Development and Metabolic Characterization of Horse Gram (Macrotyloma uniflorum Lam. (Verdc.)) Mutants for Powdery Mildew Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Material and Categorization for Powdery Mildew Resistance
2.1.1. Evolution of Genetic Materials
2.1.2. Screening of Mutants for Powdery Mildew Resistance
2.1.3. Pathogen Confirmation and Classification of Mutants
2.1.4. Confirmation of Powdery Mildew Resistance and Tagging of Mutants for Metabolomic Analysis
2.1.5. Data Analysis
2.2. Metabolite Extraction and GC-MS Analysis
3. Results and Discussion
3.1. Powdery Mildew-Induced Changes in the Yield Attributing Traits
3.2. Metabolomic Analysis through GC-MS Analysis
3.3. Biological Significance of Unique Metabolites Expressed in the Resistant Mutant
3.4. Biological Significance of Unique Metabolites Expressed in the Susceptible Mutant
3.5. Biological Significance of Expression of the Common Class of Bio-Molecules Expressed Both in Resistant and Susceptible Mutants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Purushottam, R.K.; Barman, R.; Saren, B. Improvement of Neglected Horse Gram Production for Benefit of Mankind. Int. J. Bio-Res. Environ. Agril. Sci. 2017, 3, 521–527. [Google Scholar]
- Kumar, D. Horsegram an Introduction; Scientific Publishers: Jodhpur, India, 2006; pp. 1–10. [Google Scholar]
- National Academy of Sciences. Moth bean. In Tropical Legumes: Resources for the Future; The National Academies Press: Washington, DC, USA, 1978. [Google Scholar]
- Priyanka, S.; Sudhagar, R.; Vanniarajan, C.; Ganesamurthy, K.; Souframanien, J. Induction of Genetic Variability for Quantitative Traits in Horsegram (Macrotyloma uniflorum) through Irradiation Mutagenesis. J. Environ. Biol. 2021, 42, 597–608. [Google Scholar] [CrossRef]
- Dutta, U.; Gupta, S.; Jamwal, A.; Jamwal, S. Integrated Disease Management of Horsegram (Macrotyloma uniflorum). In Diseases of Field Crops. Diagnosis and Management. Volume 2: Pulses, Oil Seeds, Narcotics, and Sugar Crops; Srivastava, J.N., Singh, A.K., Eds.; CRC Press: Boca Raton, FL, USA, 2020; pp. 91–104. [Google Scholar]
- Ullah, N.; Yüce, M.; NeslihanÖztürkGökçe, Z.; Budak, H. Comparative Metabolite Profiling of Drought Stress in Roots and Leaves of Seven Triticeae Species. BMC Genom. 2017, 18, 969. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and Metabolomic Profiling of Drought-Tolerant and Susceptible Sesame Genotypes in Response to Drought Stress. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Fountain, J.C.; Ji, P.; Ni, X.; Chen, S.; Lee, R.D.; Kemerait, R.C.; Guo, B. Deciphering Drought-Induced Metabolic Responses and Regulation in Developing Maize Kernel. Plant Biotechnol. J. 2018, 16, 1616–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golldack, D.; Luking, I.; Yang, O. Plant Tolerance to Drought and Salinity: Stress Regulating Transcription Factors and their Functional Significance in the Cellular Transcriptional Network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Rai, V. Role of Amino Acids in Plants Responses to Stresses. Biol. Plant. 2002, 45, 481–487. [Google Scholar] [CrossRef]
- Pal, A.B.; Brahmappa, H.S.; Rawal, R.D.; Ullasa, A. Field Resistance of Pea Germplasm to Powdery Mildew (Erysiphe polygoni) and Rust (Uromyces fabae). Plant Dis. 1980, 64, 1085–1086. [Google Scholar] [CrossRef]
- Banyal, D.K.; Tyagi, P.D. Comparison of Screening Techniques for Evaluation of Resistance among Pea Genotypes for Powdery Mildew. In Natural Resource Management for Sustainable Hill Agriculture, 2nd ed.; Ghabroo, S.K., Bhagat, R.M., Kapoor, A.C., Eds.; HPKV: Palampur, India, 1998; pp. 340–346. [Google Scholar]
- Banyal, D.K.; Singh, A.; Tyagi, P.D. Pathogenic Variability in Erysiphe pisi Causing Pea Powdery Mildew. Himachal J. Agric. Res. 2005, 32, 87–92. [Google Scholar]
- Fiehn, O.; Kopka, J.; Dormann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite Profiling for Plant Functional Genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Epstein, E.; Bloom, A.J. Mineral Nutrition of Plants: Principles and Perspectives, 2nd ed.; Sinauer Associates Inc.: Sunderland, UK, 2005; p. 48. [Google Scholar]
- Guerriero, G.; Hausman, J.F.; Legay, S. Silicon and the Plant Extracellular Matrix. Front. Plant Sci. 2016, 7, 463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauteux, F.; Remus-Borel, W.; Menzies, J.G.; Belanger, R.R. Silicon and Plant Disease Resistance against Pathogenic Fungi. FEMS Microbiol. Lett. 2005, 249, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remus-Borel, W.; Menzies, J.G.; Bélanger, R.R. Silicon Induces Antifungal Compounds in Powdery Mildew-Infected Wheat. Physiol. Mol. Plant Pathol. 2005, 66, 108–115. [Google Scholar] [CrossRef]
- Rahman, A.; Wallis, C.M.; Uddin, W. Silicon-Induced Systemic Defense Responses in Perennial Ryegrass against Infection by Magnaportheoryzae. Phytopathology 2015, 105, 748–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulbert, P.B.; Beuding, E.; Robinson, C.H. Structure and Antisehistosomal Activity in the Nitrofuran Series. J. Med. Chem. 1973, 16, 72. [Google Scholar] [CrossRef]
- Spiteller, G. Furan Fatty Acids: Occurrence, Synthesis, and Reactions. Are Furan Fatty Acids Responsible for the Cardioprotective Effects of a Fish Diet? Lipids 2005, 40, 755–771. [Google Scholar] [CrossRef]
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic Acid: A Likely Endogenous Signal in the Resistance Response of Tobacco to Viral Infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef] [Green Version]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, A Multifaceted Hormone to Combat Disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [Green Version]
- Mutui, T.M.; Mibus, H.; Serek, M. Effect of Meta-Topolin on Leaf Senescence and Rooting in Pelargonium × Hortorum Cuttings. Postharvest Biol. Technol. 2012, 63, 107–110. [Google Scholar] [CrossRef]
- Geckeler, K.E.; Eberhardt, W. Biogenic Organochlorine Compounds-Occurrence, Function and Ecological Relevance. Naturwissenschaften 1995, 82, 2–11. [Google Scholar] [CrossRef]
- Siuda, J.F.; DeBernardis, J.F. Naturally Occurring Halogenated Organic Compounds. Lloydia 1973, 36, 107–143. [Google Scholar]
- Monde, K.; Satoh, H.; Nakamura, M.; Tamura, M.; Takasugi, M. Organochlorine Compounds from a Terrestrial Higher Plant: Structures and Origin of Chlorinated Orcinol Derivatives from Diseased Bulbs of Lilium maximowiczii. J. Nat. Prod. 1998, 61, 913–921. [Google Scholar] [CrossRef]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Stahl, B. Diversity and Distribution of Floran Scent. Bot. Rev. 2006, 71, 1–120. [Google Scholar] [CrossRef]
- Irmisch, S.; McCormick, A.C.; Boeckler, G.A.; Schmidt, A.; Reichelt, M.; Schneider, B.; Block, K.; Schnitzler, J.P.; Gershenzon, J.; Unsicker, S.B.; et al. Two Herbivore-Induced Cytochrome P450 Enzymes CYP79D6 and CYP79D7 Catalyze the Formation of Volatile Aldoximes Involved in Poplar Defense. Plant Cell 2013, 25, 4737–4754. [Google Scholar] [CrossRef] [Green Version]
- Raguso, R.A. Wake up and Smell the Roses: The Ecology and Evolution of Floral Scent. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 549–569. [Google Scholar] [CrossRef]
- Sakurada, K.; Ikegaya, H.; Ohta, H.; Fukushima, H.; Akutsu, T.; Watanabe, K. Effects of Oximes on Mitochondrial Oxidase Activity. Toxicol. Lett. 2009, 189, 110–114. [Google Scholar] [CrossRef]
- Møller, B.L. Plant Science. Dynamic Metabolons. Science 2010, 330, 1328–1329. [Google Scholar] [CrossRef]
- Bak, S.; Olsen, C.E.; Halkier, B.A.; Møller, B.L. Transgenic Tobacco and Arabidopsis Plants Expressing the Two Multifunctional Sorghum Cytochrome P450 Enzymes, CYP79A1 and CYP71E1, are Cyanogenic and Accumulate Metabolites Derived from Intermediates in Dhurrin Biosynthesis. Plant Physiol. 2000, 123, 1437–1448. [Google Scholar] [CrossRef] [Green Version]
- Blomstedt, C.K.; Gleadow, R.M.; O’Donnell, N.; Naur, P.; Jensen, K.; Laursen, T.; Olsen, C.E.; Stuart, P.; Hamill, J.D.; Møller, B.L.; et al. A Combined Biochemical Screen and TILLING Approach Identifies Mutations in Sorghum bicolor L. Moench Resulting in Acyanogenic Forage Production. Plant Biotechnol. J. 2012, 10, 54–66. [Google Scholar] [CrossRef]
- Clausen, M.; Kannangara, R.M.; Olsen, C.E.; Blomstedt, C.K.; Gleadow, R.M.; Jorgensen, K.; Bak, S.; Motawie, M.S.; Møller, B.L. The Bifurcation of the Cyanogenic Glucoside and Glucosinolate Biosynthetic Pathways. Plant J. 2015, 84, 558–573. [Google Scholar] [CrossRef]
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Volatile C6-Aldehydes and Allo-Ocimene Activate Defense Genes and Induce Resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1093–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansjakob, A.; Bischof, S.; Bringmann, G.; Riederer, M.; Hildebrandt, U. Very-Long-Chain Aldehydes Promote Vitro in Prepenetration Processes of Blumeriagraminis in a Dose and Chain Length-Dependent Manner. New Phytol. 2010, 188, 1039–1054. [Google Scholar] [CrossRef] [PubMed]
- Pinakoulaki, E.; Koutsoupakis, C.; Sawai, H.; Pavlou, A.; Kato, Y.; Asano, Y.; Aono, S. Aldoxime Dehydratase: Probing the Heme Environment Involved in the Synthesis of the Carbon-Nitrogen Triple Bond. J. Phys. Chem. B 2011, 115, 13012–13018. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Tawara, J.N.; Boeckl, M.; Pomeroy, M.; Foderaro, T.A.; Todd, F.G. Piperidine Alkaloid Content of Picea (spruce) and Pinus (pine). Phytochemistry 1994, 35, 951–953. [Google Scholar] [CrossRef]
- Shtykova, L.; Masuda, M.; Eriksson, C.; Sjödin, K.; Marling, E.; Schlyter, F.; Nydén, M. Latex Coatings Containing Antifeedants: Formulation, Characterization, and Application for Protection of Conifer Seedlings against Pine Weevil Feeding. Prog. Org. Coat. 2008, 63, 160–166. [Google Scholar] [CrossRef]
- Oppel, C.B.; Dussourd, D.E.; Garimella, U. Visualizing a Plant Defense and Insect Counterploy: Alkaloid Distribution in Lobelia Leaves Trenched by a Plusiine Caterpillar. J. Chem. Ecol. 2009, 35, 625–634. [Google Scholar] [CrossRef]
- Castells, E.; Berenbaum, M.R. Resistance of the Generalist Moth Trichoplusiani (Noctuidae) to a Novel Chemical Defense in the Invasive Plant Conium maculatum. Chemoecology 2007, 18, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Willamil, J.; Creus, E.; Pe’rez, J.F.; Mateu, E.; Martin-Orúe, S.M. Effect of a Microencapsulated Feed Additive of Lactic and Formic Acid on the Prevalence of Salmonella in Pigs Arriving at the Abattoir. Arch. Anim. Nutr. 2011, 65, 431–444. [Google Scholar] [CrossRef]
- Lu, T.; Jiang, M.; Jiang, Z.; Hui, D.; Wang, Z.; Zhou, Z. Effect of Surface Modification of Bamboo Cellulose Fibers on Mechanical Properties of Cellulose/Epoxy Composites. Compos. Part B Eng. 2013, 51, 28–34. [Google Scholar] [CrossRef]
- Roessingh, P.; Stadler, E.; Baur, R.; Hurter, J.; Ramp, T. Tarsal Chemoreceptors and Oviposition Behaviour of the Cabbage Root Fly (Delia radicum) Sensitive to Fractions and New Compounds of Host-Leaf Surface Extracts. Physiol. Entomol. 1997, 22, 140–148. [Google Scholar] [CrossRef]
- Rodrigues, F.A.; Resende, R.S.; Dallagnol, L.J.; Datnoff, L.E. Silicon Potentiates Host Defense Mechanisms against Infection by Plant Pathogens; Springer: Cham, Switzerland, 2015; pp. 109–138. [Google Scholar]
- Sakr, N. The Role of Silicon (Si) in Increasing Plant Resistance against Fungal Diseases. Hell. Plant Prot. J. 2016, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lu, H. Dissection of Salicylic Acid-Mediated Defense Signaling Networks. Plant Signal Behav. 2009, 4, 713–717. [Google Scholar] [CrossRef] [Green Version]
- Elwaki, M.A.; El-Metwally, M.A. Hydroquinone, a Promising Antioxidant for Managing Seed Borne Pathogenic Fungi of Peanut. Pak. J. Biol. Sci. 2000, 3, 374–375. [Google Scholar]
- Rasmann, S.; Hiltpold, I.; Ali, J. The Role of Root-Produced Volatile Secondary Metabolites in Mediating Soil Interactions. In Advances in Selected Plant Physiology Aspects; Montanaro, G., Bartolomeo, D., Eds.; InTech: Rijeka, Croatia, 2012; pp. 269–290. [Google Scholar]
- Mumm, R.; Posthumus, M.A.; Dicke, M. Significance of Terpenoids in Induced Indirect Plant Defence against Herbivorous Arthropods. Plant Cell Environ. 2008, 31, 575–585. [Google Scholar] [CrossRef]
- Prisic, S.; Xu, M.M.; Wilderman, P.R.; Peters, R.J. Rice contains Two Disparate Ent-Copalyl Diphosphate Synthases with Distinct Metabolic Functions. Plant Physiol. 2004, 136, 4228–4236. [Google Scholar] [CrossRef] [Green Version]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid Metabolism in Plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [Green Version]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylides, C.; Havaux, M. Carotenoid Oxidation Products are Stress Signals that Mediate Gene Responses to Singlet Oxygen in Plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef] [Green Version]
- Van Norman, J.M.; Zhang, J.; Cazzonelli, C.I.; Pogson, B.J.; Harrison, P.J.; Bugg, T.D.H.; Chan, K.X.; Thompson, A.J.; Benfey, P.N. Periodic Root Branching in Arabidopsis requires Synthesis of an Uncharacterized Carotenoid Derivative. Proc. Natl. Acad. Sci. USA 2014, 111, E1300–E1309. [Google Scholar] [CrossRef] [Green Version]
- Von Wettstein-Knowles, P. Elongases and Epicuticular Wax Biosynthesis. Physiol. Veg. 1982, 20, 797–809. [Google Scholar]
- Bernard, A.; Joubès, J. Arabidopsis Cuticular Waxes: Advances in Synthesis, Export and Regulation. Prog. Lipid Res. 2013, 52, 110–129. [Google Scholar] [CrossRef]
- Gonzales-Vigil, E.; Hefer, C.A.; Von Loessl, M.E.; La Mantia, J.; Mansfield, S.D. Exploiting Natural Variation To uncover an Alkene Biosynthetic Enzyme in Poplar. Plant Cell 2017, 29, 2000–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittstock, U.; Kurzbach, E.; Herfurth, A.M.; Stauber, E. Glucosinolate Breakdown. In Advances in Botanical Research; Kopriva, S., Ed.; Academic Press: London, UK, 2016; pp. 125–169. [Google Scholar]
- Sajid, M.; Khan, M.R.; Hammad Ismail, H.; Latif, S.; Rahim, A.A.; Mehboob, R.; Shah, S.A. Antidiabetic and Antioxidant Potential of Alnus nitida Leaves in Alloxan Induced Diabetic Rats. J. Ethnopharmacol. 2020, 251, 112544. [Google Scholar] [CrossRef] [PubMed]
- Soffietti, R.; Trevisan, E.; Rudà, R. Handbook of Clinical Neurology. In Neurologic Complications of Chemotherapy and Other Newer and Experimental Approaches; Biller, J., Ferro, J.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1199–1218. [Google Scholar]
- Herbers, K.; Meuwly, P.; Frommer, W.B.; Metraux, J.P.; Sonnewald, U. Systemic Acquired Resistance Mediated by the Ectopic Expression of Invertase: Possible Hexose Sensing in the Secretory Pathway. Plant Cell 1996, 8, 793–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usadel, B.; Bläsing, O.E.; Gibon, Y.; Retzlaff, K.; Höhne, M.; Günther, M.; Stitt, M. Global Transcript Levels Respond to Small Changes of the Carbon Status during Progressive Exhaustion of Carbohydrates in Arabidopsis Rosettes. Plant Physiol. 2008, 146, 1834–1861. [Google Scholar] [CrossRef] [Green Version]
- Baena-González, E.; Sheen, J. Convergent Energy and Stress Signalling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Watson, M.B.; Malmberg, R.L. Arginine Decarboxylase (Polyamine Synthesis) Mutants of Arabidopsis thaliana Exhibit Altered Root Growth. Plant J. 1998, 13, 231–239. [Google Scholar] [CrossRef]
- Evans, P.T.; Malmberg, R.L. Do Polyamines Have Roles in Plant Development? Annu. Rev. Plant Physiol. 1989, 40, 235–269. [Google Scholar] [CrossRef]
- Walters, D.R. Polyamines in Plant-Microbe Interactions. Physiol. Mol. Plant Path. 2000, 57, 137–146. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; Torres, C.; Campos, L.; Lisón, P.; Rodrigo, I.; Bellés, J.M.; Conejero, V. Identification of Defence Metabolites in Tomato Plants Infected by the Bacterial Pathogen Pseudomonas syringae. Environ. Exp. Bot. 2011, 74, 216–228. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing Plant Surfaces: Cuticular Wax Formation by Epidermal Cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [Green Version]

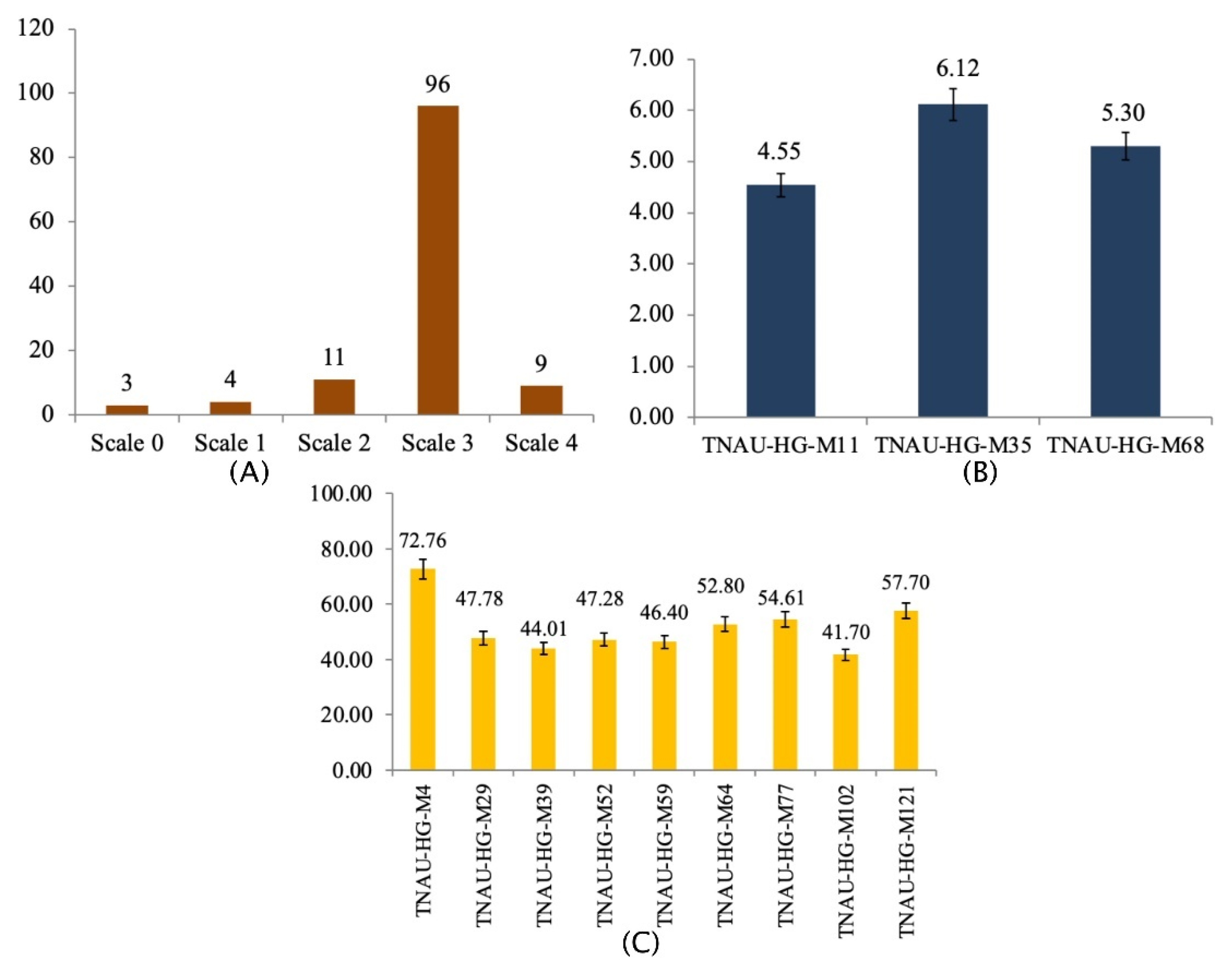


| Description | Score | Disease Reaction |
|---|---|---|
| No symptoms of powdery mildew growth | 0 | R |
| Small and sparse mycelial growth. Visualization of non-significant sporulation | 1 | R |
| Macroscopic: Slight mycelia growth Microscopic: Slight to moderate mycelia growth with conidiophores | 2 | R |
| Macroscopic: Moderate mycelial growth Microscopic: Moderate mycelial development with moderate to heavy sporulation | 3 | S |
| Macroscopic: Heavy mycelia growth Microscopic: Heavy mycelia growth with copious sporulation | 4 | S |
| Traits | Filed Condition | Shade-Net Condition | ||||||
|---|---|---|---|---|---|---|---|---|
| Inoculated | Control | Inoculated | Control | |||||
| SM | RM | SM | RM | SM | RM | SM | RM | |
| 1 | DuG | DaG | DaG | DaG | DuG | DaG | DaG | DaG |
| 2 | 0.32 ± 0.01 | 0.88 ± 0.01 | 0.86 ± 0.01 | 0.89 ± 0.01 | 0.33 ± 0.02 | 0.79 ± 0.02 | 0.77 ± 0.01 | 0.79 ± 0.01 |
| 3 | 73.80 ± 0.66 | 78.20 ± 1.04 | 74.40 ± 0.61 | 82.00 ± 0.63 | 56.20 ± 0.96 | 72.80 ± 0.52 | 74.80 ± 0.34 | 79.60 ± 0.46 |
| 4 | 44.00 ± 0.75 | 144.40 ± 1.00 | 135.20 ± 0.52 | 153.40 ± 0.67 | 61.20 ± 0.77 | 132.60 ± 0.46 | 130.40 ± 0.47 | 141.60 ± 0.61 |
| 5 | 1.40 ± 0.22 | 5.60 ± 0.22 | 5.20 ± 0.18 | 5.40 ± 0.22 | 1.20 ± 0.18 | 5.00 ± 0.28 | 4.80 ± 0.34 | 5.60 ± 0.22 |
| 6 | 74.00 ± 1.10 | 119.00 ± 0.63 | 121.00 ± 0.52 | 124.40 ± 0.61 | 72.40 ± 0.73 | 116.60 ± 0.46 | 117.80 ± 0.52 | 119.00 ± 0.63 |
| 7 | 14.12 ± 0.73 | 50.40 ± 0.92 | 52.20 ± 0.77 | 52.80 ± 0.77 | 14.60 ± 0.46 | 46.80 ± 0.59 | 50.20 ± 0.77 | 50.60 ± 0.61 |
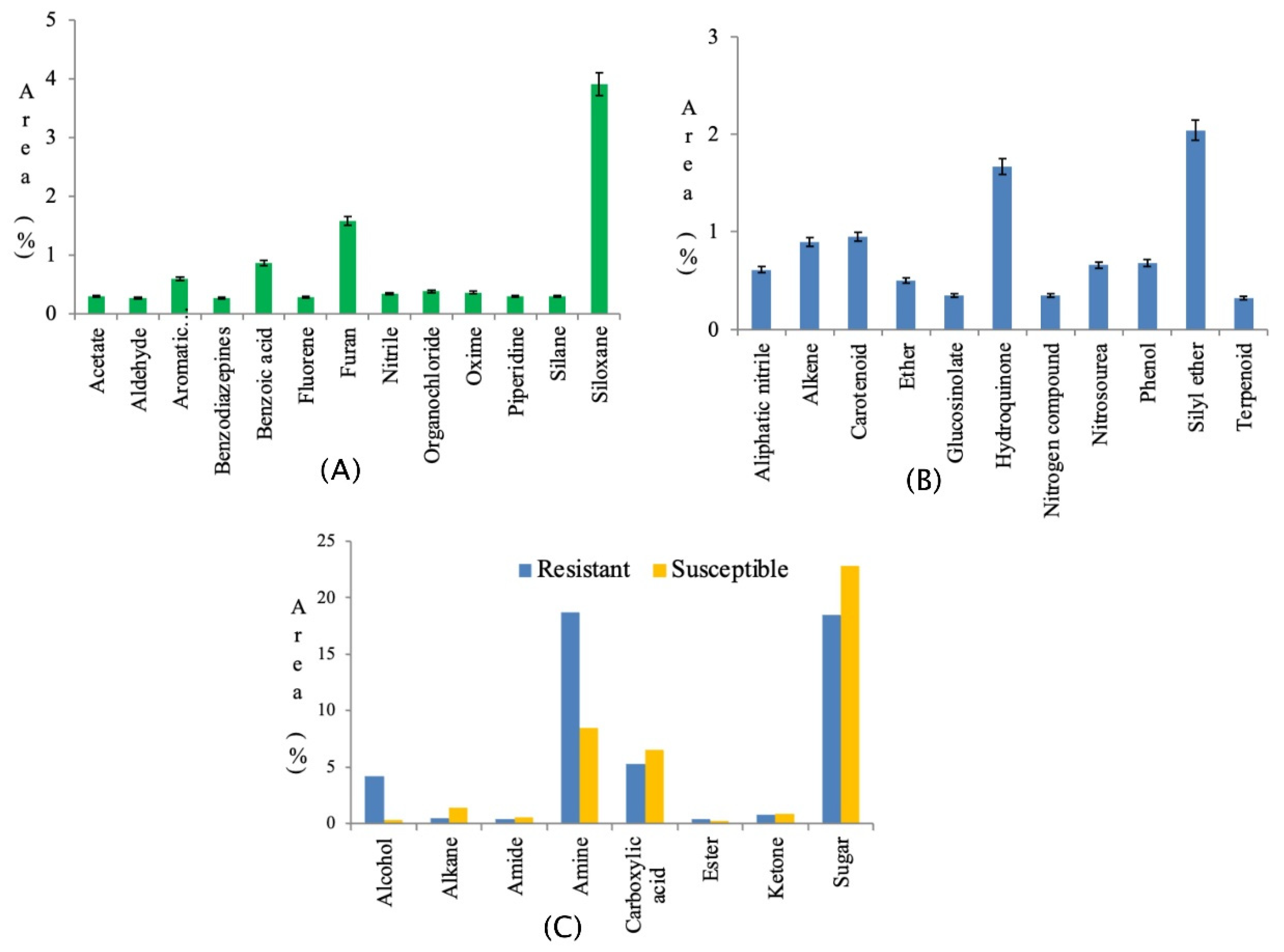
| S. No | Class | Peak Area (%) | S. No | Class | Peak Area (%) | ||
|---|---|---|---|---|---|---|---|
| R | S | R | S | ||||
| 1 | Acetate | 0.29 | - | 17 | Furan | 1.58 | - |
| 2 | Alcohol | 4.18 | 0.30 | 18 | Glucosinolate | - | 0.35 |
| 3 | Aldehyde | 0.27 | - | 19 | Hydroquinone | - | 1.67 |
| 4 | Aliphatic nitrile | - | 0.61 | 20 | Ketone | 0.79 | 0.86 |
| 5 | Alkane | 0.49 | 1.42 | 21 | Nitrile | 0.34 | - |
| 6 | Alkene | - | 0.90 | 22 | Nitrogen compound | - | 0.35 |
| 7 | Amide | 0.43 | 0.56 | 23 | Nitrosourea | - | 0.66 |
| 8 | Amine | 18.70 | 8.46 | 24 | Organochloride | 0.38 | - |
| 9 | Aromatic dicarboxylic acid | 0.60 | - | 25 | Oxime | 0.36 | - |
| 10 | Benzodiazepines | 0.27 | - | 26 | Phenol | - | 0.68 |
| 11 | Benzoic acid | 0.86 | - | 27 | Piperidine | 0.30 | - |
| 12 | Carboxylic acid | 5.30 | 6.5 | 28 | Silane | 0.30 | - |
| 13 | Carotenoid | - | 0.95 | 29 | Siloxane | 3.91 | - |
| 14 | Ester | 0.37 | 0.26 | 30 | Silyl ether | - | 2.04 |
| 15 | Ether | - | 0.50 | 31 | Sugar | 18.47 | 22.79 |
| 16 | Fluorene | 0.28 | - | 32 | Terpenoid | - | 0.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sudhagar, R.; Priyanka, S.; Chockalingam, V.; Sendhilvel, V.; Souframanien, J.; Raja, K.; Kanagarajan, S. Development and Metabolic Characterization of Horse Gram (Macrotyloma uniflorum Lam. (Verdc.)) Mutants for Powdery Mildew Resistance. Agronomy 2022, 12, 800. https://doi.org/10.3390/agronomy12040800
Sudhagar R, Priyanka S, Chockalingam V, Sendhilvel V, Souframanien J, Raja K, Kanagarajan S. Development and Metabolic Characterization of Horse Gram (Macrotyloma uniflorum Lam. (Verdc.)) Mutants for Powdery Mildew Resistance. Agronomy. 2022; 12(4):800. https://doi.org/10.3390/agronomy12040800
Chicago/Turabian StyleSudhagar, Rajaprakasam, Shanmugavel Priyanka, Vanniarajan Chockalingam, Vaithiyanathan Sendhilvel, Jegadeesan Souframanien, Kalimuthu Raja, and Selvaraju Kanagarajan. 2022. "Development and Metabolic Characterization of Horse Gram (Macrotyloma uniflorum Lam. (Verdc.)) Mutants for Powdery Mildew Resistance" Agronomy 12, no. 4: 800. https://doi.org/10.3390/agronomy12040800






