Comparative Transcriptional Analysis of Two Contrasting Rice Genotypes in Response to Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Salt Treatment
2.2. cDNA Library Preparation and Sequencing
2.3. RNA-Seq Data Analysis
2.4. Co-Expressed Gene Network Analysis
2.5. qRT-PCR Analysis
3. Results
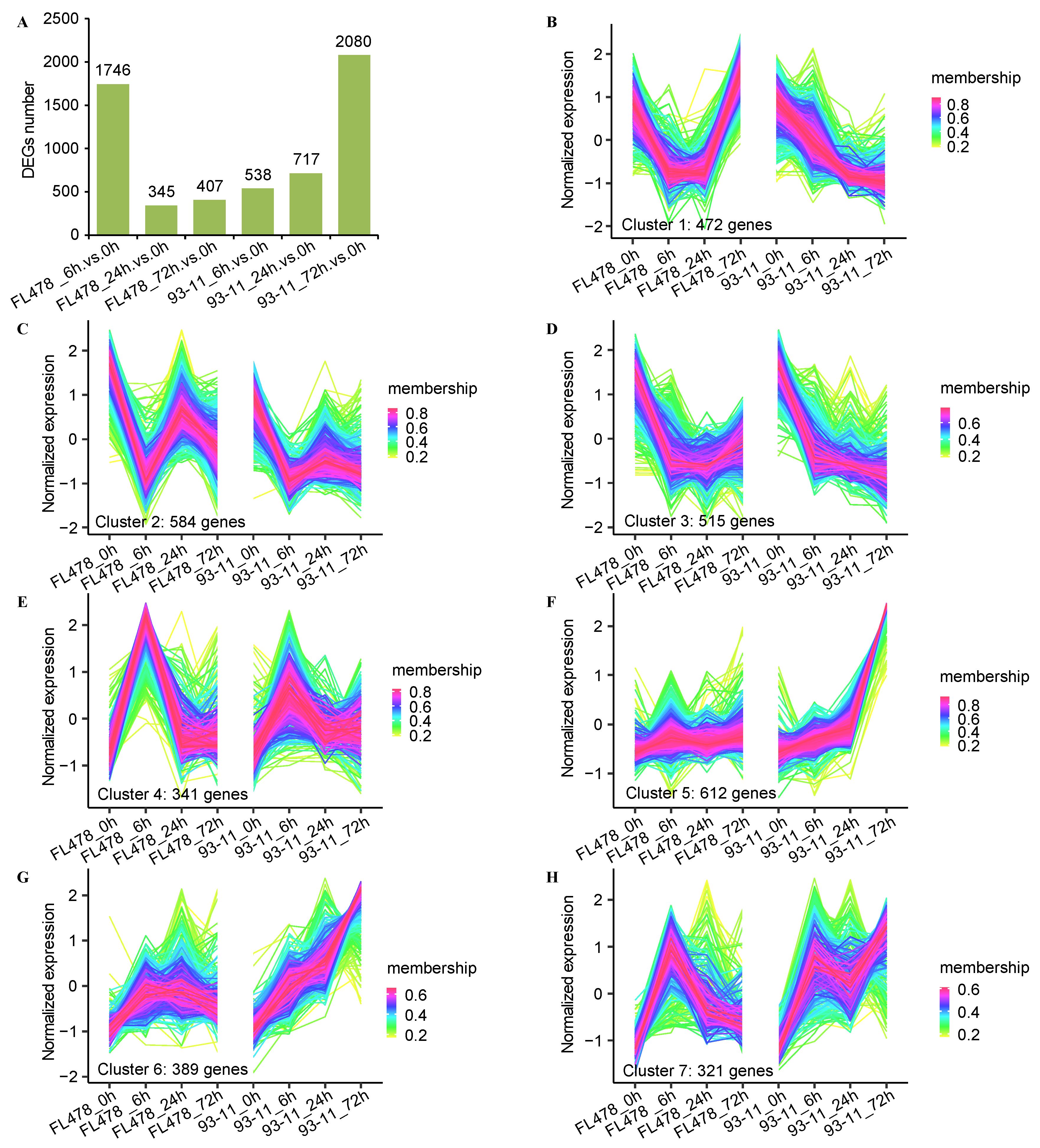
3.1. DEGs Identification and Cluster Analysis
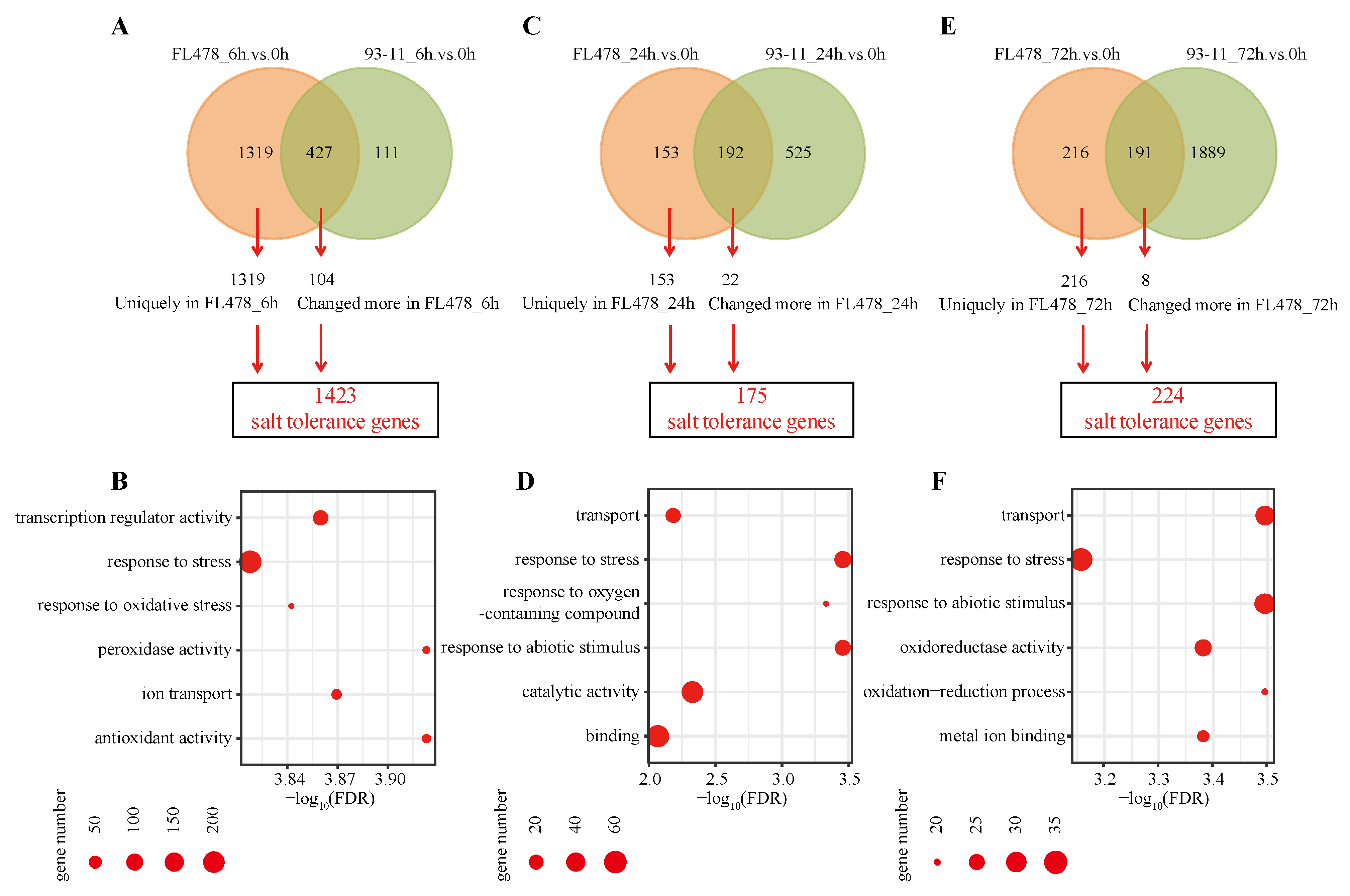
3.2. Salt Tolerance Genes Play Pivotal Roles in FL478 Compared with 93-11
3.3. Enhanced Transport Ability Is Important for the Adaption to Salt Stress
3.4. Differential Expression of Transcription Factors and ROS Scavenging Genes Is Correlated to Salt Tolerance
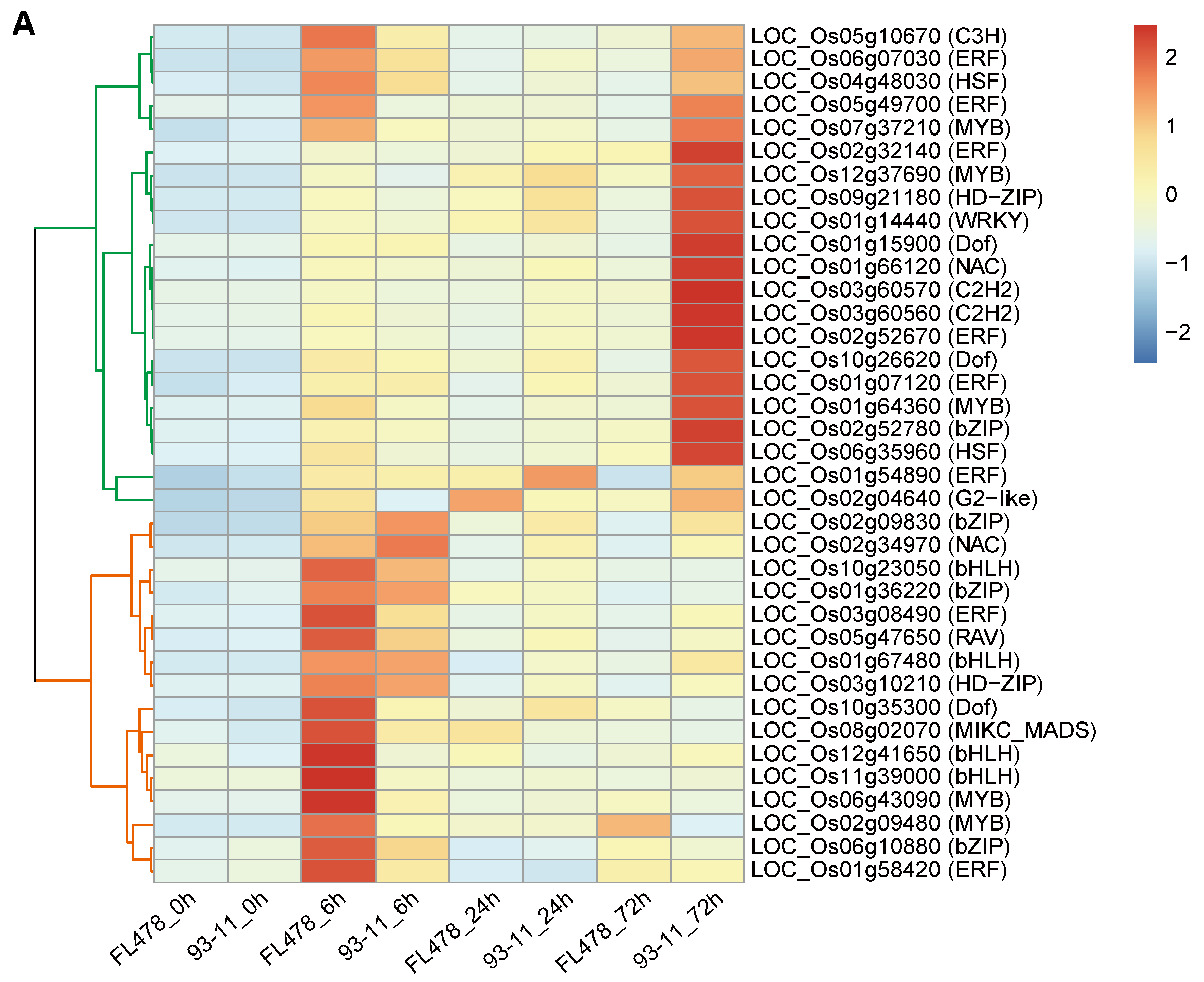

3.5. Interaction Network of Key Genes Relevant to Salt Tolerance
3.6. Validation of the DEGs by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010, 37, 613–620. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 29, 403–433. [Google Scholar]
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Li, Y.; Zhou, Z.; Xiang, X.; Liu, X.; Wang, J.; Hu, Z.; Xiang, S.; Li, W.; Xiao, Q.; et al. Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidase NtRbohE expression. Plant Physiol. 2021, 186, 1706–1720. [Google Scholar] [CrossRef]
- He, L.; Bian, J.; Xu, J.; Yang, K. Novel Maize NAC Transcriptional Repressor ZmNAC071 Confers Enhanced Sensitivity to ABA and Osmotic Stress by Downregulating Stress-Responsive Genes in Transgenic Arabidopsis. J. Agric. Food Chem. 2019, 67, 8905–8918. [Google Scholar] [CrossRef]
- Cotsaftis, O.; Plett, D.; Johnson, A.A.; Walia, H.; Wilson, C.; Ismail, A.M.; Close, T.J.; Tester, M.; Baumann, U. Root-Specific Transcript Profiling of Contrasting Rice Genotypes in Response to Salinity Stress. Mol. Plant 2011, 4, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Senadheera, P.; Singh, R.K.; Maathuis, F.J.M. Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. J. Exp. Bot. 2009, 60, 2553–2563. [Google Scholar] [CrossRef] [Green Version]
- Walia, H.; Wilson, C.; Condamine, P.; Liu, X.; Ismail, A.M.; Zeng, L.; Wanamaker, S.I.; Mandal, J.; Xu, J.; Cui, X.; et al. Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes under Salinity Stress during the Vegetative Growth Stage. Plant Physiol. 2005, 139, 822–835. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Fung-Leung, W.-P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and Microarray in Transcriptome Profiling of Activated T Cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Ye, X.; Guo, P.; Hu, Z.; Qi, G.; Cui, F.; Liu, S. An S-ribonuclease binding protein EBS1 and brassinolide signaling are specifically required for Arabidopsis tolerance to bicarbonate. J. Exp. Bot. 2021, 72, 1449–1459. [Google Scholar] [CrossRef]
- Ye, X.; Zhao, X.; Sun, Y.; Zhang, M.; Feng, S.; Zhou, A.; Wu, W.; Ma, S.; Liu, S. The underlying molecular conservation and diversification of dioecious flower and leaf buds provide insights into the development, dormancy breaking, flowering, and sex association of willows. Plant Physiol. Biochem. 2021, 167, 651–664. [Google Scholar] [CrossRef]
- Ye, X.; Wang, H.; Cao, X.; Jin, X.; Cui, F.; Bu, Y.; Liu, H.; Wu, W.; Takano, T.; Liu, S. Transcriptome profiling of Puccinellia tenuiflora during seed germination under a long-term saline-alkali stress. BMC Genom. 2019, 20, 589. [Google Scholar] [CrossRef]
- Jahan, N.; Lv, Y.; Song, M.; Zhang, Y.; Shang, L.; Lu, Y.; Ye, G.; Qian, Q.; Gao, Z.; Guo, L. Transcriptomic Analysis of Short-Term Salt-Stress Response in Mega Hybrid Rice Seedlings. Agronomy 2021, 11, 1328. [Google Scholar] [CrossRef]
- Zhou, X.; Dou, Y.; Huang, X.; Li, G.; Zhang, H.; Jiang, D.; Fan, J.; Condori-Apfata, J.; Liu, X.; Condori-Pacsi, S.; et al. Using Principal Component Analysis and RNA-Seq to Identify Candidate Genes That Control Salt Tolerance in Garlic (Allium sativum L.). Agronomy 2021, 11, 691. [Google Scholar] [CrossRef]
- Mansuri, R.M.; Shobbar, Z.-S.; Jelodar, N.B.; Ghaffari, M.R.; Nematzadeh, G.; Asari, S. Dissecting molecular mechanisms underlying salt tolerance in rice: A comparative transcriptional profiling of the contrasting genotypes. Rice 2019, 12, 13. [Google Scholar] [CrossRef]
- Cartagena, J.A.; Yao, Y.; Mitsuya, S.; Tsuge, T. Comparative transcriptome analysis of root types in salt tolerant and sensitive rice varieties in response to salinity stress. Physiol. Plant. 2021, 173, 1629–1642. [Google Scholar] [CrossRef]
- Gregorio, G.; Senadhir, D.; Mendoza, R. Screening Rice for Salinity Tolerance; IRRI Discussion Paper Series; IRRI: Los Baños, Philippines, 1997; Volume 22. [Google Scholar]
- Zhao, L.; Lin, J.; Meng, J.; Ming-Li, G.; Ying-Hao, C.; Li-Juan, L.; Zhen-Wei, C.; Li-Yun, L.; Guo-Zhen, L. Expression on profiling of rice PP2Ac type phosphatase proteins in seedlings under salt-stressed conditions. Sci. Agric. Sin. 2012, 45, 2339–2345. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Ding, Z.; Tie, W.; Yan, Y.; Liu, Y.; Wu, C.; Liu, J.; Wang, J.; Peng, M.; Xu, B.; et al. Comparative physiological and transcriptomic analyses provide integrated insight into osmotic, cold, and salt stress tolerance mechanisms in banana. Sci. Rep. 2017, 7, 43007. [Google Scholar] [CrossRef]
- Wu, M.; Gu, L. TCseq: Time Course Sequencing Data Analysis. R Package Version 1.16.0. 2021. Available online: http://bioconductor.riken.jp/packages/release/bioc/html/TCseq.html (accessed on 25 January 2022).
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Ding, Z.; Han, B.; Hu, W.; Li, Y.; Zhang, J. Physiological Investigation and Transcriptome Analysis of Polyethylene Glycol (PEG)-Induced Dehydration Stress in Cassava. Int. J. Mol. Sci. 2016, 17, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Caverzan, A.; Casassola, A.; Brammer, S.P. Antioxidant responses of wheat plants under stress. Genet. Mol. Biol. 2016, 39, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Cellier, F.; Conejero, G.; Ricaud, L.; Luu, D.T.; Lepetit, M.; Gosti, F.; Casse, F. Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J. 2004, 39, 834–846. [Google Scholar] [CrossRef]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-Driven Potassium Uptake by the Plant Potassium Transporter HKT1 and Mutations Conferring Salt Tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Y.; Wang, Y.; Gao, C. Vacuolar membrane H+-ATPase c subunit gene (ThVHAc 1) from Tamarix hispida Willd improves salt stress tolerance. Plant Physiol. Biochem. 2020, 157, 370–378. [Google Scholar] [CrossRef]
- Kabała, K.; Janicka-Russak, M.; Reda, M.; Migocka, M. Transcriptional regulation of the V-ATPase subunit c and V-PPase isoforms in Cucumis sativus under heavy metal stress. Physiol. Plantarum. 2014, 150, 32–45. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, X.; Song, Y.; Zhu, L.; Yu, Z.; Gan, L.; Zhou, S.; Liu, H.; Wen, F.; Zhu, C. NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 2018, 8, 8873. [Google Scholar] [CrossRef] [Green Version]
- Dhar, N.; Caruana, J.; Erdem, I.; Raina, R. An Arabidopsis disease related nonspecific lipid transfer protein 1 is required for resistance against various phytopathogens and tolerance to salt stress. Gene 2020, 753, 144802. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.-C. Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein 15 affects seed coat permeability in Arabidopsis. Plant J. 2018, 96, 1206–1217. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H.; et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 2021, 21, 54. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. OsNAC45 is Involved in ABA Response and Salt Tolerance in Rice. Rice 2020, 13, 79. [Google Scholar] [CrossRef]
- Kim, S.Y. The role of ABF family bZIP class transcription factors in stress response. Physiol. Plant. 2006, 126, 519–527. [Google Scholar] [CrossRef]
- Piao, W.; Sakuraba, Y.; Paek, N.C. Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thaliana. BMB Rep. 2019, 52, 653–658. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, L.; Zhou, K.; Zhang, Y.; Han, X.; Din, Y.; Ge, X.; Qin, W.; Wang, P.; Li, F.; et al. GhWRKY6 Acts as a Negative Regulator in Both Transgenic Arabidopsis and Cotton during Drought and Salt Stress. Front. Genet. 2019, 10, 392. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Yu, F.; Hu, B.; Jia, Y.; Sha, H.; Zhao, H. Differential activity of the antioxidant defence system and alterations in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Sci. Rep. 2019, 9, 8543. [Google Scholar] [CrossRef]
- Moradi, F.; Ismail, A.M. Responses of Photosynthesis, Chlorophyll Fluorescence and ROS-Scavenging Systems to Salt Stress During Seedling and Reproductive Stages in Rice. Ann. Bot. 2007, 99, 1161–1173. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, X.; Tie, W.; Xu, J.; Ding, Z.; Hu, W. Comparative Transcriptional Analysis of Two Contrasting Rice Genotypes in Response to Salt Stress. Agronomy 2022, 12, 1163. https://doi.org/10.3390/agronomy12051163
Ye X, Tie W, Xu J, Ding Z, Hu W. Comparative Transcriptional Analysis of Two Contrasting Rice Genotypes in Response to Salt Stress. Agronomy. 2022; 12(5):1163. https://doi.org/10.3390/agronomy12051163
Chicago/Turabian StyleYe, Xiaoxue, Weiwei Tie, Jianlong Xu, Zehong Ding, and Wei Hu. 2022. "Comparative Transcriptional Analysis of Two Contrasting Rice Genotypes in Response to Salt Stress" Agronomy 12, no. 5: 1163. https://doi.org/10.3390/agronomy12051163




