0.97 | 0.92 | 0.65 | 1.42 | 11.56 | 0.57 | 1.05 | 0.32 | 0.24 | G55 | 0.46 | 0.46 | 0.46 | 0.33 | 1.23 | 9.19 | 0.92 | 0.62 | 0.99 | 0.54 |
| G21 | 1.01 | 0.95 | 0.95 | 0.56 | 1.22 | 10.70 | 0.41 | 0.63 | 0.19 | 0.29 | G56 | 0.84 | 0.55 | 0.19 | 0.08 | 3.52 | 20.12 | 0.36 | 0.66 | 0.10 | 0.15 |
| G22 | 0.99 | 0.95 | 0.81 | 0.43 | 1.61 | 11.47 | 0.52 | 0.25 | 0.78 | 0.24 | G57 | 0.78 | 0.51 | 0.59 | 0.36 | 1.91 | 11.44 | 0.25 | 0.26 | 0.22 | 0.18 |
| G23 | 1.03 | 0.95 | 0.99 | 0.71 | 1.15 | 10.68 | 0.62 | 1.33 | 0.64 | 0.41 | G58 | 0.57 | 0.40 | 0.44 | 0.33 | 1.71 | 11.84 | 0.95 | 0.77 | 0.70 | 0.31 |
| G24 | 0.97 | 0.95 | 1.04 | 1.04 | 0.99 | 9.42 | 0.87 | 1.05 | 0.70 | 0.27 | G59 | 0.77 | 0.51 | 0.69 | 0.31 | 1.45 | 11.64 | 0.58 | 0.56 | 0.30 | 0.44 |
| G25 | 1.07 | 0.94 | 0.71 | 0.59 | 1.84 | 10.32 | 0.83 | 0.51 | 0.65 | 0.06 | G60 | 0.53 | 0.40 | 0.40 | 0.20 | 2.10 | 13.13 | 1.15 | 0.50 | 0.85 | 0.64 |
| G26 | 1.05 | 0.95 | 1.16 | 0.84 | 0.93 | 9.77 | 0.89 | 0.82 | 0.71 | 0.39 | G61 | 0.58 | 0.34 | 0.17 | 0.11 | 2.21 | 14.02 | 0.95 | 0.47 | 0.88 | 0.44 |
| G27 | 0.93 | 0.93 | 0.84 | 0.51 | 1.35 | 10.76 | 0.42 | 0.70 | 0.49 | 1.04 | G62 | 0.31 | 0.31 | 0.07 | 0.07 | 3.08 | 12.71 | 0.79 | 1.00 | 1.01 | 0.28 |
| G28 | 0.89 | 0.86 | 0.19 | 0.17 | 2.88 | 15.17 | 0.26 | 0.91 | 0.06 | 0.17 | G63 | 0.75 | 0.38 | 0.46 | 0.23 | 2.17 | 12.08 | 0.81 | 0.51 | 0.29 | 0.26 |
| G29 | 0.98 | 0.98 | 0.96 | 0.80 | 1.49 | 8.50 | 0.72 | 0.72 | 0.56 | 0.35 | G64 | 0.59 | 0.33 | 0.33 | 0.14 | 2.57 | 14.95 | 0.27 | 0.55 | 0.03 | 0.16 |
| G30 | 0.86 | 0.85 | 0.91 | 0.80 | 1.05 | 9.41 | 1.02 | 1.07 | 1.33 | 1.38 | G65 | 0.44 | 0.36 | 0.21 | 0.20 | 2.59 | 15.90 | 1.03 | 1.04 | 0.85 | 1.07 |
| G31 | 0.96 | 0.92 | 0.83 | 0.81 | 1.35 | 11.49 | 0.42 | 0.52 | 0.40 | 0.26 | G66 | 0.53 | 0.40 | 0.41 | 0.34 | 1.79 | 9.13 | 0.94 | 0.74 | 0.54 | 0.50 |
| G32 | 0.92 | 0.92 | 0.49 | 0.42 | 2.86 | 13.04 | 0.81 | 0.66 | 0.53 | 0.50 | G67 | 1.07 | 0.38 | 0.78 | 0.18 | 1.99 | 13.97 | 0.54 | 0.62 | 0.21 | 0.33 |
| G33 | 1.11 | 0.89 | 0.55 | 0.30 | 3.52 | 17.15 | 0.41 | 0.96 | 0.39 | 0.89 | G68 | 0.27 | 0.20 | 0.17 | 0.07 | 2.10 | 16.36 | 0.97 | 1.30 | 0.87 | 0.28 |
| G34 | 1.01 | 0.89 | 0.88 | 0.44 | 1.26 | 11.47 | 0.23 | 0.42 | 0.13 | 0.21 | G69 | 0.65 | 0.18 | 0.63 | 0.06 | 1.32 | 10.13 | 0.77 | 0.99 | 0.69 | 0.35 |
| G35 | 1.01 | 0.87 | 1.17 | 0.79 | 0.78 | 8.69 | 0.54 | 0.69 | 0.39 | 0.18 | G70 | 0.34 | 0.14 | 0.07 | 0.03 | 1.47 | 8.38 | 0.59 | 0.61 | 0.17 | 0.22 |
| Traits | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
|---|---|---|---|---|
| Genotypes No. | 27 | 28 | 7 | 8 |
| G% (S1) | 0.99 | 0.94 | 0.75 | 0.75 |
| G% (S2) | 0.93 | 0.78 | 0.56 | 0.54 |
| GI (S1) | 1.01 | 0.76 | 0.34 | 0.48 |
| GI (S2) | 0.82 | 0.48 | 0.20 | 0.25 |
| MGT (S1) | 1.14 | 1.59 | 2.86 | 2.26 |
| MGT (S2) | 9.10 | 11.20 | 16.45 | 13.11 |
| SL (S1) | 0.69 | 0.54 | 0.50 | 0.86 |
| RL (S1) | 0.72 | 0.68 | 0.82 | 0.86 |
| SFW (S1) | 0.51 | 0.45 | 0.34 | 0.78 |
| RFW (S1) | 0.39 | 0.38 | 0.40 | 0.56 |
| Gen. | GS | SS | Sum | Rank | FTD | Gen. | GS | SS | Sum | Rank | FTD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| G18 | 1 | 1 | 2 | 1 | T | G15 | 1 | 4 | 5 | 3 | MS |
| G19 | 1 | 1 | 2 | 1 | T | G16 | 1 | 4 | 5 | 3 | MS |
| G30 | 1 | 1 | 2 | 1 | T | G17 | 1 | 4 | 5 | 3 | MS |
| G1 | 1 | 2 | 3 | 1 | T | G22 | 2 | 3 | 5 | 3 | MS |
| G3 | 1 | 2 | 3 | 1 | T | G35 | 1 | 4 | 5 | 3 | MS |
| G24 | 1 | 2 | 3 | 1 | T | G37 | 2 | 3 | 5 | 3 | MS |
| G27 | 2 | 1 | 3 | 1 | T | G38 | 1 | 4 | 5 | 3 | MS |
| G41 | 2 | 1 | 3 | 1 | T | G49 | 2 | 3 | 5 | 3 | MS |
| G45 | 2 | 1 | 3 | 1 | T | G52 | 1 | 4 | 5 | 3 | MS |
| G69 | 1 | 2 | 3 | 1 | T | G53 | 1 | 4 | 5 | 3 | MS |
| G4 | 1 | 3 | 4 | 2 | MT | G54 | 4 | 1 | 5 | 3 | MS |
| G5 | 1 | 3 | 4 | 2 | MT | G58 | 2 | 3 | 5 | 3 | MS |
| G6 | 1 | 3 | 4 | 2 | MT | G68 | 3 | 2 | 5 | 3 | MS |
| G8 | 2 | 2 | 4 | 2 | MT | G70 | 1 | 4 | 5 | 3 | MS |
| G11 | 1 | 3 | 4 | 2 | MT | G10 | 2 | 4 | 6 | 4 | S |
| G20 | 2 | 2 | 4 | 2 | MT | G21 | 2 | 4 | 6 | 4 | S |
| G23 | 2 | 2 | 4 | 2 | MT | G31 | 2 | 4 | 6 | 4 | S |
| G25 | 1 | 3 | 4 | 2 | MT | G34 | 2 | 4 | 6 | 4 | S |
| G26 | 1 | 3 | 4 | 2 | MT | G42 | 2 | 4 | 6 | 4 | S |
| G29 | 1 | 3 | 4 | 2 | MT | G44 | 2 | 4 | 6 | 4 | S |
| G33 | 3 | 1 | 4 | 2 | MT | G46 | 2 | 4 | 6 | 4 | S |
| G36 | 1 | 3 | 4 | 2 | MT | G50 | 2 | 4 | 6 | 4 | S |
| G39 | 2 | 2 | 4 | 2 | MT | G51 | 2 | 4 | 6 | 4 | S |
| G40 | 1 | 3 | 4 | 2 | MT | G57 | 2 | 4 | 6 | 4 | S |
| G43 | 1 | 3 | 4 | 2 | MT | G59 | 2 | 4 | 6 | 4 | S |
| G47 | 2 | 2 | 4 | 2 | MT | G62 | 4 | 2 | 6 | 4 | S |
| G55 | 1 | 3 | 4 | 2 | MT | G63 | 2 | 4 | 6 | 4 | S |
| G65 | 3 | 1 | 4 | 2 | MT | G28 | 3 | 4 | 7 | 4 | S |
| G66 | 1 | 3 | 4 | 2 | MT | G32 | 4 | 3 | 7 | 4 | S |
| G2 | 1 | 4 | 5 | 3 | MS | G48 | 3 | 4 | 7 | 4 | S |
| G7 | 1 | 4 | 5 | 3 | MS | G56 | 3 | 4 | 7 | 4 | S |
| G9 | 1 | 4 | 5 | 3 | MS | G60 | 4 | 3 | 7 | 4 | S |
| G12 | 1 | 4 | 5 | 3 | MS | G61 | 4 | 3 | 7 | 4 | S |
| G13 | 1 | 4 | 5 | 3 | MS | G64 | 3 | 4 | 7 | 4 | S |
| G14 | 1 | 4 | 5 | 3 | MS | G67 | 4 | 4 | 8 | 4 | S |
| Traits | T | MT | MS | S |
|---|---|---|---|---|
| G% (C) | 90.17 | 91.67 | 87.58 | 86.23 |
| G% (S1) | 83.83 | 81.75 | 80.67 | 76.78 |
| G% (S2) | 74.67 | 73.60 | 70.83 | 58.33 |
| GI (C) | 17.24 | 17.11 | 16.22 | 14.17 |
| GI (S1) | 13.78 | 12.34 | 12.84 | 9.78 |
| GI (S2) | 10.01 | 9.35 | 9.22 | 5.75 |
| MGT (C) | 1.11 | 1.17 | 1.15 | 1.37 |
| MGT (S1) | 1.59 | 1.94 | 1.76 | 2.22 |
| MGT (S2) | 13.23 | 13.29 | 13.20 | 13.90 |
| SL (C) | 35.03 | 34.91 | 33.77 | 31.10 |
| SL (S1) | 20.60 | 25.20 | 19.84 | 17.52 |
| RL (C) | 13.19 | 12.94 | 11.90 | 11.52 |
| RL (S1) | 9.96 | 10.58 | 7.97 | 7.26 |
| SFW (C) | 1.35 | 1.10 | 1.05 | 1.09 |
| SFW (S1) | 0.65 | 0.72 | 0.51 | 0.36 |
| RFW (C) | 0.86 | 0.67 | 0.61 | 0.68 |
| RFW (S1) | 0.38 | 0.33 | 0.24 | 0.20 |
| Primer Comb. | AN | PBN | P% | TP | DP% | PIC |
|---|---|---|---|---|---|---|
| 1 | 8 | 8 | 100 | 352 | 11.94 | 0.82 |
| 2 | 7 | 7 | 100 | 324 | 10.45 | 0.80 |
| 3 | 7 | 3 | 43 | 422 | 4.48 | 0.80 |
| 4 | 11 | 8 | 73 | 582 | 11.94 | 0.90 |
| 5 | 13 | 11 | 85 | 336 | 16.42 | 0.86 |
| 6 | 6 | 6 | 100 | 279 | 8.96 | 0.81 |
| 7 | 7 | 7 | 100 | 350 | 10.45 | 0.84 |
| 8 | 6 | 11 | 183 | 263 | 16.42 | 0.80 |
| 9 | 6 | 6 | 100 | 303 | 8.96 | 0.40 |
| Total | 71 | 67 | 884 | 3211 | 100 | 7.11 |
| Avg/Primer | 7.89 | 7.44 | 98 | 356.78 | 11.11 | 0.79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, M.M.; Al-Doss, A.A.; Tahir, M.U.; Khan, M.A.; El-Hendawy, S. Assessing the Suitability of Selection Approaches and Genetic Diversity Analysis for Early Detection of Salt Tolerance of Barley Genotypes. Agronomy 2022, 12, 3217. https://doi.org/10.3390/agronomy12123217
Javed MM, Al-Doss AA, Tahir MU, Khan MA, El-Hendawy S. Assessing the Suitability of Selection Approaches and Genetic Diversity Analysis for Early Detection of Salt Tolerance of Barley Genotypes. Agronomy. 2022; 12(12):3217. https://doi.org/10.3390/agronomy12123217
Chicago/Turabian StyleJaved, Muhammad Matloob, Abdullah A. Al-Doss, Muhammad Usman Tahir, Muhammad Altaf Khan, and Salah El-Hendawy. 2022. "Assessing the Suitability of Selection Approaches and Genetic Diversity Analysis for Early Detection of Salt Tolerance of Barley Genotypes" Agronomy 12, no. 12: 3217. https://doi.org/10.3390/agronomy12123217
APA StyleJaved, M. M., Al-Doss, A. A., Tahir, M. U., Khan, M. A., & El-Hendawy, S. (2022). Assessing the Suitability of Selection Approaches and Genetic Diversity Analysis for Early Detection of Salt Tolerance of Barley Genotypes. Agronomy, 12(12), 3217. https://doi.org/10.3390/agronomy12123217







