Utilizing Genomic Selection for Wheat Population Development and Improvement
Abstract
:1. Wheat Breeding
2. Genomic Selection
3. Product Development
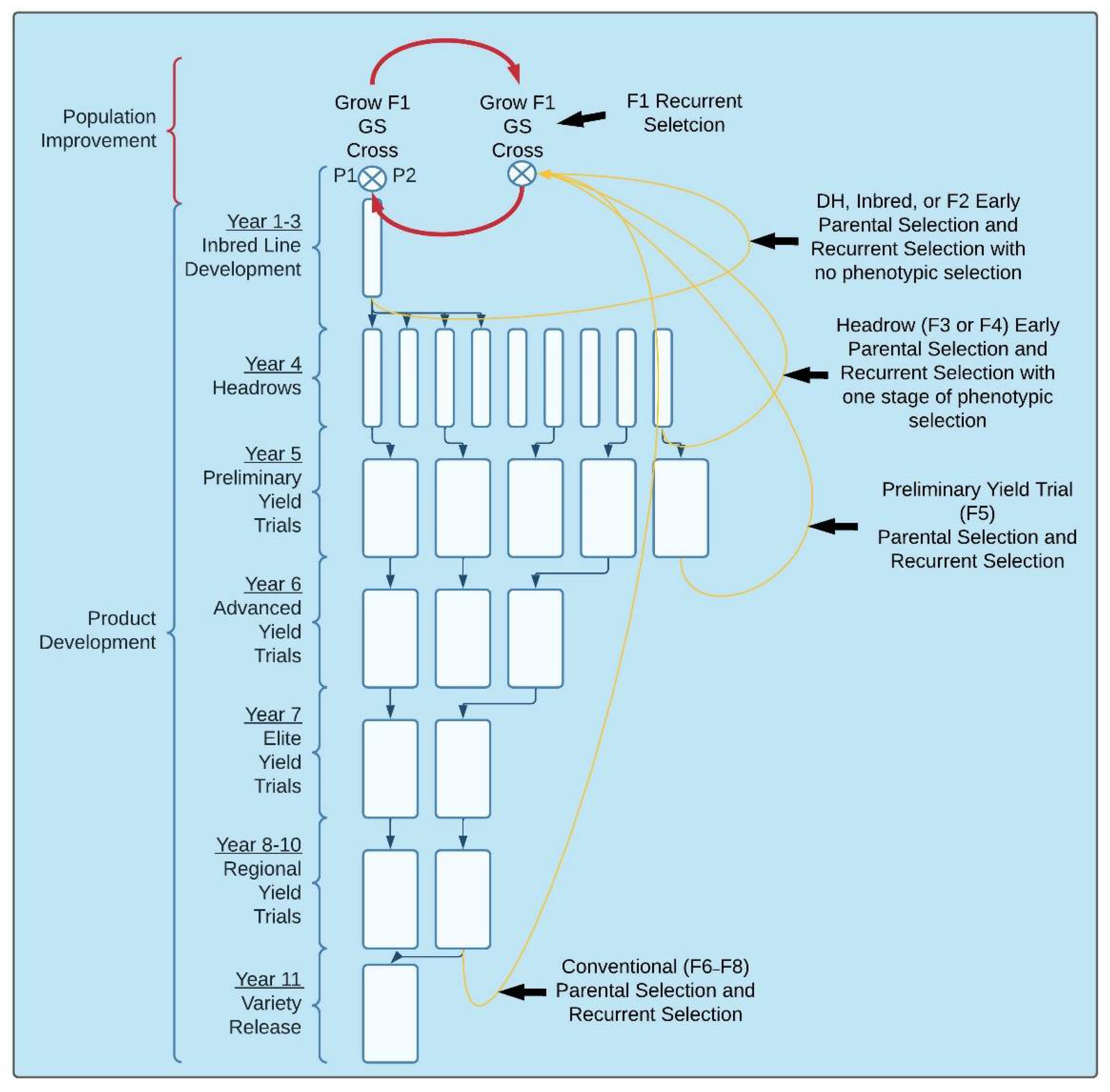
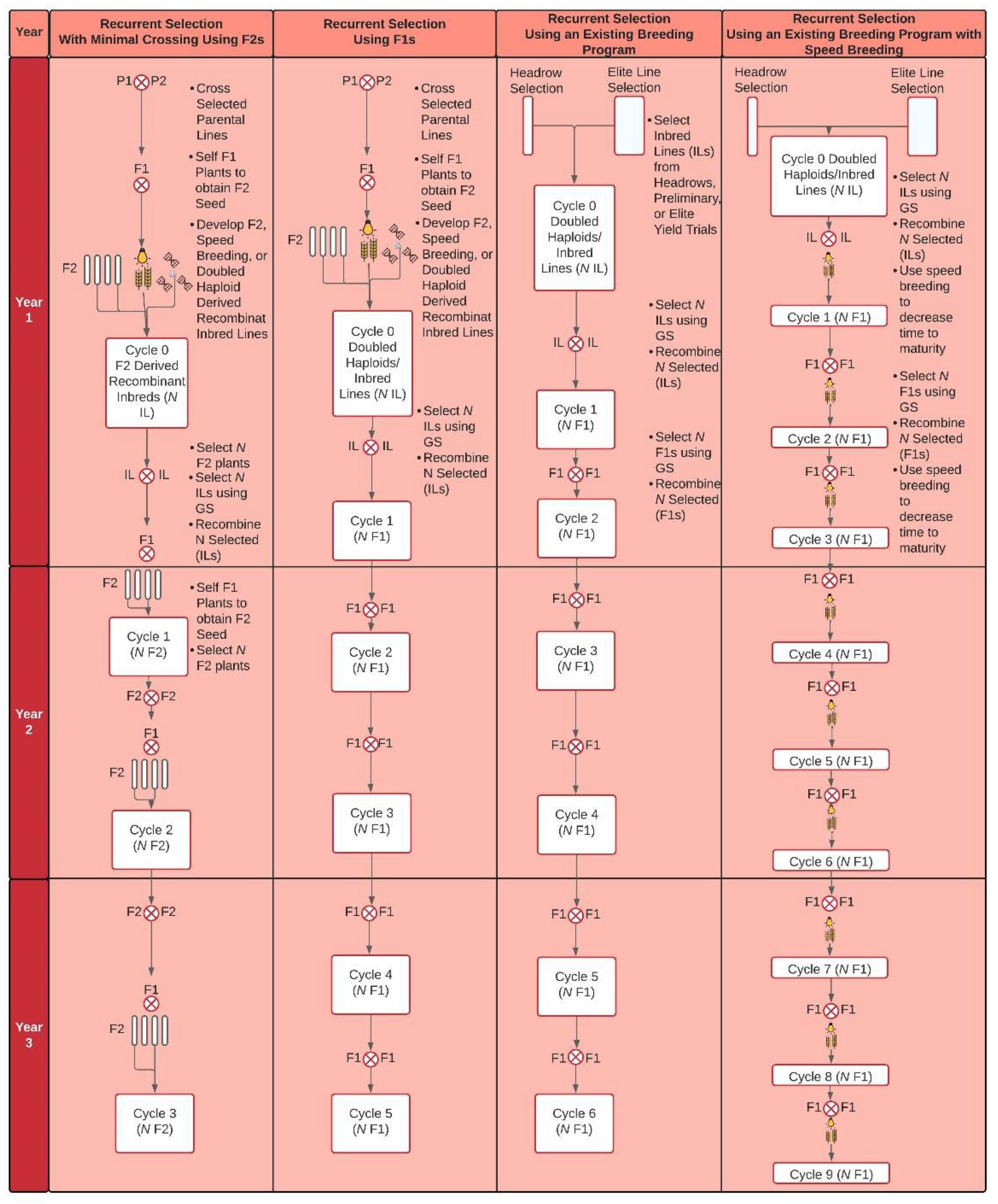
3.1. Implementation of GS for Recurrent and Parental Selection
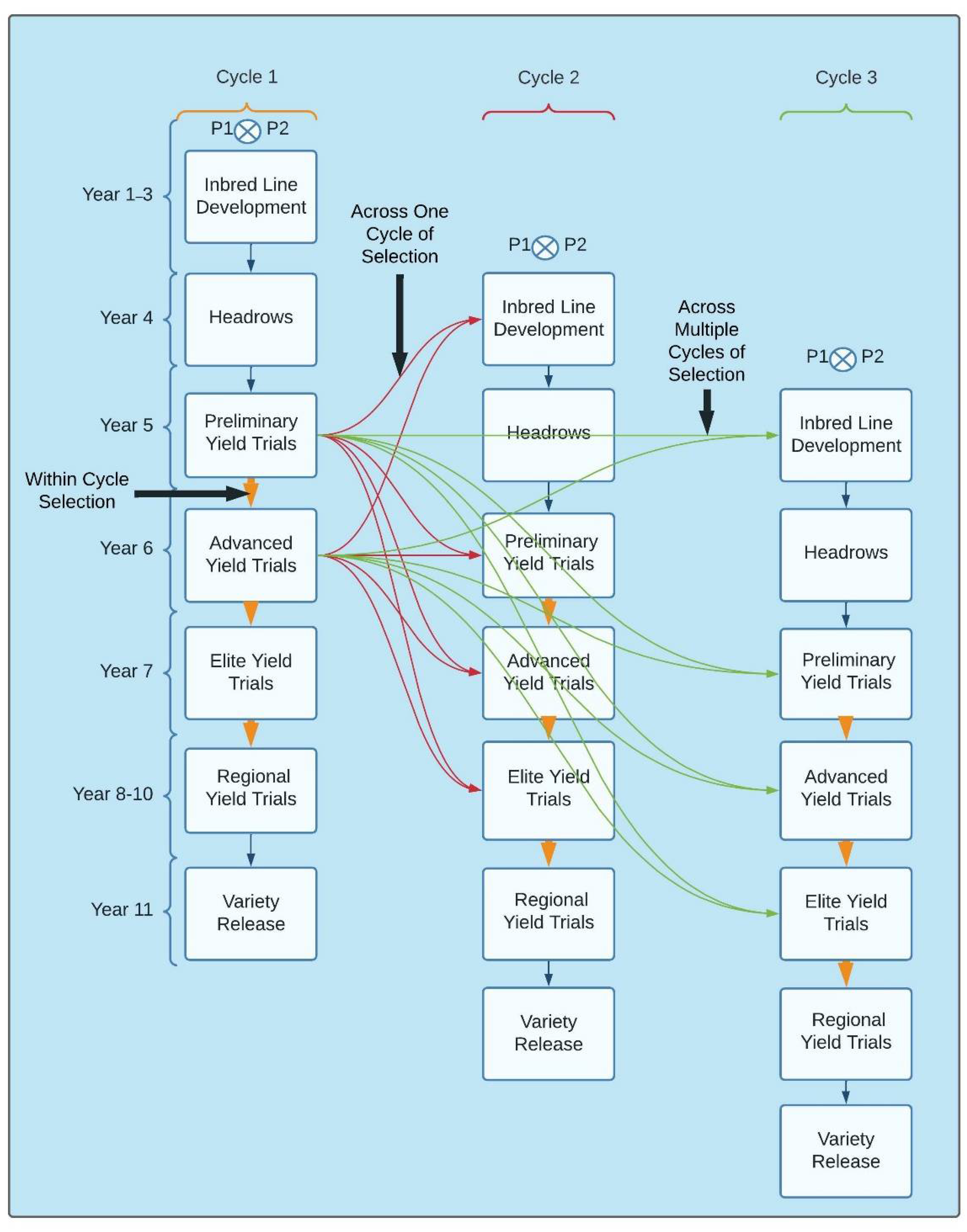
3.2. Implementation of GS for within and across Breeding Cycles
4. Population Improvement
4.1. Selection Scheme
4.2. Integration of Germplasm and Maintaining Genetic Variance
4.3. Optimal Cross-Prediction
5. Real-World Applications
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Acquaah, G. Principles of Plant Genetics and Breeding, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2012. [Google Scholar]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 10 February 2022).
- Varshney, R.K.; Roorkiwal, M.; Sorrells, M.E. Genomic Selection for Crop Improvement: New Molecular Breeding Strategies for Crop Improvement; Springer: Cham, Switzerland, 2017; ISBN 3-319-63170-5. [Google Scholar]
- Carver, B.F. Wheat: Science and Trade; Wiley-Blackwell: Ames, IA, USA, 2009. [Google Scholar]
- Massman, J.M.; Jung, H.-J.G.; Bernardo, R. Genomewide Selection versus Marker-Assisted Recurrent Selection to Improve Grain Yield and Stover-Quality Traits for Cellulosic Ethanol in Maize. Crop Sci. 2013, 53, 58. [Google Scholar] [CrossRef] [Green Version]
- Rutkoski, J.; Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Genetic Gain from Phenotypic and Genomic Selection for Quantitative Resistance to Stem Rust of Wheat. Plant Genome 2015, 8, 2–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J.-L. Genomic Selection for Crop Improvement. Crop Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Poland, J.A.; Rife, T.W. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome J. 2012, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, P.E.; Ruperao, P.; Mason, A.S.; Stiller, J.; Chan, C.-K.K.; Hayashi, S.; Long, Y.; Meng, J.; Sutton, T.; Visendi, P.; et al. High-Resolution Skim Genotyping by Sequencing Reveals the Distribution of Crossovers and Gene Conversions in Cicer Arietinum and Brassica Napus. Theor. Appl. Genet. 2015, 128, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Buckler, E.S.; Ilut, D.C.; Wang, X.; Kretzschmar, T.; Gore, M.; Mitchell, S.E. rAmpSeq: Using Repetitive Sequences for Robust Genotyping. BioRxiv 2016, 096628. [Google Scholar] [CrossRef]
- Campbell, N.R.; Harmon, S.A.; Narum, S.R. Genotyping-in-Thousands by Sequencing (GT-Seq): A Cost Effective SNP Genotyping Method Based on Custom Amplicon Sequencing. Mol. Ecol. Resour. 2015, 15, 855–867. [Google Scholar] [CrossRef]
- Heffner, E.L.; Lorenz, A.J.; Jannink, J.-L.; Sorrells, M.E. Plant Breeding with Genomic Selection: Gain per Unit Time and Cost. Crop Sci. 2010, 50, 1681. [Google Scholar] [CrossRef]
- Michel, S.; Ametz, C.; Gungor, H.; Akgöl, B.; Epure, D.; Grausgruber, H.; Löschenberger, F.; Buerstmayr, H. Genomic Assisted Selection for Enhancing Line Breeding: Merging Genomic and Phenotypic Selection in Winter Wheat Breeding Programs with Preliminary Yield Trials. Theor. Appl. Genet. 2017, 130, 363–376. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics; Longmans Green: Harlow, UK, 1996; Volume 3. [Google Scholar]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the Rate of Genetic Gain in Public-Sector Plant Breeding Programs: Lessons from the Breeder’s Equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef] [Green Version]
- Lorenzana, R.E.; Bernardo, R. Accuracy of Genotypic Value Predictions for Marker-Based Selection in Biparental Plant Populations. Theor. Appl. Genet. 2009, 120, 151–161. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Toledo, F.H.; Pérez-Hernández, O.; Eskridge, K.M.; Rutkoski, J. A Genomic Bayesian Multi-Trait and Multi-Environment Model. G3 Genes Genomes Genet. 2016, 6, 2725–2744. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Jannink, J.-L. Multiple-Trait Genomic Selection Methods Increase Genetic Value Prediction Accuracy. Genetics 2012, 192, 1513–1522. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Zhao, Y.; Rodemann, B.; Plieske, J.; Kollers, S.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Ling, J. Potential and Limits to Unravel the Genetic Architecture and Predict the Variation of Fusarium Head Blight Resistance in European Winter Wheat (Triticum aestivum L.). Heredity 2015, 114, 318–326. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Khan, J.; Pradhan, S.; Shahi, D.; Khan, N.; Avci, M.; Mcbreen, J.; Harrison, S.; Brown-Guedira, G.; Murphy, J.P.; et al. Multi-Trait Genomic Prediction of Yield-Related Traits in US Soft Wheat under Variable Water Regimes. Genes 2020, 11, 1270. [Google Scholar] [CrossRef]
- Bernardo, R. Breeding for Quantitative Traits in Plants, 3rd ed.; Stemma Press: Woodbury, MN, USA, 2020; ISBN 978-0-9720724-3-4. [Google Scholar]
- Arruda, M.P.; Lipka, A.E.; Brown, P.J.; Krill, A.M.; Thurber, C.; Brown-Guedira, G.; Dong, Y.; Foresman, B.J.; Kolb, F.L. Comparing Genomic Selection and Marker-Assisted Selection for Fusarium Head Blight Resistance in Wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 84. [Google Scholar] [CrossRef]
- Belamkar, V.; Guttieri, M.J.; Hussain, W.; Jarquín, D.; El-basyoni, I.; Poland, J.; Lorenz, A.J.; Baenziger, P.S. Genomic Selection in Preliminary Yield Trials in a Winter Wheat Breeding Program. G3 Genes Genomes Genet. 2018, 8, 2735–2747. [Google Scholar] [CrossRef] [Green Version]
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding Schemes for the Implementation of Genomic Selection in Wheat (Triticum spp.). Plant Sci. 2016, 242, 23–36. [Google Scholar] [CrossRef]
- Robertsen, C.D.; Hjortshøj, R.L.; Janss, L.L. Genomic Selection in Cereal Breeding. Agronomy 2019, 9, 95. [Google Scholar] [CrossRef] [Green Version]
- Larkin, D.L.; Lozada, D.N.; Mason, R.E. Genomic Selection—Considerations for Successful Implementation in Wheat Breeding Programs. Agronomy 2019, 9, 479. [Google Scholar] [CrossRef] [Green Version]
- Gaynor, R.C.; Gorjanc, G.; Bentley, A.R.; Ober, E.S.; Howell, P.; Jackson, R.; Mackay, I.J.; Hickey, J.M. A Two-Part Strategy for Using Genomic Selection to Develop Inbred Lines. Crop Sci. 2017, 57, 2372–2386. [Google Scholar] [CrossRef] [Green Version]
- Rutkoski, J.E.; Poland, J.A.; Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Barbier, H.; Rouse, M.N.; Jannink, J.-L.; Sorrells, M.E. Genomic Selection for Quantitative Adult Plant Stem Rust Resistance in Wheat. Plant Genome 2014, 7, 3–6. [Google Scholar] [CrossRef] [Green Version]
- Endelman, J.B.; Atlin, G.N.; Beyene, Y.; Semagn, K.; Zhang, X.; Sorrells, M.E.; Jannink, J.-L. Optimal Design of Preliminary Yield Trials with Genome-Wide Markers. Crop Sci. 2014, 54, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Hoefler, R.; González-Barrios, P.; Bhatta, M.; Nunes, J.A.R.; Berro, I.; Nalin, R.S.; Borges, A.; Covarrubias, E.; Diaz-Garcia, L.; Quincke, M.; et al. Do Spatial Designs Outperform Classic Experimental Designs? J. Agric. Biol. Environ. Stat. 2020, 25, 523–552. [Google Scholar] [CrossRef]
- Longin, C.F.H.; Mi, X.; Würschum, T. Genomic Selection in Wheat: Optimum Allocation of Test Resources and Comparison of Breeding Strategies for Line and Hybrid Breeding. Theor. Appl. Genet. 2015, 128, 1297–1306. [Google Scholar] [CrossRef]
- Michel, S.; Ametz, C.; Gungor, H.; Epure, D.; Grausgruber, H.; Löschenberger, F.; Buerstmayr, H. Genomic Selection across Multiple Breeding Cycles in Applied Bread Wheat Breeding. Theor. Appl. Genet. 2016, 129, 1179–1189. [Google Scholar] [CrossRef] [Green Version]
- Verges, V.L.; Van Sanford, D.A. Genomic Selection at Preliminary Yield Trial Stage: Training Population Design to Predict Untested Lines. Agronomy 2020, 10, 60. [Google Scholar] [CrossRef] [Green Version]
- Michel, S.; Kummer, C.; Gallee, M.; Hellinger, J.; Ametz, C.; Akgöl, B.; Epure, D.; Löschenberger, F.; Buerstmayr, H. Improving the Baking Quality of Bread Wheat by Genomic Selection in Early Generations. Theor. Appl. Genet. 2018, 131, 477–493. [Google Scholar] [CrossRef]
- Tiede, T.; Smith, K.P. Evaluation and Retrospective Optimization of Genomic Selection for Yield and Disease Resistance in Spring Barley. Mol. Breed. 2018, 38, 1–16. [Google Scholar] [CrossRef]
- Auinger, H.-J.; Schönleben, M.; Lehermeier, C.; Schmidt, M.; Korzun, V.; Geiger, H.H.; Piepho, H.-P.; Gordillo, A.; Wilde, P.; Bauer, E.; et al. Model Training across Multiple Breeding Cycles Significantly Improves Genomic Prediction Accuracy in Rye (Secale cereale L.). Theor. Appl. Genet. 2016, 129, 2043–2053. [Google Scholar] [CrossRef] [Green Version]
- Merrick, L.F.; Carter, A.H. Comparison of Genomic Selection Models for Exploring Predictive Ability of Complex Traits in Breeding Programs. Plant Genome 2021, 14, e20158. [Google Scholar] [CrossRef]
- Merrick, L.F.; Burke, A.B.; Chen, X.; Carter, A.H. Breeding With Major and Minor Genes: Genomic Selection for Quantitative Disease Resistance. Front. Plant Sci. 2021, 12, 1599. [Google Scholar] [CrossRef]
- Podlich, D.W.; Cooper, M. QU-GENE: A Simulation Platform for Quantitative Analysis of Genetic Models. Bioinformatics 1998, 14, 632–653. [Google Scholar] [CrossRef] [Green Version]
- Faux, A.-M.; Gorjanc, G.; Gaynor, C.; Battagin, M.; Edwards, S.M.; Wilson, D.L.; Hearne, S.; Gonen, S.; Hickey, J.M. AlphaSim: Software for Breeding Program Simulation. Plant Genome 2016, 9, 13. [Google Scholar] [CrossRef] [Green Version]
- Jahufer, M.Z.Z.; Luo, D. DeltaGen: A Comprehensive Decision Support Tool for Plant Breeders. Crop Sci. 2018, 58, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Gaynor, R.C.; Gorjanc, G.; Hickey, J.M. AlphaSimR: An R Package for Breeding Program Simulations. G3 2021, 11, jkaa017. [Google Scholar] [CrossRef]
- Bernardo, R.; Yu, J. Prospects for Genomewide Selection for Quantitative Traits in Maize. Crop Sci. 2007, 47, 1082. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, R. Genomewide Selection with Minimal Crossing in Self-Pollinated Crops. Crop Sci. 2010, 50, 624. [Google Scholar] [CrossRef]
- Crain, J.; Bajgain, P.; Anderson, J.; Zhang, X.; DeHaan, L.; Poland, J. Enhancing Crop Domestication through Genomic Selection, a Case Study of Intermediate Wheatgrass. Front. Plant Sci. 2020, 11, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeHaan, L.R.; Wang, S.; Larson, S.; Cattani, D.J.; Zhang, X.I.; Kantarski, T.; Batello, C.; Wade, L.; Cox, S.; Pogna, N. Current Efforts to Develop Perennial Wheat and Domesticate Thinopyrum Intermedium as a Perennial Grain. In Proceedings of the Perennial Crops for Food Security: Proceedings of the FAO Expert Workshop, Logan, UT, USA, 1 May 2014. [Google Scholar]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Hickey, L.T.N.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuwissen, T.H.E. Maximizing the Response of Selection with a Predefined Rate of Inbreeding. J. Anim. Sci. 1997, 75, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Woolliams, J.A.; Berg, P.; Dagnachew, B.S.; Meuwissen, T.H.E. Genetic Contributions and Their Optimization. J. Anim. Breed. Genet. 2015, 132, 89–99. [Google Scholar] [CrossRef]
- Dreisigacker, S.; Crossa, J.; Pérez-Rodríguez, P.; Montesinos-L֯ópez, O.A.; Rosyara, U.; Juliana, P.; Mondal, S.; Crespo-Herrera, L.; Govindan, V.; Singh, R.P. Implementation of Genomic Selection in the CIMMYT Global Wheat Program, Findings from the Past 10 Years. Crop Breed. Genet. Genom. 2021, 3, 2. [Google Scholar] [CrossRef]
- Gorjanc, G.; Gaynor, R.C.; Hickey, J.M. Optimal Cross Selection for Long-Term Genetic Gain in Two-Part Programs with Rapid Recurrent Genomic Selection. Theor. Appl. Genet. 2018, 131, 1953–1966. [Google Scholar] [CrossRef] [Green Version]
- Gorjanc, G.; Hickey, J.M. AlphaMate: A Program for Optimizing Selection, Maintenance of Diversity and Mate Allocation in Breeding Programs. Bioinformatics 2018, 34, 3408–3411. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, M.; Tiede, T.; Smith, K.P. PopVar: A Genome-Wide Procedure for Predicting Genetic Variance and Correlated Response in Biparental Breeding Populations. Crop Sci. 2015, 55, 2068. [Google Scholar] [CrossRef]
- Lado, B.; Battenfield, S.D.; Guzmán, C.; Quincke, M.; Singh, R.P.; Dreisigacker, S.; Peña-Bautista, R.J.; Fritz, A.K.; Silva, P.; Poland, J. Strategies for Selecting Crosses Using Genomic Prediction in Two Wheat Breeding Programs. Plant Genome 2017, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Zhao, D.; Chen, X.; Zhang, Y.; Wang, J. Use of Genomic Selection and Breeding Simulation in Cross Prediction for Improvement of Yield and Quality in Wheat (Triticum aestivum L.). Crop J. 2018, 6, 353–365. [Google Scholar] [CrossRef]
- Bernardo, R. Genomewide Selection of Parental Inbreds: Classes of Loci and Virtual Biparental Populations. Crop Sci. 2014, 54, 2586–2595. [Google Scholar] [CrossRef] [Green Version]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package RrBLUP. Plant Genome J. 2011, 4, 250. [Google Scholar] [CrossRef] [Green Version]
- Covarrubias-Pazaran, G. Software Update: Moving the R Package Sommer to Multivariate Mixed Models for Genome-Assisted Prediction. bioRxiv 2018, 354639. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, R. Parental Selection, Number of Breeding Populations, and Size of Each Population in Inbred Development. Theor. Appl. Genet. 2003, 107, 1252–1256. [Google Scholar] [CrossRef]
- Witcombe, J.R.; Gyawali, S.; Subedi, M.; Virk, D.S.; Joshi, K.D. Plant Breeding Can Be Made More Efficient by Having Fewer, Better Crosses. BMC Plant Biol. 2013, 13, 22. [Google Scholar] [CrossRef] [Green Version]
- Hickey, J.M.; Dreisigacker, S.; Crossa, J.; Hearne, S.; Babu, R.; Prasanna, B.M.; Grondona, M.; Zambelli, A.; Windhausen, V.S.; Mathews, K.; et al. Evaluation of Genomic Selection Training Population Designs and Genotyping Strategies in Plant Breeding Programs Using Simulation. Crop Sci. 2014, 54, 1476–1488. [Google Scholar] [CrossRef] [Green Version]
- Veenstra, L.D.; Poland, J.; Jannink, J.-L.; Sorrells, M.E. Recurrent Genomic Selection for Wheat Grain Fructans. Crop Sci. 2020, 60, 1499–1512. [Google Scholar] [CrossRef]

| Crop | Trait | Cycles | Selection Methods Compared 1 | Gain | Reference |
|---|---|---|---|---|---|
| Maize | Stover Index and Grain Yield | 3 | MARS vs. GS | GS had higher genetic gain compared to MARS | [5] |
| Wheat | Quantitative Adult Plant Stem Rust Resistance | 2 | PS vs. GS | GS had equal rates of genetic gain compared to PS | [6] |
| Wheat | Grain Yield | 1 | GS models | Reproducing kernel Hilbert spaces (RKHS) GS model had the highest realized genetic gain | [53] |
| Barley | Grain Yield and DON | 3 | TP optimization | Optimization algorithms improved accuracy compared to randomly selected TPs | [37] |
| Wheat | Wheat Grain Fructan | 2 | GS with TS vs. GS with (OCS) | OCS and TS had similar genetic gains; OCS retained greater genetic variance | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Utilizing Genomic Selection for Wheat Population Development and Improvement. Agronomy 2022, 12, 522. https://doi.org/10.3390/agronomy12020522
Merrick LF, Herr AW, Sandhu KS, Lozada DN, Carter AH. Utilizing Genomic Selection for Wheat Population Development and Improvement. Agronomy. 2022; 12(2):522. https://doi.org/10.3390/agronomy12020522
Chicago/Turabian StyleMerrick, Lance F., Andrew W. Herr, Karansher S. Sandhu, Dennis N. Lozada, and Arron H. Carter. 2022. "Utilizing Genomic Selection for Wheat Population Development and Improvement" Agronomy 12, no. 2: 522. https://doi.org/10.3390/agronomy12020522
APA StyleMerrick, L. F., Herr, A. W., Sandhu, K. S., Lozada, D. N., & Carter, A. H. (2022). Utilizing Genomic Selection for Wheat Population Development and Improvement. Agronomy, 12(2), 522. https://doi.org/10.3390/agronomy12020522







