Cellulolytic Properties of a Potentially Lignocellulose-Degrading Bacillus sp. 8E1A Strain Isolated from Bulk Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cellulase-Producing Strains’ Isolation
2.2. Bacterial Strains’ Identification
2.3. Cellulase Production
2.4. The Effect of Temperature and pH on the Cellulases’ Activity
2.5. Statistical Analyses
3. Results and Discussion
3.1. Bacterial Strains’ Identification
3.2. Cellulolytic Properties’ Detection
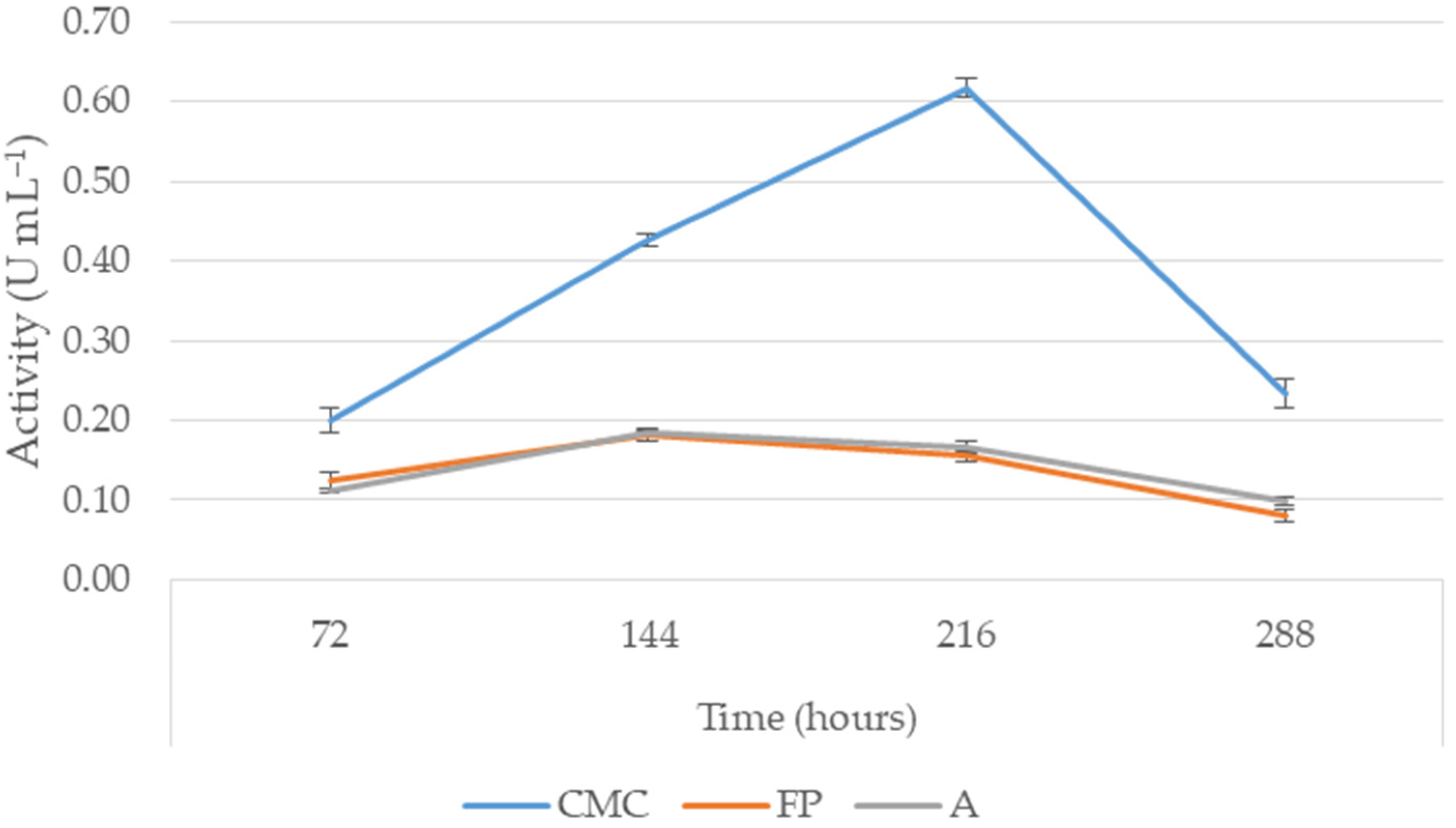
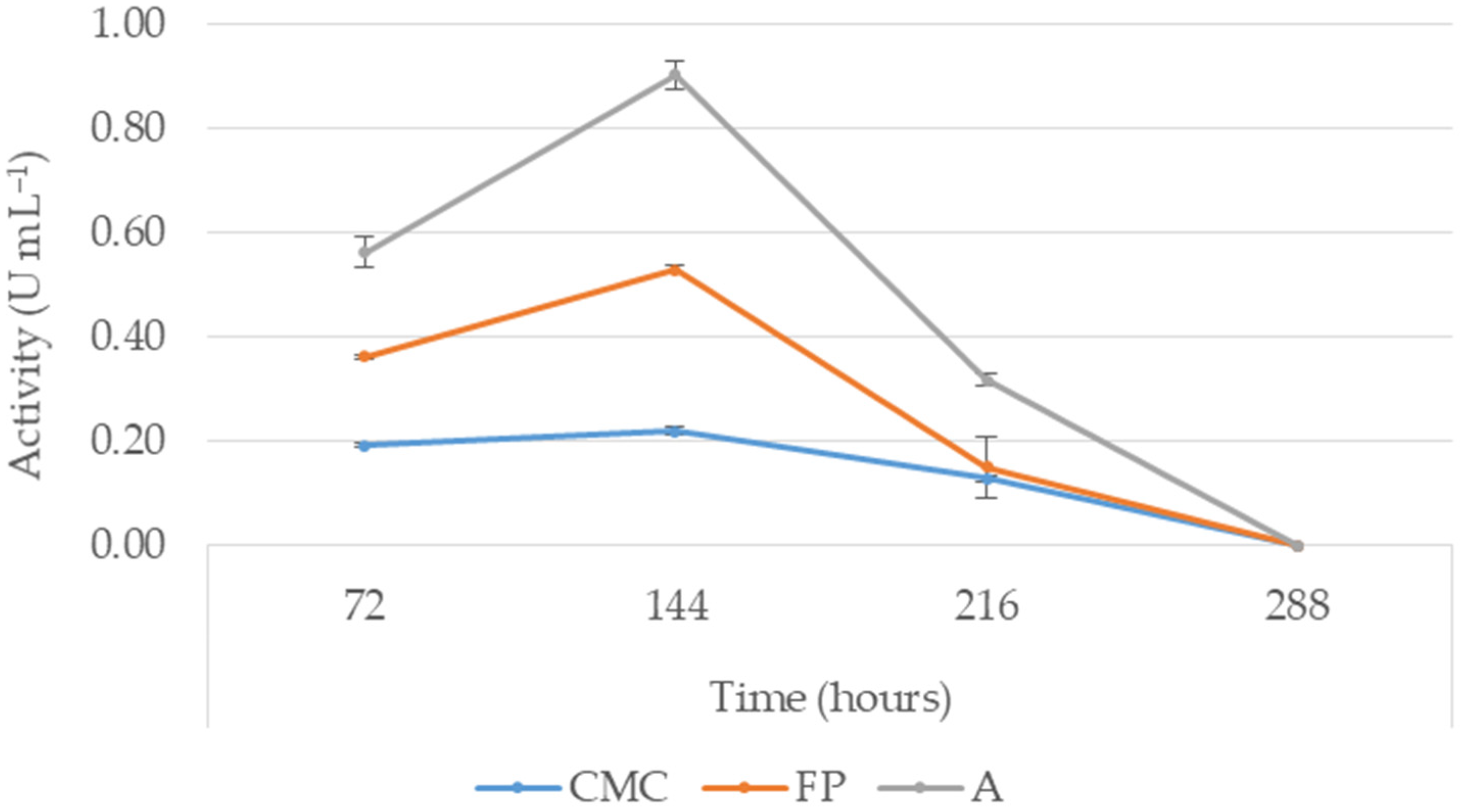
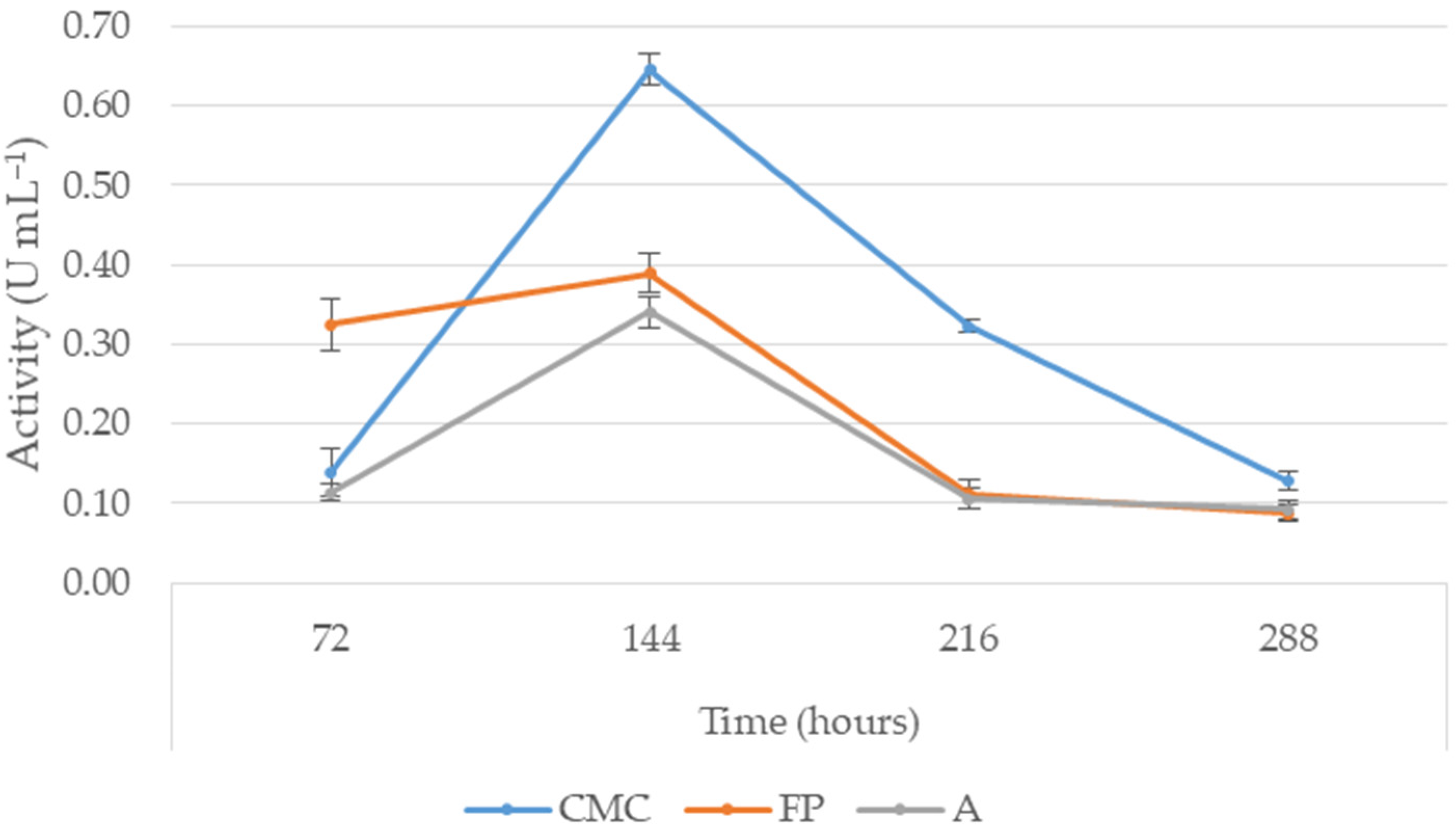
3.3. Cellulase Production
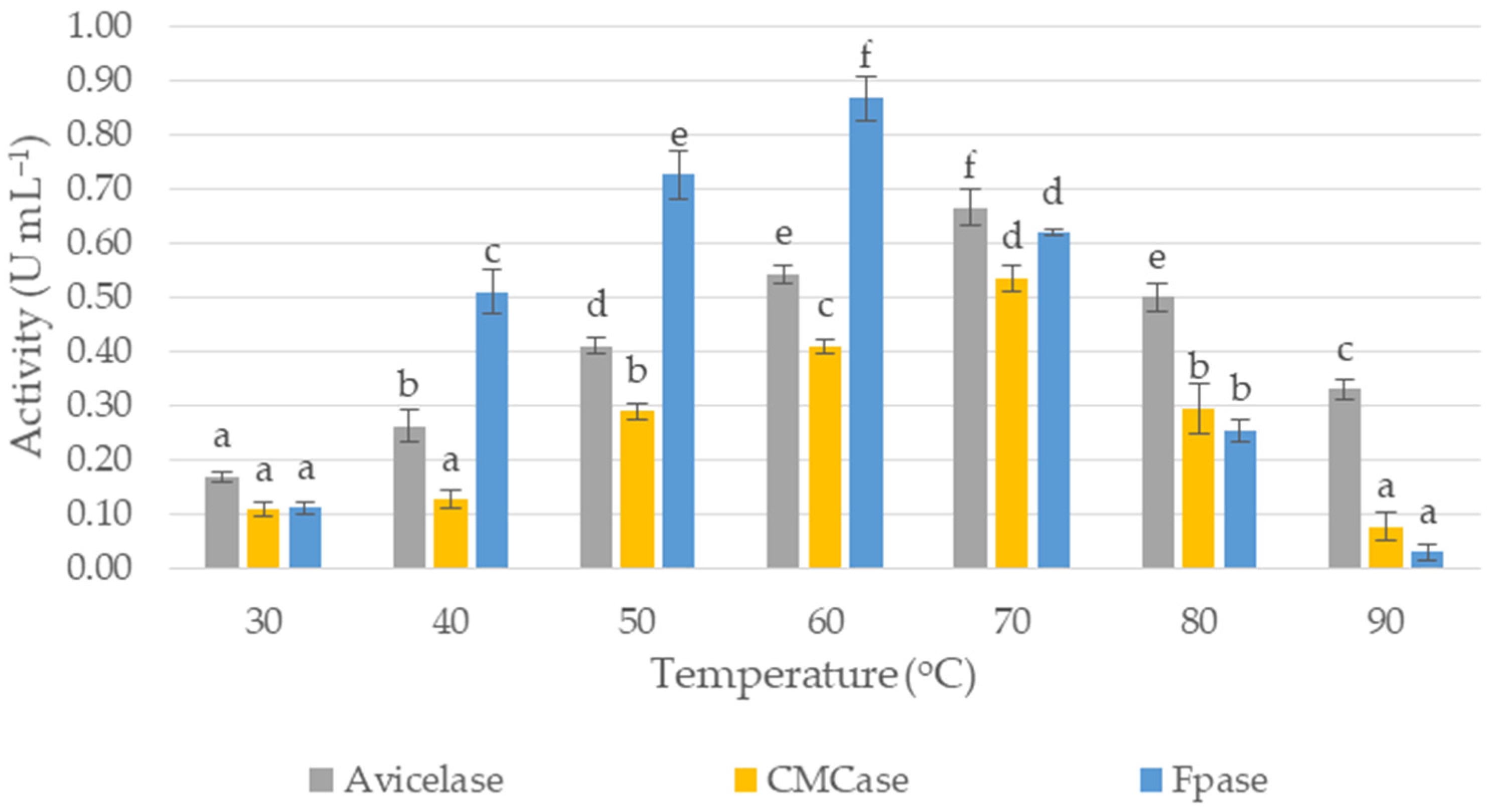
3.4. Effect of Temperature and pH on the Cellulases’ Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behr, A. Carbon Dioxide as an Alternative C1 Synthetic Unit: Activation by Transition-Metal Complexes. Angew. Chem. Int. Ed. Engl. 1988, 27, 661–678. [Google Scholar] [CrossRef]
- Wałowski, G. Development of biogas and biorafinery systems in Polish rural communities. J. Water Land Dev. 2021, 49, 156–168. [Google Scholar]
- Wang, H.; Yang, B.; Zhang, Q.; Zhu, W. Catalytic routes for the conversion of lignocellulosic biomass to aviation fuel range hydrocarbons. Renew. Sustain. Energy Rev. 2020, 120, 109612. [Google Scholar] [CrossRef]
- Voytovych, I.; Malovanyy, M.; Zhuk, V.; Mukha, O. Facilities and problems of processing organic wastes by familytype biogas plants in Ukraine. J. Water Land Dev. 2020, 45, 185–189. [Google Scholar]
- Sun, Y.; Cheng, J. Hydrolysis of lignocellulosic materials for ethanol production: A review. Bioresour. Technol. 2002, 83, 1–11. [Google Scholar] [CrossRef]
- Béguin, P.; Aubert, J.P. The biological degradation of cellulose. FEMS Microbiol. Rev. 1994, 13, 25–58. [Google Scholar] [CrossRef]
- Horn, S.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V.G. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Thapa, S.; Mishra, J.; Arora, N.; Mishra, P.; Li, H.; O′Hair, J.; Bhatti, S.; Zhou, S. Microbial cellulolytic enzymes: Diversity and biotechnology with reference to lignocellulosic biomass degradation. Rev. Environ. Sci. Biol. Technol. 2020, 19, 621–648. [Google Scholar]
- Singhania, R.R.; Ruiz, H.A.; Awasthi, M.K.; Dong, C.D.; Chen, C.W.; Patel, A.K. Challenges in cellulase bioprocess for biofuel applications. Renew. Sustain. Energy Rev. 2021, 151, 111622. [Google Scholar] [CrossRef]
- Zhang, X.-Z.; Zhang, Y.-H.P. Cellulases: Characteristics, sources, production, and applications. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; Yang, S.-T., El-Enshasy, H.A., Thongchul, N., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 131–146. [Google Scholar] [CrossRef]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 2016, 15, 106. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Anwar, Z.; Ali, A. Production and characterization of commercial cellulase produced through Aspergillus niger IMMIS1 after screening fungal species. Pak. J. Bot. 2018, 50, 1563–1570. [Google Scholar]
- Poulsen, H.V.; Willink, F.W.; Ingvorsen, K. Aerobic and anaerobic cellulase production by Cellulomonas uda. Arch. Microbiol. 2016, 198, 725–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Huang, W.C.; Liu, Y.; Li, M. Diversity of cellulolytic microorganisms and microbial cellulases. Int. Biodeterior. Biodegrad. 2021, 163, 105277. [Google Scholar] [CrossRef]
- Deka, D.; Bhargavi, P.; Sharma, A.; Goyal, D.; Jawed, M.; Goyal, A. Enhancement of cellulase activity from a new strain of Bacillus subtilis by medium optimization and analysis with various cellulosic substrates. Enzym. Res. 2011, 2011, 151656. [Google Scholar] [CrossRef] [Green Version]
- Matsubara, H.; Goto, K.; Matsumura, T.; Mochida, K.; Iwaki, M.; Niwa, M.; Yamasato, K. Alicyclobacillus acidophilus sp. nov., a novel thermo-acidophilic, omega-alicyclic fatty acid-containing bacterium isolated from acidic beverages. Int. J. Syst. Evol. Microbiol. 2005, 55, 1681–1685, Erratum in: Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 5, 2233. [Google Scholar] [CrossRef] [Green Version]
- Kusube, M.; Sugihara, A.; Moriwaki, Y.; Ueoka, T.; Shimane, Y.; Minegishi, H. Alicyclobacillus cellulosilyticus sp. nov., a thermophilic, cellulolytic bacterium isolated from steamed Japanese cedar chips from a lumbermill. Int. J. Syst. Evol. Microbiol. 2014, 64, 2257–2263. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.; Batra, N.; Sobti, R.C. Purification and characterisation of alkaline cellulase produced by a novel isolate, Bacillus sphaericus JS1. J. Ind. Microbiol. Biotechnol. 2004, 31, 51–56. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Arun, A.; Al-Dhabi, N.A.; Vincent, S.G.P.; Arasu, M.V.; Choi, K.C. Novel Bacillus subtilis IND19 cell factory for the simultaneous production of carboxy methyl cellulase and protease using cow dung substrate in solid-substrate fermentation. Biotechnol. Biofuels 2016, 9, 73. [Google Scholar] [CrossRef] [Green Version]
- Potprommanee, L.; Wang, X.-Q.; Han, Y.-J.; Nyobe, D.; Peng, Y.-P.; Huang, Q.; Liu, J.; Liao, Y.-L.; Chang, K.-L. Characterization of a thermophilic cellulase from Geobacillus sp. HTA426, an efficient cellulase-producer on alkali pre-treated of lignocellulosic biomass. PLoS ONE 2017, 12, e0175004. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbial. 2007, 57, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Khianngam, S.; Pootaeng-on, Y.; Techakriengkrai, T.; Tanasupawat, S. Screening and identification of cellulase producing bacteria isolated from oil palm meal. J. Appl. Pharm. Sci. 2014, 4, 90. [Google Scholar] [CrossRef]
- Saha, S.; Roy, R.N.; Sen, S.K.; Ray, A.K. Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquac. Res. 2006, 37, 380–388. [Google Scholar] [CrossRef]
- Doi, R.H. Cellulases of mesophilic microorganisms: Cellulosome and noncellulosome producers. Ann. N. Y. Acad. Sci. 2008, 1125, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Park, T.S.; Kwon, I.H.; Piao, M.Y.; Lee, C.H.; Ha, J.K. Characterization of cellulolytic and xylanolytic enzymes of Bacillus licheniformis JK7 isolated from the rumen of a native Korean goat. Asian-Australas. J. Anim. Sci. 2013, 26, 50–58. [Google Scholar] [CrossRef]
- Mihajlovski, K.R.; Carević, M.B.; Dević, M.L.; Šiler-Marinković, S.; Rajilić-Stojanović, M.D.; Dimitrijević-Branković, S. Lignocellulosic waste material as substrate for Avicelase production by a new strain of Paenibacillus chitinolyticus CKS1. Int. Biodeterior. Biodegrad. 2015, 104, 426–434. [Google Scholar] [CrossRef]
- Šimansky, V.; Juriga, M.; Jonczak, J.; Uzarowicz, L.; Stepien, W. How relationships between soil organic matter parameters and soil structure characteristics are affected by the long-term fertilization of a sandy soil. Geoderma 2019, 342, 75–84. [Google Scholar] [CrossRef]
- Stępień, W.; Kobiałka, M. Effect of Long-Term Organic and Mineral Fertilisation on Selected Physico-Chemical Soil Properties in Rye Monoculture and Five-Year Crop Rotation. Soil Sci. Ann. 2019, 70, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Dobrzyński, J.; Wierzchowski, P.S.; Stępień, W.; Górska, E.B. The reaction of cellulolytic and potentially cellulolytic spore-forming bacteria to various types of crop management and farmyard manure fertilization in bulk soil. Agronomy 2021, 11, 772. [Google Scholar] [CrossRef]
- Kim, B.C.; Lee, K.H.; Kim, M.N.; Kim, E.M.; Min, S.R.; Kim, H.S.; Shin, K.S. Paenibacillus pini sp. nov., a cellulolytic bacterium isolated from the rhizosphere of pine tree. J. Microbiol. 2009, 47, 699–704. [Google Scholar] [CrossRef]
- Kim, S.J.; Chun, J.; Bae, K.S.; Kim, Y.C. Polyphasic assignment of an aromatic-degrading Pseudomonas sp., strain DJ77, in the genus Sphingomonas as Sphingomonas chungbukensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 1641–1647. [Google Scholar] [CrossRef] [Green Version]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Akaracharanya, A.; Taprig, T.; Sitdhipol, J.; Tanasupawat, S. Characterization of cellulase producing Bacillus and Paenibacillus strains from Thai soils. J. Appl. Pharm. Sci. 2014, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.-L.; Zhang, Z.; Wu, M.; Wu, Y.; Feng, J.-X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27-1. BioMed Res. Int. 2014, 2014, 512497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budi, S.W.; van Tuinen, D.; Arnould, C.; Dumas-Gaudot, E.; Gianinazzi-Pearson, V.; Gianinazzi, S. Hydrolytic enzyme activity of Paenibacillus sp. strain B2 and effects of the antagonistic bacterium on cell integrity of two soil-borne pathogenic fungi. Appl. Soil Ecol. 2000, 15, 191–199. [Google Scholar] [CrossRef]
- Gupta, P.; Samant, K.; Sahu, A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012, 2012, 578925. [Google Scholar] [CrossRef] [Green Version]
- Maki, M.; Leung, K.T.; Qin, W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 2009, 5, 500–516. [Google Scholar] [CrossRef]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef] [Green Version]
- Sadhu, S.; Ghosh, P.K.; Aditya, G.; Maiti, T.K. Optimization and strain improvement by mutation for enhanced cellulase production by Bacillus sp. (MTCC10046) isolated from cow dung. J. King Saud Univ. Sci. 2014, 26, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Moholkar, V.S.; Goyal, A. Optimization of carboxymethylcellulase production from Bacillus amyloliquefaciens SS35. 3 Biotech 2014, 4, 411–424. [Google Scholar] [CrossRef] [Green Version]
- Lynd, L.R.; Weimer, P.J. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [Green Version]
- Thomas, L.; Ram, H.; Singh, V.P. Inducible cellulase production from an organic solvent tolerant Bacillus sp. SV1 and evolutionary divergence of endoglucanase in different species of the genus Bacillus. Braz. J. Microbiol. 2018, 49, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, C.H. Production and characterization of crystalline cellulose-degrading cellulase components from a thermophilic and moderately alkalophilic bacterium. J. Microbiol. Biotechnol. 1992, 2, 7–13. [Google Scholar]
- Geetha, K.; Gunasekaran, P. Optimization of nutrient medium containing agricultural waste for xylanase production by Bacillus pumilus B20. Biotechnol. Bioprocess Eng. 2010, 15, 882–889. [Google Scholar] [CrossRef]
- Saratale, G.D.; Oh, S.E. Production of thermotolerant and alkalotolerant cellulolytic enzymes by isolated Nocardiopsis sp. KNU. Biodegradation 2011, 22, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Ladeira, S.A.; Cruz, E.; Delatorre, A.B.; Barbosa, J.B.; Martins, M.L.L. Cellulase production by thermophilic Bacillus sp. SMIA-2 and its detergent compatibility. Electron. J. Biotechnol. 2015, 18, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, G.; Bhalla, A.; Adhikari, A.; Bischoff, K.M.; Hughes, S.R.; Christopher, L.P.; Sani, R.K. Characterization of thermostable cellulases produced by Bacillus and Geobacillus strains. Bioresour. Technol. 2010, 101, 8798–8806. [Google Scholar] [CrossRef]
- Bhuyan, P.M.; Sandilya, S.P.; Nath, P.K.; Gandotra, S.; Subramanian, S.; Kardong, D.; Gogoi, D.K. Optimization and characterization of extracellular cellulase produced by Bacillus pumilus MGB05 isolated from midgut of muga silkworm (Antheraea assamensis Helfer). J. Asia-Pac. Entomol. 2018, 21, 1171–1181. [Google Scholar] [CrossRef]
- Kazeem, M.O.; Shah, U.K.M.; Baharuddin, A.S.; Abdul Rahman, N.A. Enhanced cellulase production by a novel thermophilic Bacillus licheniformis 2D55: Characterization and application in lignocellulosic saccharification. Bioresources 2016, 11, 5404–5423. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, N.; Gupta, V.; Kumar, M.; Kumari, P.; Reddy, C.R.K.; Jha, B. An alkali-halotolerant cellulase from Bacillus flexus isolated from green seaweed Ulva lactuca. Carbohydr. Polym. 2011, 83, 891–897. [Google Scholar] [CrossRef]
- Li, W.; Zhang, W.-W.; Yang, M.-M.; Chen, Y.-L. Cloning of the thermostable cellulase gene from newly isolated Bacillus subtilis and its expression in Escherichia coli. Mol. Biotechnol. 2008, 40, 195–201. [Google Scholar] [CrossRef]
- Tai, S.-K.; Lin, H.-P.P.; Kuo, J.; Liu, J.-K. Isolation and characterization of a cellulolytic Geobacillus thermoleovorans T4 strain from sugar refinery wastewater. Extremophiles 2004, 8, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Talamantes, D.; Biabini, N.; Dang, H.; Abdoun, K.; Berlemont, R. Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol. Biofuels 2016, 9, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No. | Biochemical Characteristic | Strain Number | ||||||
|---|---|---|---|---|---|---|---|---|
| 8E1A | 14AV1 | 14AV2 | 15AV1 | 15AV2 | 15E1A1 | 24DV | ||
| 1. | Glycerol | + | + | - | - | - | - | - |
| 2. | Erythritol | - | - | - | - | - | - | - |
| 3. | D-Arabinose | - | - | - | - | - | - | - |
| 4. | L-Arabinose | + | + | + | - | + | + | + |
| 5. | D-Ribose | + | + | + | + | + | + | + |
| 6. | D-Xylose | + | + | + | + | + | + | + |
| 7. | L-Xylose | - | - | - | - | - | - | - |
| 8. | D-Adonithol | - | - | - | - | - | - | - |
| 9. | Methyl-β-D-xylopyranoside | - | - | - | - | + | - | - |
| 10. | D-Galactose | + | + | + | + | + | + | + |
| 11. | D-Glucose | + | + | + | + | + | + | + |
| 12. | D-Fructose | + | + | + | + | + | + | + |
| 13. | D-Mannose | + | + | + | - | + | + | + |
| 14. | L-Sorbose | - | - | - | - | - | - | - |
| 15. | L-Rhamnose | - | + | - | - | - | - | - |
| 16. | Dulcitol | - | - | - | - | - | - | - |
| 17. | Inositol | + | + | - | - | + | - | + |
| 18. | D-Mannitol | + | + | + | + | + | + | + |
| 19. | D-Sorbitol | + | - | - | - | + | - | + |
| 20. | Methyl-α-D-mannopyranoside | - | - | - | - | - | - | - |
| 21. | Methyl-α-D-glucopyranoside | + | + | - | - | - | - | - |
| 22. | N-acetyl-glucosamine | - | - | + | + | + | + | + |
| 23. | Amygdalin | + | + | + | + | + | + | + |
| 24. | Arbutin | + | + | - | - | - | + | - |
| 25. | Esculin | + | + | + | + | + | + | + |
| 26. | Salicin | + | + | - | + | - | + | + |
| 27. | D-Cellobiose | + | + | + | + | + | + | + |
| 28. | D-Maltose | + | + | + | + | + | + | + |
| 29. | D-Lactose | + | + | + | + | + | + | + |
| 30. | D-Melibiose | + | + | + | + | + | + | + |
| 31. | D-Sucrose | + | + | + | + | + | + | + |
| 32. | D-Trehalose | - | - | + | + | + | - | + |
| 33. | Inulin | - | - | - | - | - | - | - |
| 34. | D-Melizitose | + | + | - | - | - | - | - |
| 35. | D-Raffinose | - | + | + | - | + | + | + |
| 36. | Starch | + | + | + | + | + | + | + |
| 37. | Glycogen | + | + | + | + | + | - | + |
| 38. | Xylitol | + | + | - | - | - | - | - |
| 39. | Gentibiose | - | + | - | + | - | - | - |
| 40. | D-Turanose | + | + | - | - | - | - | - |
| 41. | D-Lyxose | - | - | - | - | - | - | - |
| 42. | D-Tagatose | - | - | - | - | - | - | - |
| 43. | D-Fucose | - | - | - | - | - | - | - |
| 44. | L-Fucose | - | - | - | - | - | - | - |
| 45. | D-Arabitol | - | - | - | - | - | - | - |
| 46. | L-Arabitol | - | - | - | - | - | - | - |
| 47. | Potassium gluconate | - | - | - | - | - | - | - |
| 48. | Potassium 2-ketogluconate | - | - | - | - | - | - | - |
| 49. | Potassium 5-ketogluconate | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrzyński, J.; Wróbel, B.; Górska, E.B. Cellulolytic Properties of a Potentially Lignocellulose-Degrading Bacillus sp. 8E1A Strain Isolated from Bulk Soil. Agronomy 2022, 12, 665. https://doi.org/10.3390/agronomy12030665
Dobrzyński J, Wróbel B, Górska EB. Cellulolytic Properties of a Potentially Lignocellulose-Degrading Bacillus sp. 8E1A Strain Isolated from Bulk Soil. Agronomy. 2022; 12(3):665. https://doi.org/10.3390/agronomy12030665
Chicago/Turabian StyleDobrzyński, Jakub, Barbara Wróbel, and Ewa Beata Górska. 2022. "Cellulolytic Properties of a Potentially Lignocellulose-Degrading Bacillus sp. 8E1A Strain Isolated from Bulk Soil" Agronomy 12, no. 3: 665. https://doi.org/10.3390/agronomy12030665






