CRISPR/Cas9-Mediated Mutagenesis of GmFAD2-1A and/or GmFAD2-1B to Create High-Oleic-Acid Soybean
Abstract
1. Introduction
2. Materials and Methods
2.1. sgRNA Design and Plasmid Construction
2.2. Soybean Transformation
2.3. DNA Extraction and Mutant Identification
2.4. Fatty Acid Analysis
2.5. Phenotype Identification
3. Results
3.1. Generation of fad2-1a, fad2-1b, and fad2-1a/fad2-1b Mutants with the CRISPR/Cas9 System
3.2. Fatty Acid Profiles of Mutant Seeds
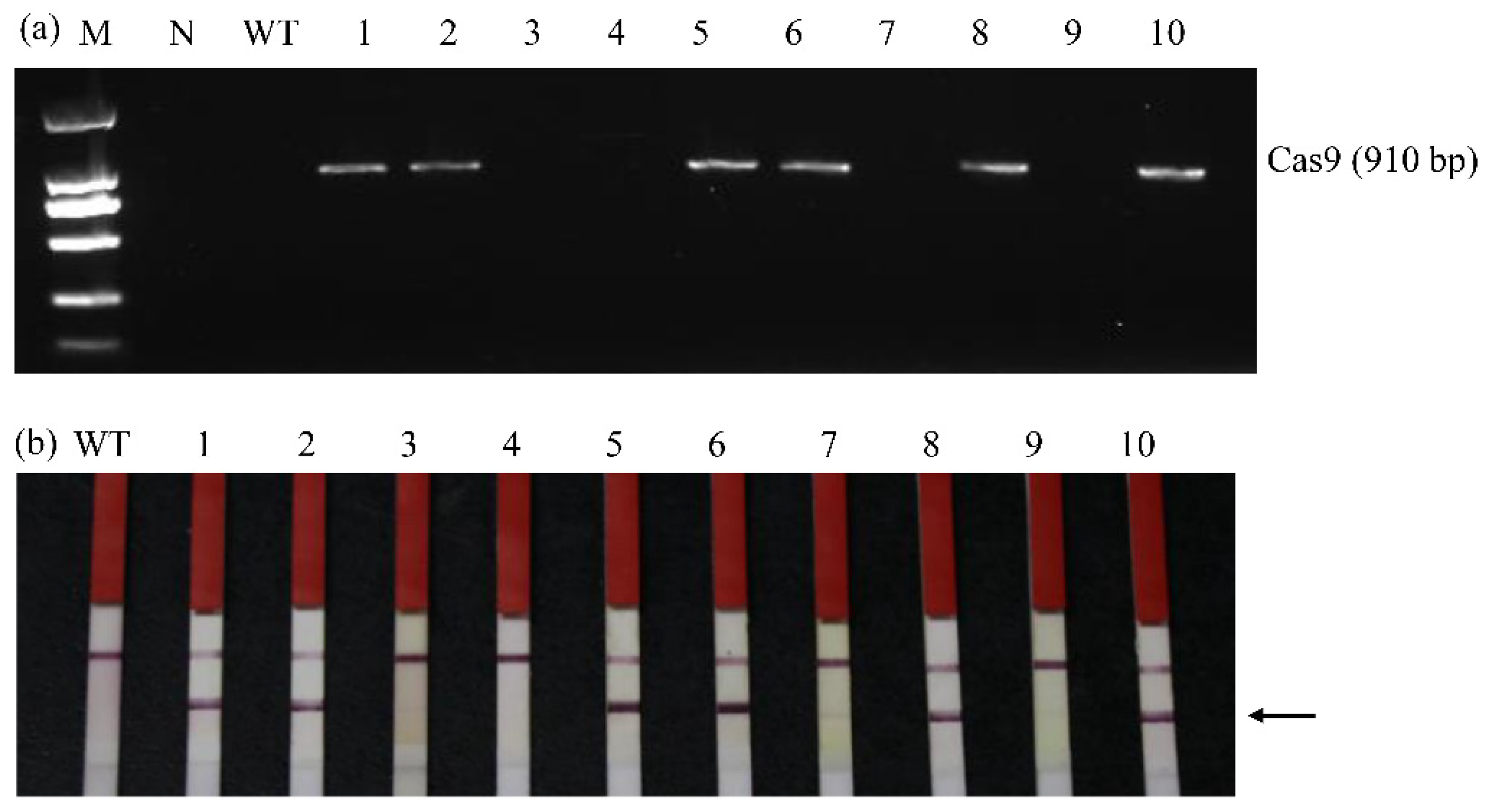
3.3. Identification of Transgene-Free fad2-1a/fad2-1b Mutant Plants
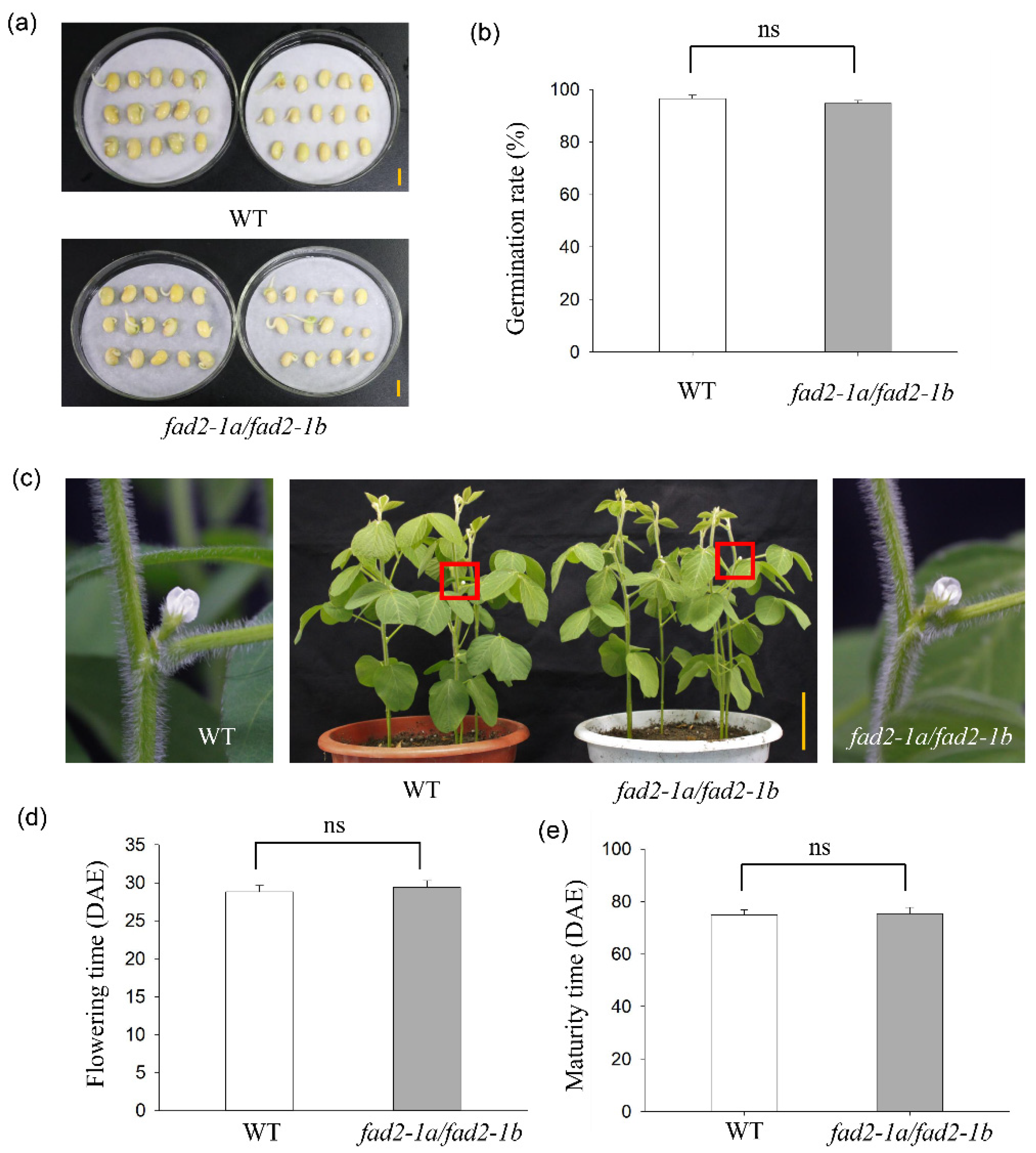
3.4. No Adverse Phenotyping of fad2-1a/fad2-1b Mutant Plants
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clemente, T.E.; Cahoon, E.B. Soybean Oil: Genetic Approaches for Modification of Functionality and Total Content. Plant Physiol. 2009, 151, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Koehler, K.M. Probabilistic Quantitative Assessment of Coronary Heart Disease Risk from Dietary Exposure to Industrially-Produced Trans Fatty Acids in Partially Hydrogenated Oils. Toxicol. Sci. Off. J. Soc. Toxicol. 2019, 172, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.; Vianna, G.R.; Machado, A.M.; da Cunha, N.B.; Coelho, C.M.; Lacerda, V.A.; Coelho, M.C.; Rech, E.L. Mass Spectrometry Characterisation of Fatty Acids from Metabolically Engineered Soybean Seeds. Anal. Bioanal. Chem. 2014, 406, 2873–2883. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.R.; Haas, M.J.; Winkler, J.K.; Jackson, M.A.; Erhan, S.Z.; List, G.R. Evaluation of Partially Hydrogenated Methyl Esters of Soybean Oil as Biodiesel. Eur. J. Lipid Sci. 2007, 109, 17–24. [Google Scholar] [CrossRef]
- Wilson, R.F. The Role of Genomics and Biotechnology in Achieving Global Food Security for High-Oleic Vegetable Oil. J. Oleo Sci. 2012, 61, 357–367. [Google Scholar] [CrossRef]
- Ohlrogge, J.B. Design of New Plant Products: Engineering of Fatty Acid Metabolism. Plant Physiol. 1994, 104, 821–826. [Google Scholar] [CrossRef]
- Schlueter, J.A.; Vasylenko-Sanders, I.F.; Deshpande, S.; Yi, J.; Siegfried, M.; Roe, B.A.; Schlueter, S.D.; Scheffler, B.E.; Shoemaker, R.C. Fad2 Gene Family of Soybean: Insights into the Structural and Functional Divergence of a Paleopolyploid Genome. Crop Sci. 2007, 47, 14. [Google Scholar] [CrossRef]
- Lee, J.M.; Lee, H.; Kang, S.; Park, W.J. Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances. Nutrients 2016, 8, 23. [Google Scholar] [CrossRef]
- Heppard, E.P.; Kinney, A.J.; Stecca, K.L.; Miao, G.H. Developmental and Growth Temperature Regulation of Two Different Microsomal Omega-6 Desaturase Genes in Soybeans. Plant Physiol. 1996, 110, 311–319. [Google Scholar] [CrossRef]
- Bachlava, E.; Dewey, R.E.; Burton, J.W.; Cardinal, A.J. Mapping Candidate Genes for Oleate Biosynthesis and Their Association with Unsaturated Fatty Acid Seed Content in Soybean. Mol. Breed. 2009, 23, 337–347. [Google Scholar] [CrossRef]
- Pham, A.T.; Lee, J.D.; Shannon, J.G.; Bilyeu, K.D. Mutant Alleles of Fad2-1a and Fad2-1b Combine to Produce Soybeans with the High Oleic Acid Seed Oil Trait. BMC Plant Biol. 2010, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis Fad2 Gene Encodes the Enzyme That Is Essential for Polyunsaturated Lipid Synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [PubMed]
- Kinney, A.J.; Cahoon, E.B.; Hitz, W.D. Manipulating Desaturase Activities in Transgenic Crop Plants. Biochem. Soc. Trans. 2002, 30, 1099. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.T.; Lee, J.D.; Shannon, J.G.; Bilyeu, K.D. A Novel Fad2-1 a Allele in a Soybean Plant Introduction Offers an Alternate Means to Produce Soybean Seed Oil with 85% Oleic Acid Content. Theor. Appl. Genet. 2011, 123, 793–802. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, X.D.; Zhang, Y.Y.; Yang, J.; Qi, G.X.; Guo, D.Q.; Xing, G.J.; Yao, Y.; Xu, W.J.; Li, H.Y.; et al. Changes in Oleic Acid Content of Transgenic Soybeans by Antisense Rna Mediated Posttranscriptional Gene Silencing. Int. J. Genom. 2014, 921950. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.; Mroczka, A.; Roberts, P.D.; Schreckengost, W.; Voelker, T. Rnai Trigger Fragment Truncation Attenuates Soybean Fad2-1 Transcript Suppression and Yields Intermediate Oil Phenotypes. Plant Biotechnol. J. 2011, 9, 723–728. [Google Scholar] [CrossRef]
- Yang, J.; Xing, G.; Lu, N.; He, H.; Guo, D.; Du, Q.; Qian, X.; Yao, Y.; Li, H.; Zhong, X.; et al. Improved Oil Quality in Transgenic Soybean Seeds by Rnai-Mediated Knockdown of Gmfad2-1b. Transgenic Res. 2018, 27, 155–166. [Google Scholar] [CrossRef]
- Haun, W.; Coffman, A.; Clasen, B.M.; Demorest, Z.L.; Lowy, A.; Ray, E.; Retterath, A.; Stoddard, T.; Juillerat, A.; Cedrone, F.; et al. Improved Soybean Oil Quality by Targeted Mutagenesis of the Fatty Acid Desaturase 2 Gene Family. Plant Biotechnol. J. 2014, 12, 934–940. [Google Scholar] [CrossRef]
- Demorest, Z.L.; Coffman, A.; Baltes, N.J.; Stoddard, T.J.; Clasen, B.M.; Luo, S.; Retterath, A.; Yabandith, A.; Gamo, M.E.; Bissen, J.; et al. Direct Stacking of Sequence-Specific Nuclease-Induced Mutations to Produce High Oleic and Low Linolenic Soybean Oil. BMC Plant Biol. 2016, 16, 225. [Google Scholar] [CrossRef]
- Do, P.T.; Nguyen, C.X.; Bui, H.T.; Tran, L.T.N.; Stacey, G.; Gillman, J.D.; Zhang, Z.J.; Stacey, M.G. Demonstration of Highly Efficient Dual Grna Crispr/Cas9 Editing of the Homeologous Gmfad2-1a and Gmfad2-1b Genes to Yield a High Oleic, Low Linoleic and Alpha-Linolenic Acid Phenotype in Soybean. BMC Plant Biol. 2019, 19, 311. [Google Scholar] [CrossRef]
- Cai, Y.P.; Chen, L.; Liu, X.J.; Sun, S.; Wu, C.X.; Jiang, B.J.; Han, T.F.; Hou, W.S. Crispr/Cas9-Mediated Genome Editing in Soybean Hairy Roots. PLoS ONE 2015, 10, e0136064. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. Crispr-P: A Web Tool for Synthetic Single-Guide Rna Design of Crispr-System in Plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cai, Y.; Liu, X.; Yao, W.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. Improvement of Soybean Agrobacterium-Mediated Transformation Efficiency by Adding Glutamine and Asparagine into the Culture Media. Int. J. Mol. Sci. 2018, 19, 3039. [Google Scholar] [CrossRef] [PubMed]
- Abdelghany, A.M.; Zhang, S.R.; Azam, M.; Shaibu, A.; Sun, J.M. Profiling of Seed Fatty Acid Composition in 1025 Chinese Soybean Accessions from Diverse Ecoregions. Crop J. 2019, 8, 635–644. [Google Scholar] [CrossRef]
- Cai, Y.P.; Wang, L.; Chen, L.; Wu, T.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Yuan, S.; Han, T.; et al. Mutagenesis of Gmft2a and Gmft5a Mediated by Crispr/Cas9 Contributes for Expanding the Regional Adaptability of Soybean. Plant Biotechnol. J. 2020, 18, 298–309. [Google Scholar] [CrossRef]
- Yang, C.H.; Zhang, Y.; Huang, C.F. Reduction in Cadmium Accumulation in Japonica Rice Grains by Crispr/Cas9-Mediated Editing of Osnramp5. J. Integr. Agric. 2019, 18, 210–219. [Google Scholar] [CrossRef]
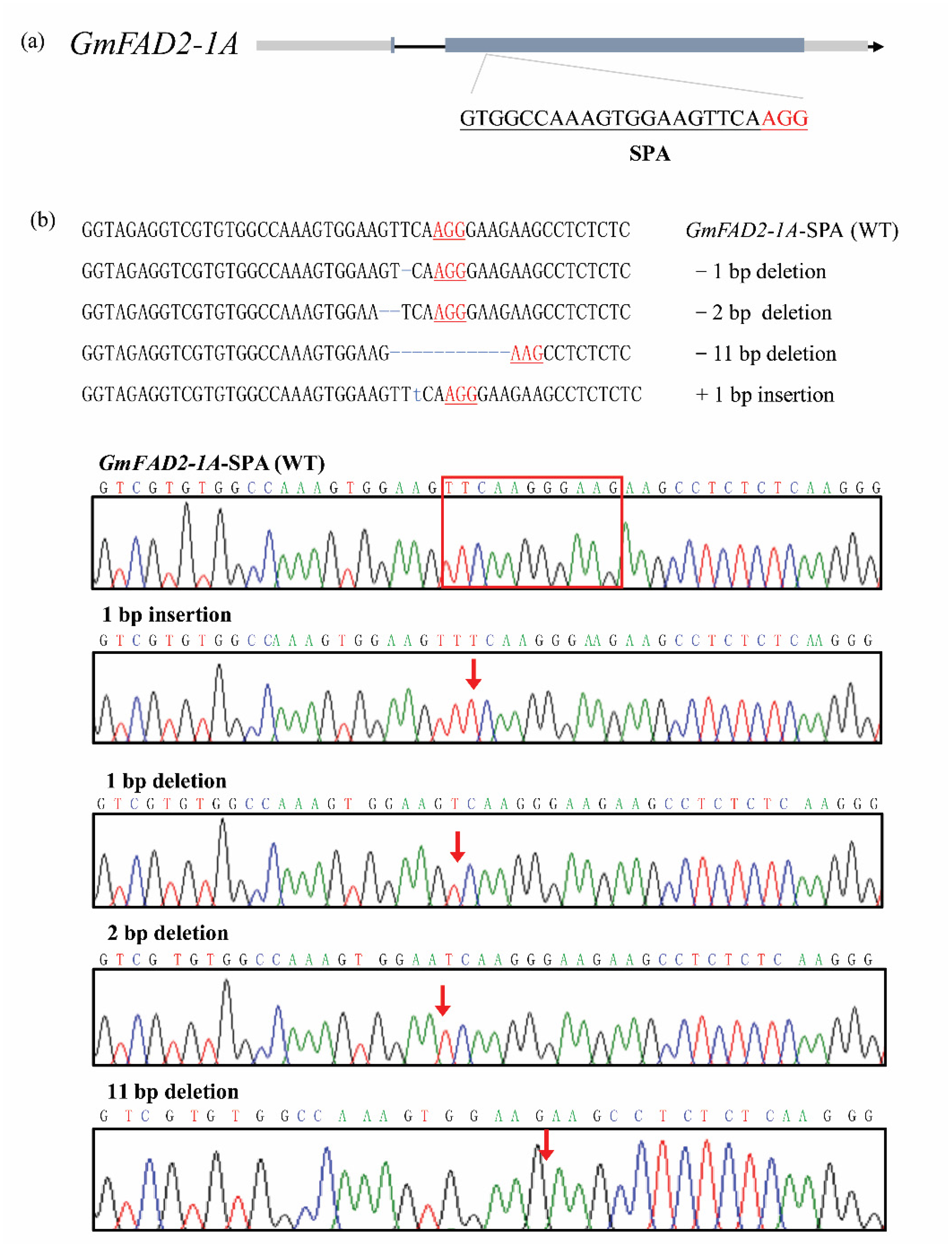

| Mutant Lines | Editing Types |
|---|---|
| SEA-73 | fad2-1a (−2 bp) |
| SEA-81 | fad2-1a (+1 bp) |
| SEA-122 | fad2-1a (−1 bp) |
| PB-41 | fad2-1b (−7 bp) |
| PB-86 | fad2-1b (+1 bp) |
| PB-116 | fad2-1b (−17 bp) |
| PB-154 | fad2-1b (−4 bp) |
| JM-72 | fad2-1a (−5 bp)/fad2-1b (−4 bp) |
| JM-90 | fad2-1a (−5 bp)/fad2-1b (−8 bp) |
| JM-264 | fad2-1a (−5 bp)/fad2-1b (+1 bp) |
| Mutant | Palmitic (%) | Stearic (%) | Oleic (%) | Linoleic (%) | Linolenic (%) |
|---|---|---|---|---|---|
| WT | 11.14 ± 0.22 | 4.29 ± 0.32 | 21.69 ± 0.84 | 57.22 ± 1.14 | 5.66 ± 0.37 |
| SEA-73 | 10.32 ± 0.03 * | 3.1 ± 0.02 ** | 35.64 ± 0.15 ** | 41.97 ± 0.08 ** | 8.97 ± 0.06 ** |
| SEA-81 | 9.68 ± 0.06 ** | 3.48 ± 0.07 ** | 50.11 ± 0.28 ** | 31.12 ± 0.22 ** | 5.61 ± 0.08 |
| SEA-122 | 11.12 ± 0.15 | 4.25 ± 0.07 | 38.28 ± 0.43 ** | 40.9 ± 0.23 ** | 5.44 ± 0.07 |
| PB-41 | 8.63 ± 0.03 ** | 4.52 ± 0.02 | 54.08 ± 0.33 ** | 27.52 ± 0.27 ** | 5.25 ± 0.02 |
| PB-86 | 8.51 ± 0.06 ** | 4.60 ± 0.03 | 53.30 ± 0.46 ** | 28.27 ± 0.38 ** | 5.32 ± 0.02 |
| PB-116 | 10.44 ± 0.07 * | 3.79 ± 0.06 * | 39.95 ± 0.13 ** | 38.98 ± 0.18 ** | 6.84 ± 0.09 * |
| PB-154 | 8.88 ± 0.05 ** | 4.39 ± 0.11 | 50.67 ± 0.41 ** | 30.92 ± 0.49 ** | 5.14 ± 0.05 |
| JM-72 | 6.11 ± 0.02 ** | 4.23 ± 0.01 | 84.55 ± 0.05 ** | 2.35 ± 0.01 ** | 2.76 ± 0.05 ** |
| JM-90 | 6.13 ± 0.03 ** | 3.50 ± 0.04 ** | 85.42 ± 0.06 ** | 2.36 ± 0.06 ** | 2.59 ± 0.03 ** |
| JM-264 | 6.70 ± 0.01 ** | 2.78 ± 0.03 ** | 84.69 ± 0.12 ** | 2.78 ± 0.05 ** | 3.04 ± 0.09 ** |
| fad2-1a/fad2-1b Mutant Lines | T-DNA in the T3 Mutants | No. of Progeny Plants Identified | No. of T4 “Transgene-Free” Mutants |
|---|---|---|---|
| fad2-1-SPD-JM-72 | T-DNA-free | 7 | 7 |
| fad2-1-SPD-JM-264 | T-DNA-free | 8 | 8 |
| fad2-1-SPD-JM-90 | T-DNA-positive | 10 | 3 |
| fad2-1-SPD-JM-95 | T-DNA-positive | 9 | 0 |
| fad2-1-SPD-JM-113 | T-DNA-positive | 8 | 1 |
| fad2-1-SPD-JM-196 | T-DNA-positive | 12 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, M.; Chen, L.; Cai, Y.; Su, Q.; Chen, Y.; Hou, W. CRISPR/Cas9-Mediated Mutagenesis of GmFAD2-1A and/or GmFAD2-1B to Create High-Oleic-Acid Soybean. Agronomy 2022, 12, 3218. https://doi.org/10.3390/agronomy12123218
Fu M, Chen L, Cai Y, Su Q, Chen Y, Hou W. CRISPR/Cas9-Mediated Mutagenesis of GmFAD2-1A and/or GmFAD2-1B to Create High-Oleic-Acid Soybean. Agronomy. 2022; 12(12):3218. https://doi.org/10.3390/agronomy12123218
Chicago/Turabian StyleFu, Mingxue, Li Chen, Yupeng Cai, Qiang Su, Yingying Chen, and Wensheng Hou. 2022. "CRISPR/Cas9-Mediated Mutagenesis of GmFAD2-1A and/or GmFAD2-1B to Create High-Oleic-Acid Soybean" Agronomy 12, no. 12: 3218. https://doi.org/10.3390/agronomy12123218
APA StyleFu, M., Chen, L., Cai, Y., Su, Q., Chen, Y., & Hou, W. (2022). CRISPR/Cas9-Mediated Mutagenesis of GmFAD2-1A and/or GmFAD2-1B to Create High-Oleic-Acid Soybean. Agronomy, 12(12), 3218. https://doi.org/10.3390/agronomy12123218








