Throwing Copper Around: How Plants Control Uptake, Distribution, and Accumulation of Copper
Abstract
:1. Introduction
2. Copper in Soils
3. Plant Responses to Copper Toxicity
3.1. Effects of Copper Excess on Seed Germination
3.2. Effects of Copper Excess on Anatomical/Morphological Changes
3.3. Effects of Copper Excess on Photosynthetic Apparatus
3.4. Effects of Copper Excess on the Uptake and Accumulation of Other Mineral Nutrients
3.5. Plant Tolerance to Copper Toxicity
4. Copper Homeostasis in Model Plant Species
4.1. Regulation of Copper Uptake in Roots
4.2. Copper Uptake from the Soil
4.3. COPT Transporters in Arabidopsis thaliana
4.4. COPT Proteins Characterized in Other Species
4.5. A Possible Role of ZIPs in Copper Homeostasis
4.6. PIB-Type ATPase/Heavy Metal-Associated (HMA) Proteins and Their Role in Copper Homeostasis in Arabidopsis thaliana
4.7. HMA Copper Transporters in Other Species
4.8. Copper Chaperones in Model Species
4.9. The Role of Yellow Stripe-like (YSL) Transporters in Copper Homeostasis
5. Iron and Copper Crosstalk
5.1. Iron and Copper Crosstalk in Arabidopsis thaliana
5.2. Iron and Copper Crosstalk in Rice
6. Copper-Hyperaccumulating Plants
7. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Aguirre, G.; Pilon, M. Copper Delivery to Chloroplast Proteins and Its Regulation. Front. Plant Sci. 2016, 6, 1250. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Pandita, S.; Preet, G.S.S.; Sharma, A.; Khanna, K.; Kaur, P.; Shreeya, A.; Setia, R. Copper Bioavailability, Uptake, Toxicity and Tolerance in Plants: A Comprehensive Review. Chemosphere 2021, 262, 127810. [Google Scholar] [CrossRef] [PubMed]
- Ogunkunle, C.O.; Bornmann, B.; Wagner, R.; Fatoba, P.O.; Frahm, R.; Lützenkirchen-Hecht, D. Copper Uptake, Tissue Partitioning and Biotransformation Evidence by XANES in Cowpea (Vigna unguiculata L) Grown in Soil Amended with Nano-Sized Copper Particles. Environ. Nanotechnol. Monit. Manag. 2019, 12, 100231. [Google Scholar] [CrossRef]
- Printz, B.; Lutts, S.; Hausman, J.; Sergeant, K. Copper Trafficking in Plants and Its Implication on Cell Wall Dynamics. Front. Plant Sci. 2016, 7, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabbir, Z.; Sardar, A.; Shabbir, A.; Abbas, G.; Shamshad, S. Copper Uptake, Essentiality, Toxicity, Detoxification and Risk Assessment in Soil-Plant Environment. Chemosphere 2020, 259, 127436. [Google Scholar] [CrossRef] [PubMed]
- Whitby, H.; Posacka, A.M.; Maldonado, M.T.; van den Berg, C.M.G. Copper-Binding Ligands in the NE Pacific. Mar. Chem. 2018, 204, 36–48. [Google Scholar] [CrossRef]
- Cohu, C.; Pilon, M. Cell Biology of Copper. In Cell Biology of Metals and Nutrients-Plant Cell Monographs; Rüdiger, H., Mendel, R.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 55–74. [Google Scholar]
- Ghazaryan, K.; Movsesyan, H.; Ghazaryan, N.; Watts, B.A. Copper Phytoremediation Potential of Wild Plant Species Growing in the Mine Polluted Areas of Armenia. Environ. Pollut. 2019, 249, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Festa, R.A.; Thiele, D.J. Copper: An Essential Metal in Biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, B.; van der Ent, A.; Baker, A.J.M.; Echevarria, G.; Mahy, G.; Malaisse, F.; Meerts, P.; Pourret, O.; Verbruggen, N.; Faucon, M.P. Copper and Cobalt Accumulation in Plants: A Critical Assessment of the Current State of Knowledge. New Phytol. 2017, 213, 537–551. [Google Scholar] [CrossRef]
- Hänsch, R.; Mendel, R.R. Physiological Functions of Mineral Micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef]
- Ravet, K.; Pilon, M. Copper and Iron Homeostasis in Plants: The Challenges of Oxidative Stress. Antioxid. Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, L.; Welchen, E.; Gonzalez, D.H. Mitochondria and Copper Homeostasis in Plants. Mitochondrion 2014, 19, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Peñarrubia, L.; Andrés-Colás, N.; Moreno, J.; Puig, S. Regulation of Copper Transport in Arabidopsis thaliana: A Biochemical Oscillator? J. Biol. Inorg. Chem. 2010, 15, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper Homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef]
- Palmer, C.M.; Guerinot, M.L. Facing the Challenges of Cu, Fe and Zn Homeostasis in Plants. Nat. Chem. Biol. 2009, 5, 333–340. [Google Scholar] [CrossRef] [Green Version]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and Metal Sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef]
- Andresen, E.; Peiter, E.; Küpper, H. Trace Metal Metabolism in Plants. J. Exp. Bot. 2018, 69, 909–954. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, X.; Ma, J.; Yang, H.; Zhang, W.; Li, C. Genotypic Differences and Glutathione Metabolism Response in Wheat Exposed to Copper. Environ. Exp. Bot. 2019, 157, 250–259. [Google Scholar] [CrossRef]
- Orłowska, R.; Zimny, J.; Bednarek, P.T. Copper Ions Induce DNA Sequence Variation in Zygotic Embryo Culture-Derived Barley Regenerants. Front. Plant Sci. 2021, 11, 614837. [Google Scholar] [CrossRef]
- Tavladoraki, P.; Cona, A.; Angelini, R. Copper-Containing Amine Oxidases and FAD-Dependent Polyamine Oxidases Are Key Players in Plant Tissue Differentiation and Organ Development. Front. Plant Sci. 2016, 7, 824. [Google Scholar] [CrossRef] [Green Version]
- Fraudentali, I.; Rodrigues-Pousada, R.A.; Angelini, R.; Ghuge, S.A.; Cona, A. Plant Copper Amine Oxidases: Key Players in Hormone Signaling Leading to Stress-Induced Phenotypic Plasticity. Int. J. Mol. Sci. 2021, 22, 5136. [Google Scholar] [CrossRef] [PubMed]
- Rather, B.A.; Masood, A.; Sehar, Z.; Majid, A.; Anjum, N.A.; Khan, N.A. Mechanisms and Role of Nitric Oxide in Phytotoxicity-Mitigation of Copper. Front. Plant Sci. 2020, 11, 375. [Google Scholar] [CrossRef] [PubMed]
- Komárek, M.; Cadková, E.; Chrastný, V.; Bordas, F.; Bollinger, J.-C. Contamination of Vineyard Soils with Fungicides: A Review of Environmental and Toxicological Aspects. Environ. Int. 2010, 36, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E.; Anwar, M.; Sajad, A. Phytoremediation of Heavy Metals—Concepts and Applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef]
- Miotto, A.; Ceretta, C.A.; Girotto, E.; Trentin, G.; Kaminski, J.; De Conti, L.; Moreno, T.; Elena, B.; Brunetto, G. Copper Accumulation and Availability in Sandy, Acid, Vineyard Soils. Commun. Soil Sci. Plant Anal. 2017, 48, 1167–1183. [Google Scholar] [CrossRef]
- Ballabio, C.; Panagos, P.; Lugato, E.; Huang, J.-H.; Orgiazzi, A.; Jones, A.; Fernández-Ugalde, O.; Borrelli, P.; Montanarella, L. Copper Distribution in European Topsoils: An Assessment Based on LUCAS Soil Survey. Sci. Total Environ. 2018, 636, 282–298. [Google Scholar] [CrossRef]
- Brunetto, G.; De Melo, G.W.B.; Terzano, R.; De Buonol, D.; Astolfi, S.; Tomasi, N.; Pii, Y.; Mimmo, T.; Cesco, S. Copper Accumulation in Vineyard Soils: Rhizosphere Processes and Agronomic Practices to Limit Its Toxicity. Chemosphere 2016, 162, 293–307. [Google Scholar] [CrossRef] [Green Version]
- De Conti, L.; Ceretta, C.A.; Melo, G.W.B.; Tiecher, T.L.; Silva, L.O.S.; Garlet, L.P.; Mimmo, T.; Cesco, S.; Brunetto, G. Intercropping of Young Grapevines with Native Grasses for Phytoremediation of Cu-Contaminated Soils. Chemosphere 2019, 216, 147–156. [Google Scholar] [CrossRef] [Green Version]
- McBride, M.B. Environmental Chemistry of Soils 1994; Oxford Press: New York, NY, USA, 1994. [Google Scholar]
- Sparks, D. Environmental Soil Chemistry, 1st ed.; Academic Press: San Diego, CA, USA, 1995. [Google Scholar]
- Xu, L.; Xing, X.; Peng, J.; Ji, M. Effects of in situ Remediation on Copper Distribution and Soil Aggregate Adsorption–Desorption Characteristics in Smelter-Impacted Soil. Front. Environ. Sci. 2022, 10, 151. [Google Scholar] [CrossRef]
- Bradl, H.B. Adsorption of Heavy Metal Ions on Soils and Soils Constituents. J. Colloid Interface Sci. 2004, 277, 187. [Google Scholar] [CrossRef]
- Alleoni, L.R.F.; Iglesias, C.S.M.; de Mello, S.C.; Camargo, O.A.; Casagrande, J.C.; Lavorenti, N.A. Atributos Do Solo Relacionados à Adsorção de Cádmio e Cobre Em Solos Tropicais. Acta Sci. 2005, 27, 729–737. [Google Scholar] [CrossRef] [Green Version]
- Brunetto, G.; Miotto, A.; Ceretta, C.A.; Eugênio, D.; Heinzen, J.; De Moraes, M.P.; Canton, L.; Tiecher, L.; Comin, J.J.; Girotto, E.; et al. Mobility of Copper and Zinc Fractions in Fungicide-Amended Vineyard Sandy Soils. Arch. Agron. Soil Sci. 2014, 60, 609–624. [Google Scholar] [CrossRef]
- Park, J.H.; Lamb, D.; Paneerselvam, P.; Choppala, G.; Bolan, N.; Chung, J.-W. Role of Organic Amendments on Enhanced Bioremediation of Heavy Metal (Loid) Contaminated Soils. J. Hazard. Mater. 2011, 185, 549–574. [Google Scholar] [CrossRef] [PubMed]
- Nolan, A.L.; Mclaughlin, M.J.; Mason, S.D. Chemical Speciation of Zn, Cd, Cu, and Pb in Pore Waters of Agricultural and Contaminated Soils Using Donnan Dialysis. Environ. Sci. Technol. 2003, 37, 90–98. [Google Scholar] [CrossRef] [PubMed]
- De Conti, L.; Ceretta, C.A.; Tiecher, T.L.; da Silva, L.O.S.; Tassinari, A.; Somavilla, L.M.; Mimmo, T.; Cesco, S.; Brunetto, G. Growth and Chemical Changes in the Rhizosphere of Black Oat (Avena strigosa) Grown in Soils Contaminated with Copper. Ecotoxicol. Environ. Saf. 2018, 163, 19–27. [Google Scholar] [CrossRef] [PubMed]
- De Conti, L.; Ceretta, C.A.; Ferreira, P.A.A.; Lourenzi, C.R.; Girotto, E.; Lorensini, F.; Tiecher, T.L.; Marchezan, C.; Anchieta, M.G.; Brunetto, G. Soil Solution Concentrations and Chemical Species of Copper and Zinc in a Soil with a History of Pig Slurry Application and Plant Cultivation. Agric. Ecosyst. Environ. 2016, 216, 374–386. [Google Scholar] [CrossRef]
- Babcsányi, I.; Chabaux, F.; Granet, M.; Meite, F.; Payraudeau, S.; Duplay, J.; Imfeld, G. Copper in Soil Fractions and Runoff in a Vineyard Catchment: Insights from Copper Stable Isotopes. Sci. Total Environ. 2016, 557–558, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Angelaki, A.; Dionysidis, A.; Sihag, P.; Golia, E.E. Assessment of Contamination Management Caused by Copper and Zinc Cations Leaching and Their Impact on the Hydraulic Properties of a Sandy and a Loamy Clay Soil. Land 2022, 11, 290. [Google Scholar] [CrossRef]
- Pérez-Esteban, J.; Escolástico, C.; Masaguer, A.; Vargas, C.; Moliner, A. Soluble Organic Carbon and pH of Organic Amendments Affect Metal Mobility and Chemical Speciation in Mine Soils. Chemosphere 2014, 103, 164–171. [Google Scholar] [CrossRef]
- Kim, K.; Owens, G.; Naidu, R.; Kwon, S. Influence of Plant Roots on Rhizosphere Soil Solution Composition of Long-Term Contaminated Soils. Geoderma 2010, 155, 86–92. [Google Scholar] [CrossRef]
- Dresler, S.; Hanaka, A.; Bednarek, W.; Maksymiec, W. Accumulation of Low-Molecular-Weight Organic Acids in Roots and Leaf Segments of Zea mays Plants Treated with Cadmium and Copper. Acta Physiol. Plant. 2014, 36, 1565–1575. [Google Scholar] [CrossRef] [Green Version]
- de Conti, L.; Cesco, S.; Mimmo, T.; Pii, Y.; Valentinuzzi, F.; Melo, G.W.B.; Ceretta, C.A.; Trentin, E.; Marques, A.C.R.; Brunetto, G. Iron Fertilization to Enhance Tolerance Mechanisms to Copper Toxicity of Ryegrass Plants Used as Cover Crop in Vineyards. Chemosphere 2020, 243, 125298. [Google Scholar] [CrossRef] [PubMed]
- Chmielowska-Bak, J.; Deckert, J. Plant Recovery after Metal Stress—A Review. Plants 2021, 10, 450. [Google Scholar] [CrossRef] [PubMed]
- Küpper, H.; Andresen, E. Mechanisms of Metal Toxicity in Plants. Metallomics 2016, 8, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Khoudi, H. Significance of Vacuolar Proton Pumps and Metal/H+ Antiporters in Plant Heavy Metal Tolerance. Physiol. Plant. 2021, 173, 384–393. [Google Scholar] [CrossRef]
- Yadav, V.; Arif, N.; Kováč, J.; Singh, V.P.; Tripathi, D.K.; Chauhan, D.K.; Vaculík, M. Structural Modifications of Plant Organs and Tissues by Metals and Metalloids in the Environment: A Review. Plant Physiol. Biochem. 2021, 159, 100–112. [Google Scholar] [CrossRef]
- Adrees, M.; Ali, S.; Rizwan, M.; Ibrahim, M.; Abbas, F.; Farid, M.; Zia-ur-Rehman, M.; Irshad, M.K.; Bharwana, S.A. The Effect of Excess Copper on Growth and Physiology of Important Food Crops: A Review. Environ. Sci. Pollut. Res. 2015, 22, 8148–8162. [Google Scholar] [CrossRef]
- Lux, A.; Martinka, M.; Vaculík, M.; White, P.J. Root Responses to Cadmium in the Rhizosphere: A Review. J. Exp. Bot. 2011, 62, 21–37. [Google Scholar] [CrossRef] [Green Version]
- Krzesłowska, M. The Cell Wall in Plant Cell Response to Trace Metals: Polysaccharide Remodeling and Its Role in Defense Strategy. Acta Physiol. Plant. 2011, 33, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.F.; Lei, G.J.; Wang, Z.W.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Coordination between Apoplastic and Symplastic Detoxification Confers Plant Aluminum Resistance. Plant Physiol. 2013, 162, 1947–1955. [Google Scholar] [CrossRef] [Green Version]
- O’Lexy, R.; Kasai, K.; Clark, N.; Fujiwara, T.; Sozzani, R.; Gallagher, K.L. Exposure to Heavy Metal Stress Triggers Changes in Plasmodesmatal Permeability via Deposition and Breakdown of Callose. J. Exp. Bot. 2018, 69, 3715–3728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.L. Cellular Mechanisms for Heavy Metal Detoxification and Tolerance. J. Exp. Bot. 2002, 53, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.R.; Pichtel, J.; Hayat, S. Copper: Uptake, Toxicity and Tolerance in Plants and Management of Cu-Contaminated Soil. BioMetals 2021, 34, 737–759. [Google Scholar] [CrossRef] [PubMed]
- Baruah, N.; Mondal, S.C.; Farooq, M.; Gogoi, N. Influence of Heavy Metals on Seed Germination and Seedling Growth of Wheat, Pea, and Tomato. Water Air Soil Pollut. 2019, 230, 173. [Google Scholar] [CrossRef]
- Sethy, S.K.; Ghosh, S. Effect of Heavy Metals on Germination of Seeds. J. Nat. Sci. Biol. Med. 2013, 4, 272–275. [Google Scholar] [CrossRef] [Green Version]
- Hogan, C. Heavy Metal. In Encyclopedia of Earth; Monosson, E., Cleveland, C., Eds.; National Council for Science and the Environment: Washington, DC, USA, 2010; pp. 254–256. [Google Scholar]
- Lequeux, H.; Hermans, C.; Lutts, S.; Verbruggen, N. Response to Copper Excess in Arabidopsis thaliana: Impact on the Root System Architecture, Hormone Distribution, Lignin Accumulation and Mineral Profile. Plant Physiol. Biochem. 2010, 48, 673–682. [Google Scholar] [CrossRef]
- Kovác, J.; Lux, A.; Vaculík, M. Formation of a Subero-Lignified Apical Deposit in Root Tip of Radish (Raphanus sativus) as a Response to Copper Stress. Ann. Bot. 2018, 122, 823–831. [Google Scholar] [CrossRef]
- Panou-Filotheou, H.; Bosabalidis, A.M. Root Structural Aspects Associated with Copper Toxicity in Oregano (Origanum vulgare Subsp. hirtum). Plant Sci. 2004, 166, 1497–1504. [Google Scholar] [CrossRef]
- Gowayed, S.M.H.; Almaghrabi, O.A. Effect of Copper and Cadmium on Germination and Anatomical Structure of Leaf and Root Seedling in Maize (Zea mays L.). Aust. J. Basic Appl. Sci. 2013, 7, 548–555. [Google Scholar]
- Bouazizi, H.; Jouili, H.; Geitmann, A.; El Ferjani, E. Cell Wall Accumulation of Cu Ions and Modulation of Lignifying Enzymes in Primary Leaves of Bean Seedlings Exposed to Excess Copper. Biol. Trace Elem. Res. 2011, 139, 97–107. [Google Scholar] [CrossRef]
- Kasim, W.A. Changes Induced by Copper and Cadmium Stress in the Anatomy and Grain Yield of Sorghum bicolor (L.) Moench. Int. J. Agric. Biol. 2006, 15, 123–128. [Google Scholar]
- Panou-Filotheou, H.; Bosabalidis, A.M.; Karataglis, S. Effects of Copper Toxicity on Leaves of Oregano (Origanum vulgare Subsp. hirtum). Ann. Bot. 2001, 88, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Gajewska, E.; Skłodowska, M. Differential Effect of Equal Copper, Cadmium and Nickel Concentration on Biochemical Reactions in Wheat Seedlings. Ecotoxicol. Environ. Saf. 2010, 73, 996–1003. [Google Scholar] [CrossRef]
- Metwali, E.M.R.; Gowayed, S.M.H.; Al-Maghrabi, O.A.; Mosleh, Y.Y. Evaluation of Toxic Effect of Copper and Cadmium on Growth, Physiological Traits and Protein Profile of Wheat (Triticum aestivium L.), Maize (Zea mays L.) and Sorghum (Sorghum bicolor L.). World Appl. Sci. J. 2013, 21, 301–314. [Google Scholar] [CrossRef]
- Wheeler, D.M.; Power, I.L. Comparison of Plant Uptake and Plant Toxicity of Various Ions in Wheat. Plant Soil 1995, 172, 167–173. [Google Scholar] [CrossRef]
- Trentin, E.; Facco, D.B.; Hammerschmitt, R.K.; Avelar Ferreira, P.A.; Morsch, L.; Belles, S.W.; Ricachenevsky, F.K.; Nicoloso, F.T.; Ceretta, C.A.; Tiecher, T.L.; et al. Potential of Vermicompost and Limestone in Reducing Copper Toxicity in Young Grapevines Grown in Cu-Contaminated Vineyard Soil. Chemosphere 2019, 226, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Trentin, E.; Cesco, S.; Pii, Y.; Valentinuzzi, F.; Celletti, S.; Feil, S.B.; Zuluaga, M.Y.A.; Ferreira, P.A.A.; Ricachenevsky, F.K.; Stefanello, L.O.; et al. Plant Species and pH Dependent Responses to Copper Toxicity. Environ. Exp. Bot. 2022, 196, 104791. [Google Scholar] [CrossRef]
- Rosa, D.J.; Ambrosini, V.G.; Kokkoris, V.; Brunetto, G.; Hart, M.; Ricachenevsky, F.; Pescador, R. Lime Protection for Young Vines Exposed to Copper Toxicity. Water Air Soil Pollut. 2020, 231, 296. [Google Scholar] [CrossRef]
- Benedet, L.; De Conti, L.; Lazzari, C.J.R.; Júnior, V.M.; Dick, D.P.; Lourenzi, C.R.; Lovato, P.E.; Comin, J.J.; Tiecher, T.L.; Ricachenevsky, F.K.; et al. Copper and Zinc in Rhizosphere Soil and Toxicity Potential in White Oats (Avena sativa) Grown in Soil with Long-Term Pig Manure Application. Water Air Soil Pollut. 2019, 230, 296. [Google Scholar] [CrossRef]
- Ambrosini, V.G.; Rosa, D.J.; Bastos de Melo, G.W.; Zalamena, J.; Cella, C.; Simão, D.G.; Souza da Silva, L.; Pessoa dos Santos, H.; Toselli, M.; Tiecher, T.L.; et al. High Copper Content in Vineyard Soils Promotes Modifications in Photosynthetic Parameters and Morphological Changes in the Root System of ‘Red Niagara’ Plantlets. Plant Physiol. Biochem. 2018, 128, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Reckova, S.; Tuma, J.; Dobrev, P.; Vankova, R. Influence of Copper on Hormone Content and Selected Morphological, Physiological and Biochemical Parameters of Hydroponically Grown Zea mays Plants. Plant Growth Regul. 2019, 89, 191–201. [Google Scholar] [CrossRef]
- Ambrosini, V.G.; Rosa, D.J.; Basso, A.; Borghezan, M.; Pescador, R.; Miotto, A.; de Melo, G.W.B.; de Soares, C.R.F.S.; Comin, J.J.; Brunetto, G. Liming as an Ameliorator of Copper Toxicity in Black Oat (Avena strigosa Schreb.). J. Plant Nutr. 2017, 40, 404–416. [Google Scholar] [CrossRef]
- Küpper, H.; Šetlík, I.; Spiller, M.; Küpper, F.C.; Prášil, O. Heavy Metal-Induced Inhibition of Photosynthesis: Targets of in vivo Heavy Metal Chlorophyll Formation. J. Phycol. 2002, 38, 429–441. [Google Scholar] [CrossRef]
- Thomas, G.; Stärk, H.J.; Wellenreuther, G.; Dickinson, B.C.; Küpper, H. Effects of Nanomolar Copper on Water Plants-Comparison of Biochemical and Biophysical Mechanisms of Deficiency and Sublethal Toxicity under Environmentally Relevant Conditions. Aquat. Toxicol. 2013, 140–141, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Thomas, G.; Andresen, E.; Mattusch, J.; Hubáček, T.; Küpper, H. Deficiency and Toxicity of Nanomolar Copper in Low Irradiance-A Physiological and Metalloproteomic Study in the Aquatic Plant Ceratophyllum demersum. Aquat. Toxicol. 2016, 177, 226–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küpper, H.; Küpper, F.C.; Spiller, M. [Heavy Metal]-Chlorophylls Formed in vivo during Heavy Metal Stress and Degradation Products Formed during Digestion, Extraction and Storage of Plant Material. In Chlorophylls and Bacteriochlorophylls; Springer: Dordrecht, The Netherlands, 2006; pp. 67–77. ISBN 1402045166. [Google Scholar]
- de Freitas, T.A.; França, M.G.C.; Almeida, A.-A.F.; Oliveira, S.J.R.; Jesus, R.M.; Souza, V.L.; dos Silva, J.V.S.; Mangabeira, P.A. Morphology, Ultrastructure and Mineral Uptake Is Affected by Copper Toxicity in Young Plants of Inga subnuda Subs. luschnathiana (Benth.) T.D. Penn. Environ. Sci. Pollut. Res. 2015, 22, 15479–15494. [Google Scholar] [CrossRef]
- Rehman, M.; Liu, L.; Wang, Q.; Saleem, M.H.; Bashir, S.; Ullah, S. Copper Environmental Toxicology, Recent Advances, and Future Outlook: A Review. Environ. Sci. Pollut. Res. 2019, 26, 18003–18016. [Google Scholar] [CrossRef]
- Sharma, P.; Sirhindi, G.; Kumar, A.; Harpreet, S. Consequences of Copper Treatment on Pigeon pea Photosynthesis, Osmolytes and Antioxidants Defense. Physiol. Mol. Biol. Plants 2017, 23, 809–816. [Google Scholar] [CrossRef]
- Quartacci, M.F.; Pinzino, C.; Sgherri, C.L.M.; Dalla, F.; Navari-izzo, F. Growth in Excess Copper Induces Changes in the Lipid Composition and Fluidity of PSII-Enriched Membranes in Wheat. Physiol. Plant. 2000, 108, 87–93. [Google Scholar] [CrossRef]
- Pospísil, P. Molecular Mechanisms of Production and Scavenging of Reactive Oxygen Species by Photosystem II. Biochim. Biophys. Acta 2012, 1817, 218–231. [Google Scholar] [CrossRef] [Green Version]
- Mateos-Naranjo, E.; Moreno, L.A.; Cambrolé, J.; Perez-Martin, A. Assessing the Effect of Copper on Growth, Copper Accumulation and Physiological Responses of Grazing Species Atriplex halimus: Ecotoxicological Implications. Ecotoxicol. Environ. Saf. 2013, 90, 136–142. [Google Scholar] [CrossRef] [Green Version]
- Kopittke, P.M.; Menzies, N.W. Effect of Cu Toxicity on Growth of Cowpea (Vigna unguiculata). Plant Soil 2006, 279, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Kroneck, P.M.H.; Küpper, H. Toxicity and Deficiency of Copper in Elsholtzia splendens Affect Photosynthesis Biophysics, Pigments and Metal Accumulation. Environ. Sci. Technol. 2013, 47, 6120–6128. [Google Scholar] [CrossRef]
- Andrés-Bordería, A.; Andrés, F.; Garcia-Molina, A.; Perea-García, A.; Domingo, C.; Puig, S.; Peñarrubia, L.; Bordería, A.A.; Andrés, F.; Molina, A.G.; et al. Copper and Ectopic Expression of the Arabidopsis Transport Protein COPT1 Alter Iron Homeostasis in Rice (Oryza sativa L.). Plant Mol. Biol. 2017, 95, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Cho, S.W.; Kwon, S.J.; Kamal, A.H.M.; Lee, D.G.; Sarker, K.; Lee, M.S.; Xin, Z.; Woo, S.H. Proteome Characterization of Copper Stress Responses in the Roots of Sorghum. BioMetals 2017, 30, 765–785. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zeng, L.; Sun, H.; Wu, H.; Chen, S. Adversity Stress-Related Responses at Physiological Attributes, Transcriptional and Enzymatic Levels after Exposure to Cu in Lycopersicum esculentm Seedlings. Sci. Hortic. 2017, 222, 213–220. [Google Scholar] [CrossRef]
- Gabriel, V.; Daniel, A.; Rosa, J.; Basso, A.; Comin, J.J. Liming as a Means of Reducing Copper Toxicity in Black Oats. Ciênc. Rural 2018, 48. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, H.; Chen, C. Proteomic Analysis of Copper-Binding Proteins in Excess Copper-Stressed Rice Roots by Immobilized Metal Affinity Chromatography and Two-Dimensional Electrophoresis. BioMetals 2014, 27, 265–276. [Google Scholar] [CrossRef]
- Osmolovskaya, N.G. The Role of Organic Acids in Heavy Metal Tolerance in Plants. Biol. Commun. 2018, 63, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Adeleke, R.; Nwangburuka, C.; Oboirien, B. Origins, Roles and Fate of Organic Acids in Soils: A Review. S. Afr. J. Bot. 2017, 108, 393–406. [Google Scholar] [CrossRef]
- Kang, W.; Bao, J.; Zheng, J.; Hu, H. Distribution and Chemical Forms of Copper in the Root Cells of Castor Seedlings and Their Tolerance to Copper Phytotoxicity in Hydroponic Culture. Environ. Sci. Pollut. Res. 2015, 22, 7726–7734. [Google Scholar] [CrossRef] [PubMed]
- Keller, C.; Rizwan, M.; Pokrovsky, O.S.; Bovet, N.; Chaurand, P.; Meunier, J.-D. Effect of Silicon on Wheat Seedlings (Triticum turgidum L.) Grown in Hydroponics and Exposed to 0 to 30 UM Cu. Planta 2014, 241, 847–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, J.M.L.; Rodríguez-Monroy, M.; Sepúlveda-Jiménez, G. Betacyanin Accumulation and Guaiacol Peroxidase Activity in Beta vulgaris L. Leaves Following Copper Stress. Acta Soc. Bot. Pol. 2012, 81, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Ryan, B.M.; Kirby, J.K.; Degryse, F.; Harris, H.; Mclaughlin, M.J.; Scheiderich, K. Copper Speciation and Isotopic Fractionation in Plants: Uptake and Translocation Mechanisms. New Phytol. 2013, 199, 367–378. [Google Scholar] [CrossRef]
- Colzi, I.; Arnetoli, M.; Gallo, A.; Doumett, S.; Del Bubba, M.; Pignattelli, S.; Gabbrielli, R.; Gonnelli, C. Copper Tolerance Strategies Involving the Root Cell Wall Pectins in Silene paradoxa L. Environ. Exp. Bot. 2012, 78, 91–98. [Google Scholar] [CrossRef]
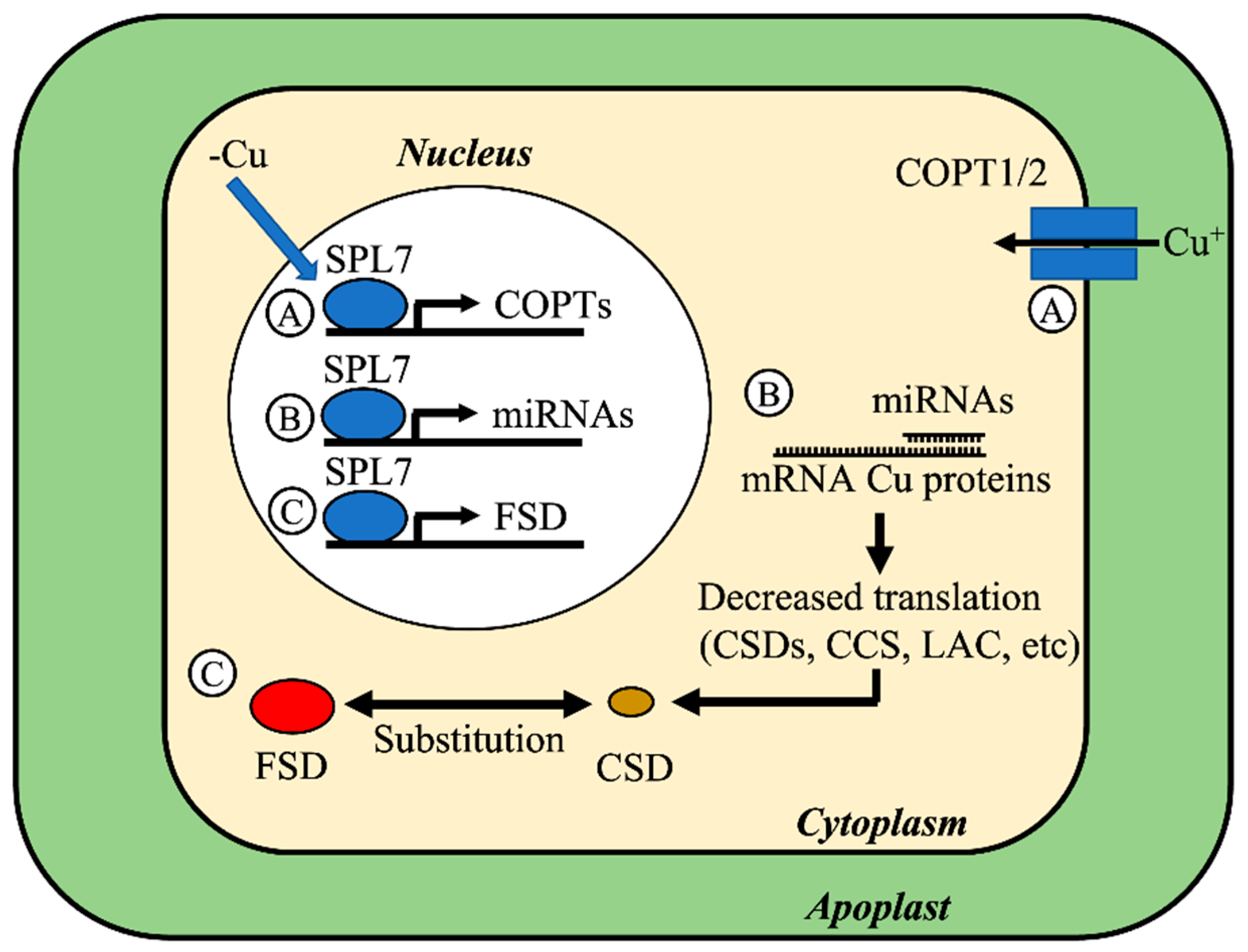
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein–Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Molina, A.; Xing, S.; Huijser, P. Functional Characterisation of Arabidopsis SPL7 Conserved Protein Domains Suggests Novel Regulatory Mechanisms in the Cu Deficiency Response. BMC Plant Biol. 2014, 14, 231. [Google Scholar] [CrossRef] [Green Version]
- Merchant, S.S.; Schmollinger, S.; Strenkert, D.; Moseley, L.; Blaby-Haas, C.E. From Economy to Luxury: Copper Homeostasis in Chlamydomonas and Other Algae. BBA Mol. Cell Res. 2020, 1867, 118822. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Xing, S.; Huijser, P. A Conserved KIN17 Curved DNA-Binding Domain Protein Assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to Adapt Arabidopsis Growth and Development to Limiting. Plant Physiol. 2014, 164, 828–840. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. MicroRNA408 Is Critical for the HY5-SPL7 Gene Network That Mediates the Coordinated Response to Light and Copper. Plant Cell 2014, 26, 4933–4953. [Google Scholar] [CrossRef] [Green Version]
- Perea-García, A.; Andrés-Bordería, A.; de Andrés, S.M.; Sanz, A.; Davis, A.M.; Davis, S.J.; Huijser, P.; Peñarrubia, L. Modulation of Copper Deficiency Responses by Diurnal and Circadian Rhythms in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 391–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, M.; Casero, D.; Singh, V.; Wilson, G.T.; Grande, A.; Yang, H.; Dodani, S.C.; Pellegrini, M.; Huijser, P.; Connolly, E.L.; et al. Transcriptome Sequencing Identifies SPL7-Regulated Copper Acquisition Genes FRO4/FRO5 and the Copper Dependence of Iron Homeostasis in Arabidopsis. Plant Cell 2012, 24, 738–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-Mediated Systemic down-Regulation of Copper Protein Expression in Response to Low Copper Availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of MiR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of Copper Homeostasis by Micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilon, M. The Copper MicroRNAs. New Phytol. 2017, 213, 1030–1035. [Google Scholar] [CrossRef]
- Navarro, B.B.; Del Frari, B.K.; da Dias, P.V.C.; Lemainski, L.E.; Mario, R.B.; Ponte, L.R.; Goergen, A.; Tarouco, C.P.; Neves, V.M.; Dressler, V.L.; et al. The Copper Economy Response Is Partially Conserved in Rice (Oryza sativa L.). Plant Physiol. Biochem. 2021, 158, 113–124. [Google Scholar] [CrossRef]
- Riaz, N.; Guerinot, M. Lou All Together Now: Regulation of the Iron Deficiency Response. J. Exp. Bot. 2021, 72, 2045–2055. [Google Scholar] [CrossRef]
- Jain, A.; Wilson, G.T.; Connolly, E.L. The Diverse Roles of FRO Family Metalloreductases in Iron and Copper Homeostasis. Front. Plant Sci. 2014, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ye, L.; Kong, Q.; Shou, H. A Vacuolar Membrane Ferric-Chelate Reductase, OsFRO1, Alleviates Fe Toxicity in Rice (Oryza sativa L.). Front. Plant Sci. 2019, 10, 700. [Google Scholar] [CrossRef] [Green Version]
- Wairich, A.; de Oliveira, B.H.N.; Arend, E.B.; Duarte, G.L.; Ponte, L.R.; Sperotto, R.A.; Ricachenevsky, F.K.; Fett, J.P.; Hur, B.; De Oliveira, N.; et al. The Combined Strategy for Iron Uptake Is Not Exclusive to Domesticated Rice (Oryza sativa). Sci. Rep. 2019, 9, 16144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancenón, V.; Puig, S.; Mira, H.; Thiele, D.J.; Peñarrubia, L.; Sancen, V.; Puig, S.; Mira, H.; Thiele, D.J.; Peñarrubia, L. Identification of a Copper Transporter Family in Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 577–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, P.; Gabrielli, A.; Sampedro, R.; Perea-García, A.; Puig, S.; Lafuente, T. Identification and Molecular Characterization of the High-Affinity Copper Transporters Family in Solanum lycopersicum. Int. J. Biol. Macromol. 2021, 192, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Li, X.; Xiao, J.; Wang, S. Molecular and Functional Analyses of COPT/Ctr- Type Copper Transporter-like Gene Family in Rice. BMC Plant Biol. 2011, 11, 69. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; Logeman, B.L.; Zhang, X.; Liu, Y.; Thiele, D.J.; Yuan, P. X-Ray Structures of the High-Affinity Copper Transporter Ctr1. Nat. Commun. 2019, 10, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig, S. Function and Regulation of the Plant COPT Family of High-Affinity Copper Transport Proteins. Adv. Bot. 2014, 2014, 476917. [Google Scholar] [CrossRef] [Green Version]
- Perea-García, A.; Garcia-Molina, A.; Andrés-Colás, N.; Vera-Sirera, F.; Pérez-Amador, M.A.; Puig, S.; Peñarrubia, L. Arabidopsis Copper Transport Protein COPT2 Participates in the Cross Talk between Iron Deficiency Responses and Low-Phosphate Signaling. Plant Physiol. 2013, 162, 180–194. [Google Scholar] [CrossRef] [Green Version]
- Andrés-Colás, N.; Perea-García, A.; Puig, S.; Peñarrubia, L. Deregulated Copper Transport Affects Arabidopsis Development Especially in the Absence of Environmental Cycles. Plant Physiol 2010, 153, 170–184. [Google Scholar] [CrossRef] [Green Version]
- Sancenón, V.; Puig, S.; Mateu-Andrés, I.; Dorcey, E.; Thiele, D.J.; Peñarrubia, L. The Arabidopsis Copper Transporter COPT1 Functions in Root Elongation and Pollen Development. J. Biol. Chem. 2004, 279, 15348–15355. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yuan, J.; Wang, H.; Zhang, H.; Zhang, H. Arabidopsis COPPER TRANSPORTER 1 Undergoes Degradation in a Proteasome-Dependent Manner. J. Exp. Bot. 2020, 71, 6174–6186. [Google Scholar] [CrossRef]
- Li, X.; Wang, Z.; Fu, Y.; Cheng, X.; Zhu, C.; Chen, Z. Two Ubiquitin-Associated ER Proteins Interact with COPT Copper Transporters and Modulate Their Accumulation. Plant Physiol. 2021, 187, 2469–2484. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, W.; Yuan, J.; Xu, J.; He, L.; Zhang, X.; Zhang, H. Ubiquitin-Independent Proteasome System Is Required for Degradation of Arabidopsis COPPER TRANSPORTER 2. Plant Sci. 2021, 304, 110825. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Gayomba, S.R.; Rutzke, M.A.; Craft, E.; Kochian, L.V.; Vatamaniuk, O.K. COPT6 Is a Plasma Membrane Transporter That Functions in Copper Homeostasis in Arabidopsis and Is a Novel Target of SQUAMOSA Promoter-Binding Protein-like 7. J. Biol. Chem. 2012, 287, 33252–33267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Molina, A.; Andrés-Colás, N.; Perea-García, A.; Neumann, U.; Dodani, S.C.; Huijser, P.; Peñarrubia, L.; Puig, S. The Arabidopsis COPT6 Transport Protein Functions in Copper Distribution under Copper-Deficient Conditions. Plant Cell Physiol. 2013, 54, 1378–1390. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Molina, A.; Andrés-Colás, N.; Perea-Garcia, A.; del Valle-tasco, S.; Peñarrubia, L.; Puig, S. The Intracellular Arabidopsis COPT5 Transport Protein Is Required for Photosynthetic Electron Transport under Severe Copper Deficiency. Plant J. 2011, 65, 848–860. [Google Scholar] [CrossRef]
- Klaumann, S.; Nickolaus, S.D.; Fürst, S.H.; Starck, S.; Schneider, S.; Neuhaus, E.H.; Trentmann, O. The Tonoplast Copper Transporter COPT5 Acts as an Exporter and Is Required for Interorgan Allocation of Copper in Arabidopsis thaliana. New Phytol. 2011, 192, 393–404. [Google Scholar] [CrossRef]
- Andrés-Colás, N.; Carrió-Seguí, A.; Abdel-Ghany, S.E.; Pilon, M.; Peñarrubia, L. Expression of the Intracellular COPT3-Mediated Cu Transport Is Temporally Regulated by the TCP16 Transcription Factor. Front. Plant Sci. 2018, 9, 910. [Google Scholar] [CrossRef] [Green Version]
- Giraud, E.; Ng, S.; Carrie, C.; Duncan, O.; Low, J.; Lee, C.P.; Van Aken, O.; Millar, A.H.; Murcha, M.; Whelan, J. TCP Transcription Factors Link the Regulation of Genes Encoding Mitochondrial Proteins with the Circadian Clock in Arabidopsis thaliana. Plant Cell 2010, 22, 3921–3934. [Google Scholar] [CrossRef] [Green Version]
- Takeda, T.; Amano, K.; Ohto, M.; Nakamura, K.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. RNA Interference of the Arabidopsis Putative Transcription Factor TCP16 Gene Results in Abortion of Early Pollen Development. Plant Mol. Biol. 2006, 61, 165–177. [Google Scholar] [CrossRef]
- Uberti-Manassero, N.G.; Coscueta, E.R.; Gonzalez, D.H. Expression of a Repressor Form of the Arabidopsis thaliana Transcription Factor TCP16 Induces the Formation of Ectopic Meristems. Plant Physiol. Biochem. 2016, 108, 57–62. [Google Scholar] [CrossRef]
- Bock, K.W.; Honys, D.; Ward, J.M.; Padmanaban, S.; Nawrocki, E.P.; Hirschi, K.D.; Twell, D.; Sze, H. Integrating Membrane Transport with Male Gametophyte Development and Function through Transcriptomics. Plant Physiol. 2006, 140, 1151–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Chia, J.; Sheng, H.; Jung, H.; Zavodna, T.; Zhang, L.; Huang, R.; Jiao, C.; Craft, E.J.; Fei, Z.; et al. Arabidopsis Pollen Fertility Requires the Transcription Factors CITF1 and SPL7 That Regulate Copper Delivery to Anthers and Jasmonic Acid Synthesis. Plant Cell 2017, 29, 3012–3029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. The Bacterial Pathogen Xanthomonas oryzae Overcomes Rice Defenses by Regulating Host Copper Redistribution. Plant Cell 2010, 22, 3164–3176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. Pathogen-Induced Expressional Loss of Function Is the Key Factor in Race-Specific Bacterial Resistance Conferred by a Recessive R Gene Xa13 in Rice. Plant Cell Physiol. 2009, 50, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Senovilla, M.; Castro-Rodríguez, R.; Abreu, I.; Escudero, V.; Kryvoruchko, I.; Udvardi, M.K.; Imperial, J.; González-Guerrero, M. Medicago truncatula Copper Transporter 1 (MtCOPT1) Delivers Copper for Symbiotic Nitrogen Fixation. New Phytol. 2018, 1, 696–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas-Santiago, P.; Pérez, K.Z.; Méndez, M.F.G.; Portillo, F.V.L.; Salas, L.J.R.; Martínez, E.C.; Maspon, A.A.; Pantoja, O. A Differential Subcellular Localization of Two Copper Transporters from the COPT Family Suggests Distinct Roles in Copper Homeostasis in Physcomitrium patens. Plant Physiol. Biochem. 2021, 167, 459–469. [Google Scholar] [CrossRef]
- Wintz, H.; Fox, T.; Wu, Y.; Feng, V.; Chen, W.; Chang, H.-S.; Zhu, T.; Vulpe, C. Expression Profiles of Arabidopsis thaliana in Mineral Deficiencies Reveal Novel Transporters Involved in Metal Homeostasis. J. Biol. Chem. 2003, 278, 47644–47653. [Google Scholar] [CrossRef] [Green Version]
- del Pozo, T.; Cambiazo, V.; González, M. Gene Expression Profiling Analysis of Copper Homeostasis in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2010, 393, 248–252. [Google Scholar] [CrossRef]
- Leng, X.; Jia, H.; Sun, X.; Shangguan, L.; Mu, Q.; Wang, B. Comparative Transcriptome Analysis of Grapevine in Response to Copper Stress. Sci. Rep. 2015, 5, 17749. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.T.; Smith, K.P.; Rosenzweig, A.C. Diversity of the Metal-Transporting P1B -Type ATPases. J. Biol. Inorg. Chem. 2014, 19, 947–960. [Google Scholar] [CrossRef] [Green Version]
- Pita-Barbosa, A.; Ricachenevsky, F.K.; Wilson, M.; Dottorini, T.; Salt, D.E. Transcriptional Plasticity Buffers Genetic Variation in Zinc Homeostasis. Sci. Rep. 2019, 9, 19482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-Type ATPase Heavy Metal Transporters with Roles in Essential Zinc Homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermand, V.; Julio, E.; de Borne, D.; Punshon, T.; Ricachenevsky, F.K.; Bellec, A.; Gosti, F.; Berthomieu, P. Inactivation of Two Newly Identified Tobacco Heavy Metal ATPases Leads to Reduced Zn and Cd Accumulation in Shoots and Reduced Pollen Germination. Metallomics 2014, 6, 1427–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrés-Colas, N.; Sancenón, V.; Rodríguez-Navarro, S.; Mayo, S.; Thiele, D.J.; Ecker, J.R.; Puig, S.; Peñarrubia, L. The Arabidopsis Heavy Metal P-Type ATPase HMA5 Interacts with Metallochaperones and Functions in Copper Detoxification of Roots. Plant J. 2006, 45, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kuroda, K.; Kimura, K.; Southron-francis, J.L.; Furuzawa, A.; Kimura, K.; Iuchi, S.; Kobayashi, M.; Taylor, G.J.; Koyama, H. Amino Acid Polymorphisms in Strictly Conserved Domains of a P-Type ATPase HMA5 Are Involved in the Mechanism of Copper Tolerance Variation in Arabidopsis. Plant Physiol. 2008, 148, 969–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikanai, T.; Müller-Moulé, P.; Munekage, Y.; Niyogi, K.K.; Pilon, M. PAA1, a P-Type ATPase of Arabidopsis, Functions in Copper Transport in Chloroplasts. Plant Cell 2003, 15, 1333–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Ghany, S.E.; Burkhead, J.L.; Gogolin, K.A.; Andrés-Colás, N.; Bodecker, J.R.; Puig, S.; Peñarrubia, L.; Pilon, M. AtCCS Is a Functional Homolog of the Yeast Copper Chaperone Ccs1/Lys7. FEBS J. 2005, 579, 2307–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaby-Haas, C.E.; Padilla-benavides, T.; Stübe, R.; Argüello, J.M.; Merchant, S.S. Evolution of a Plant-Specific Copper Chaperone Family for Chloroplast Copper Homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, E5480–E5487. [Google Scholar] [CrossRef] [Green Version]
- Seigneurin-Berny, D.; Gravot, A.; Auroy, P.; Mazard, C.; Kraut, A.; Finazzi, G.; Grunwald, D.; Rappaport, F.; Vavasseur, A.; Joyard, J.; et al. HMA1, a New Cu-ATPase of the Chloroplast Envelope, Is Essential for Growth under Adverse Light Conditions. J. Biol. Chem. 2006, 281, 2882–2892. [Google Scholar] [CrossRef] [Green Version]
- Schott-Verdugo, S.; Müller, L.; Classen, E.; Gohlke, H.; Groth, G. Structural Model of the ETR1 Ethylene Receptor Transmembrane Sensor Domain. Sci. Rep. 2019, 9, 8869. [Google Scholar] [CrossRef]
- Binder, B.M.; Rodríguez, F.I.; Bleecker, A.B. The Copper Transporter RAN1 Is Essential for Biogenesis of Ethylene Receptors in Arabidopsis. J. Biol. Chem. 2010, 285, 37263–37270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Lacey, R.F.; Ye, Y.; Lu, J.; Yeh, K.-C.; Xiao, Y.; Li, L.; Wen, C.-K.; Binder, B.M.; Zhao, Y. Triplin, a Small Molecule, Reveals Copper Ion Transport in Ethylene Signaling from ATX1 to RAN1. PLoS Genet. 2017, 13, e1006703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Kim, Y.Y.; Lee, Y.; An, G. Rice P1B-Type Heavy-Metal ATPase, OsHMA9, Is a Metal Efflux Protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, F.; Yamaji, N.; Xia, J.; Ma, J.F. A Member of the Heavy Metal P-Type ATPase OsHMA5 Is Involved in Xylem Loading of Copper in Rice. Plant Physiol. 2013, 163, 1353–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.Y.; Deng, F.; Yamaji, N.; Pinson, S.R.M.; Fujii-Kashino, M.; Danku, J.; Douglas, A.; Guerinot, M.L.; Salt, D.E.; Ma, J.F. A Heavy Metal P-Type ATPase OsHMA4 Prevents Copper Accumulation in Rice Grain. Nat. Commun. 2016, 7, 12138. [Google Scholar] [CrossRef] [Green Version]
- Ricachenevsky, F.K.; de Araújo Junior, A.T.; Fett, J.P.; Sperotto, R.A. You Shall Not Pass: Root Vacuoles as a Symplastic Checkpoint for Metal Translocation to Shoots and Possible Application to Grain Nutritional Quality. Front. Plant Sci. 2018, 9, 412. [Google Scholar] [CrossRef]
- Rae, T.D.; Schmidt, P.J.; Pufahl, R.A.; Culotta, V.C.; O’Halloran, T.V. Undetectable Intracellular Free Copper: The Requirement of a Copper Chaperone for Superoxide Dismutase. Science 1999, 284, 805–808. [Google Scholar] [CrossRef] [Green Version]
- Harrison, M.D.; Jones, C.E.; Dameron, C.T. Copper Chaperones: Function, Structure and Copper-Binding Properties. J. Biol. Inorg. Chem. 1999, 4, 145–153. [Google Scholar] [CrossRef]
- Harrison, M.D.; Jones, C.E.; Solioz, M.; Dameron, C.T. Intracellular Copper Routing: The Role of Copper Chaperones. Trends Biochem. Sci. 2000, 25, 29–32. [Google Scholar] [CrossRef]
- Shin, L.-J.; Lo, J.-C.; Yeh, K.-C. Copper Chaperone Antioxidant Protein1 Is Essential for Copper Homeostasis. Plant Physiol. 2012, 159, 1099–1110. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, K.; Zhao, F.J.; Sun, C.; Jin, C.; Shi, Y.; Sun, Y.; Li, Y.; Yang, M.; Jing, X.; et al. OsATX1 Interacts with Heavy Metal P1B-Type ATPases and Affects Copper Transport and Distribution. Plant Physiol. 2018, 178, 329–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, C.; Lee, W.; Guo, W.; Pan, S.; Chen, L.; Li, H. A Copper Chaperone for Superoxide Dismutase That Confers Three Types of Copper/Zinc Superoxide Dismutase Activity in Arabidopsis. Plant Physiol. 2005, 139, 425–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohu, C.M.; Abdel-Ghany, S.E.; Reynilds, K.A.G.; Onofrio, A.M.; Bodecker, J.R.; Kimbrel, J.A.; Niyogi, K.K.; Pilon, M. Copper Delivery by the Copper Chaperone for Chloroplast and Cytosolic Copper/Zinc-Superoxide Dismutases: Regulation and Unexpected Phenotypes in an Arabidopsis Mutant. Mol. Plant 2009, 2, 1336–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.-H.; Kuo, W.-Y.; Weiss, C.; Jinn, T.-L. Copper Chaperone-Dependent and -Independent Activation of Three Copper-Zinc Superoxide Dismutase Homologs Localized in Different Cellular Compartments in Arabidopsis. Plant Physiol. 2012, 158, 737–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobine, P.A.; Pierrel, F.; Winge, D.R. Copper Trafficking to the Mitochondrion and Assembly of Copper Metalloenzymes. Biochim. Biophys. Acta 2006, 1763, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Llases, M.-E.; Lisa, M.-N.; Morgada, M.N.; Gianini, E.; Alzari, P.M.; Vila, A.J. Arabidopsis thaliana Hcc1 Is a Sco-like Metallochaperone for CuA Assembly in Cytochrome c Oxidase. FEBS J. 2020, 287, 749–762. [Google Scholar] [CrossRef]
- Steinebrunner, I.; Landschreiber, M.; Krause-Buchholz, U.; Teichmann, J.; Rödel, G. HCC1, the Arabidopsis Homologue of the Yeast Mitochondrial Copper Chaperone SCO1, Is Essential for Embryonic Development. J. Exp. Bot. 2011, 62, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Attallah, C.V.; Welchen, E.; Martin, A.P.; Spinelli, S.V.; Bonnard, G.; Palatnik, F.; Gonzalez, D.H. Plants Contain Two SCO Proteins That Are Differentially Involved in Cytochrome c Oxidase Function and Copper and Redox Homeostasis. J. Exp. Bot. 2011, 62, 4281–4294. [Google Scholar] [CrossRef] [Green Version]
- Steinebrunner, I.; Gey, U.; Andres, M.; Garcia, L.; Gonzalez, D.H. Divergent Functions of the Arabidopsis Mitochondrial SCO Proteins: HCC1 Is Essential for COX Activity While HCC2 Is Involved in the UV-B Stress Response. Front. Plant Sci. 2014, 5, 87. [Google Scholar] [CrossRef] [Green Version]
- Mansilla, N.; Welchen, E.; Gonzalez, D.H. Arabidopsis SCO Proteins Oppositely Influence Cytochrome c Oxidase Levels and Gene Expression during Salinity Stress. Plant Cell Physiol. 2019, 60, 2769–2784. [Google Scholar] [CrossRef]
- Chai, L.-X.; Dong, K.; Liu, S.-Y.; Zhang, Z.; Zhang, X.; Tong, X.; Zhu, F.; Zou, J.-Z.; Wang, X.-B. A Putative Nuclear Copper Chaperone Promotes Plant Immunity in Arabidopsis. J. Exp. Bot. 2020, 71, 6684–6696. [Google Scholar] [CrossRef] [PubMed]
- Curie, C.; Cassin, G.; Couch, D.; Divol, F.; Higuchi, K.; Le Jean, M.; Misson, J.; Schikora, A.; Czernic, P.; Mari, S. Metal Movement within the Plant: Contribution of Nicotianamine and Yellow Stripe 1-like Transporters. Ann. Bot. 2009, 103, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Chiecko, J.C.; Kim, S.A.; Walker, E.L.; Lee, Y.; Guerinot, M.L.; An, G.; Oa, R.P.C.W.; Lee, S.; Chiecko, J.C.; et al. Disruption of OsYSL15 Leads to Iron Inefficiency in Rice Plants. Plant Physiol. 2009, 150, 786–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 Is an Iron-Regulated Iron(III)-Deoxymugineic Acid Transporter Expressed in the Roots and Is Essential for Iron Uptake in Early Growth of the Seedlings. J. Biol. Chem. 2009, 284, 3470–3479. [Google Scholar] [CrossRef] [Green Version]
- Chu, H.; Chiecko, J.; Punshon, T.; Lanzirotti, A.; Lahner, B.; Salt, D.E.; Walker, E.L. Successful Reproduction Requires the Function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 Metal-Nicotianamine Transporters in Both Vegetative and Reproductive Structures. Plant Physiol. 2010, 154, 197–210. [Google Scholar] [CrossRef] [Green Version]
- Curie, C.; Panaviene, Z.; Loulergue, C.; Dellaporta, S.L.; Briat, J.-F.; Walker, E.L. Maize Yellow Stripe1 Encodes a Membrane Protein Directly Involved in Fe(III) Uptake. Nature 2001, 409, 346. [Google Scholar] [CrossRef]
- Ueno, D.; Yamaji, N.; Ma, J.F. Further Characterization of Ferric—Phytosiderophore Transporters ZmYS1 and HvYS1 in Maize and Barley. J. Exp. Bot. 2009, 60, 3513–3520. [Google Scholar] [CrossRef] [Green Version]
- Waters, B.M.; Chu, H.; Didonato, R.J.; Roberts, L.A.; Eisley, R.B.; Lahner, B.; Salt, D.E.; Walker, E.L. Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 Reveal Their Roles in Metal Ion Homeostasis and Loading of Metal Ions in Seeds. Plant Physiol. 2006, 141, 1446–1458. [Google Scholar] [CrossRef] [Green Version]
- Didonato, R.J., Jr.; Roberts, L.A.; Sanderson, T.; Eisley, R.B.; Walker, E.L. Arabidopsis Yellow Stripe-Like2 (YSL2): A Metal-Regulated Gene Encoding a Plasma Membrane Transporter of Nicotianamine–Metal Complexes. Plant J. 2004, 2, 403–414. [Google Scholar] [CrossRef]
- Schaaf, G.; Schikora, A.; Häberle, J.; Vert, G.; Ludewig, U.; Briat, J.F.; Curie, C.; Von Wirén, N. A Putative Function for the Arabidopsis Fe-Phytosiderophore Transporter Homolog AtYSL2 in Fe and Zn Homeostasis. Plant Cell Physiol. 2005, 46, 762–774. [Google Scholar] [CrossRef] [Green Version]
- Conte, S.S.; Chu, H.H.; Chan-Rodriguez, D.; Punshon, T.; Vasques, K.A.; Salt, D.E.; Walker, E.L. Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 Localize to Internal Cellular Membranes and Are Involved in Metal Ion Homeostasis. Front. Plant Sci. 2013, 4, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Yamaji, N.; Yokosho, K.; Ma, J.F. YSL16 Is a Phloem-Localized Transporter of the Copper-Nicotianamine Complex That Is Responsible for Copper Distribution in Rice. Plant Cell 2012, 24, 3767–3782. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Lu, W.; Yang, Y.; Shen, Z.; Ma, J.F.; Zheng, L. OsYSL16 Is Required for Preferential Cu Distribution to Floral Organs in Rice. Plant Cell Physiol. 2018, 59, 2039–2051. [Google Scholar] [CrossRef]
- Jung, H.; Gayomba, S.R.; Yan, J.; Vatamaniuk, O.K. Brachypodium distachyon as a Model System for Studies of Copper Transport in Cereal Crops. Front. Plant Sci. 2014, 5, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, H.; Jiang, Y.; Rahmati, M.; Chia, J.-C.C.; Dokuchayeva, T.; Kavulych, Y.; Zavodna, T.O.T.-O.; Mendonza, P.N.; Huang, R.; Smieska, L.M.; et al. YSL3-Mediated Copper Distribution Is Required for Fertility, Seed Size and Protein Accumulation in Brachypodium. Plant Physiol. 2021, 186, 655–676. [Google Scholar] [CrossRef]
- Puig, S.; Thiele, D.J. Molecular Mechanisms of Copper Uptake and Distribution. Curr. Opin. Chem. Biol. 2002, 6, 171–180. [Google Scholar] [CrossRef]
- Puig, S.; Andrés-Colás, N.; García-Molina, A.; Peñarrubia, L. Copper and Iron Homeostasis in Arabidopsis: Responses to Metal Deficiencies, Interactions and Biotechnological Applications. Plant Cell Environ. 2007, 30, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Gulec, S.; Collins, J.F. Molecular Mediators Governing Iron-Copper Interactions. Annu. Rev. Nutr. 2014, 34, 95–116. [Google Scholar] [CrossRef] [Green Version]
- Waters, B.M.; Mcinturf, S.A.; Stein, R.J. Rosette Iron Deficiency Transcript and MicroRNA Profiling Reveals Links between Copper and Iron Homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5903–5918. [Google Scholar] [CrossRef] [Green Version]
- Waters, B.M.; Mcinturf, S.A.; Amundsen, K. Transcriptomic and Physiological Characterization of the Fefe Mutant of Melon (Cucumis melo) Reveals New Aspects of Iron—Copper Crosstalk. New Phytol. 2014, 203, 1128–1145. [Google Scholar] [CrossRef] [Green Version]
- Pilon, M. Moving Copper in Plants. New Phytol. 2011, 192, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Carrió-Seguí, À.; Romero, P.; Curie, C.; Mari, S.; Peñarrubia, L. Copper Transporter COPT5 Participates in the Crosstalk between Vacuolar Copper and Iron Pools Mobilisation. Sci. Rep. 2019, 9, 4648. [Google Scholar] [CrossRef]
- Perea-Garcia, A.; Andrés-Bordería, A.; Vera-Sirera, F.; Pérez-Amador, M.A.; Puig, S.; Peñarrubia, L. Deregulated High Affinity Copper Transport Alters Iron Homeostasis in Arabidopsis. Front. Plant Sci. 2020, 11, 1106. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, M.; Pilon, M. Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency. Plants 2019, 8, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gayomba, S.R.; Jung, H.; Yan, J.; Danku, J.; Rutzke, M.A.; Bernal, M.; Krämer, U.; Kochian, L.V.; Salt, D.E.; Vatamaniuk, O.K. The CTR/COPT-Dependent Copper Uptake and SPL7-Dependent Copper Deficiency Responses Are Required for Basal Cadmium Tolerance in A. thaliana. Metallomics 2013, 5, 1262–1275. [Google Scholar] [CrossRef]
- Colangelo, E.P.; Guerinot, M.L. The Essential Basic Helix-Loop-Helix Protein FIT1 Is Required for the Iron Deficiency Response. Plant Cell 2004, 16, 3400–3412. [Google Scholar] [CrossRef] [Green Version]
- Kastoori Ramamurthy, R.; Xiang, Q.; Hsieh, E.J.; Liu, K.; Zhang, C.; Waters, B.M. New Aspects of Iron-Copper Crosstalk Uncovered by Transcriptomic Characterization of Col-0 and the Copper Uptake Mutant Spl7 in Arabidopsis thaliana. Metallomics 2018, 10, 1824–1840. [Google Scholar] [CrossRef]
- Lanquar, V.; Lelièvre, F.; Bolte, S.; Hamès, C.; Alcon, C.; Neumann, D.; Vansuyt, G.; Curie, C.; Schröder, A.; Krämer, U.; et al. Mobilization of Vacuolar Iron by AtNRAMP3 and AtNRAMP4 Is Essential for Seed Germination on Low Iron. EMBO J. 2005, 24, 4041–4051. [Google Scholar] [CrossRef]
- Lanquar, V.; Ramos, M.S.; Lelièvre, F.; Barbier-brygoo, H.; Krieger-liszkay, A.; Krämer, U.; Thomine, S. Export of Vacuolar Manganese by AtNRAMP3 and AtNRAMP4 Is Required for Optimal Photosynthesis and Growth under Manganese Deficiency. Plant Physiol. 2010, 152, 1986–1999. [Google Scholar] [CrossRef] [Green Version]
- Brumbarova, T.; Bauer, P.; Ivanov, R. Molecular Mechanisms Governing Arabidopsis Iron Uptake. Trends Plant Sci. 2015, 20, 124–133. [Google Scholar] [CrossRef]
- Schwarz, B.; Bauer, P. FIT, a Regulatory Hub for Iron Deficiency and Stress Signaling in Roots, and FIT-Dependent and -Independent Gene Signatures. J. Exp. Bot. 2020, 71, 1694–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Li, Y.; Liang, G. FIT and BHLH Ib Transcription Factors Modulate Iron and Copper Crosstalk in Arabidopsis. Plant Cell Environ. 2021, 44, 1679–1691. [Google Scholar] [CrossRef]
- Mccaig, B.C.; Meagher, R.B.; Dean, J.F.D. Gene Structure and Molecular Analysis of the Laccase-like Multicopper Oxidase (LMCO) Gene Family in Arabidopsis thaliana. Planta 2005, 221, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Turlapati, P.V.; Kim, K.-W.; Davin, L.B.; Lewis, N.G. The Laccase Multigene Family in Arabidopsis thaliana: Towards Addressing the Mystery of Their Gene Function(S). Planta 2011, 233, 439–470. [Google Scholar] [CrossRef]
- Cai, X.; Davis, E.J.; Ballif, J.; Liang, M.; Bushman, E.; Haroldsen, V.; Torabinejad, J.; Wu, Y. Mutant Identification and Characterization of the Laccase Gene Family in Arabidopsis. J. Exp. Bot. 2006, 57, 2563–2569. [Google Scholar] [CrossRef] [Green Version]
- Buhtz, A.; Pieritz, J.; Springer, F.; Kehr, J. Phloem Small RNAs, Nutrient Stress Responses, and Systemic Mobility. BMC Plant Biol. 2010, 10, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrió-Seguí, À.; Ruiz-Rivero, O.; Villamayor-Belinchón, L.; Puig, S.; Perea-García, A.; Peñarrubia, L. The Altered Expression of MicroRNA408 Influences the Arabidopsis Response to Iron Deficiency. Front. Plant Sci. 2019, 10, 324. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L. SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 2013, 74, 98–109. [Google Scholar] [CrossRef]
- Bernal, M.; Krämer, U. Involvement of Arabidopsis Multi-Copper Oxidase-Encoding LACCASE12 in Root-to-Shoot Iron Partitioning: A Novel Example of Copper-Iron Crosstalk. Front. Plant Sci. 2021, 12, 1998. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, Y.; Feng, Y.; Zhou, Y.; Zhang, F.; Yang, Y.; Lei, M.; Zhang, Y.; Chen, Y.-Q. MiR408 Regulates Grain Yield and Photosynthesis via a Phytocyanin Protein. Plant Physiol. 2017, 175, 1175–1185. [Google Scholar] [CrossRef]
- Nobrega, M.P.; Bandeira, S.C.B.; Beers, J.; Tzagoloff, A. Characterization of COX19, a Widely Distributed Gene Required for Expression of Mitochondrial Cytochrome Oxidase. J. Biol. Chem. 2002, 277, 40206–40211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigby, K.; Zhang, L.; Cobine, P.A.; George, G.N.; Winge, D.R. Characterization of the Cytochrome c Oxidase Assembly Factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 10233–10242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attallah, C.V.; Welchen, E.; Pujol, C.; Bonnard, G.; Gonzalez, D.H. Characterization of Arabidopsis thaliana Genes Encoding Functional Homologues of the Yeast Metal Chaperone Cox19p, Involved in Cytochrome c Oxidase Biogenesis. Plant Mol. Biol. 2007, 65, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.L.; Shiba, T.; Young, L.; Harada, S.; Kita, K.; Ito, K. Unraveling the Heater: New Insights into the Structure of the Alternative Oxidase. Annu. Rev. Plant Biol. 2013, 64, 637–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clifton, R.; Millar, A.H.; Whelan, J. Alternative Oxidases in Arabidopsis: A Comparative Analysis of Differential Expression in the Gene Family Provides New Insights into Function of Non-Phosphorylating Bypasses. Biochim. Biophys. Acta 2006, 1757, 730–741. [Google Scholar] [CrossRef] [Green Version]
- Garcia, L.; Mansilla, N.; Ocampos, N.; Pagani, M.A.; Welchen, E.; Gonzalez, D.H. The Mitochondrial Copper Chaperone COX19 Influences Copper and Iron Homeostasis in Arabidopsis. Plant Mol. Biol. 2019, 99, 621–638. [Google Scholar] [CrossRef]
- Gomez-Casati, D.F.; Busi, M.V.; Pagani, M.A. Plant Frataxin in Metal Metabolism. Front. Plant Sci. 2018, 9, 1706. [Google Scholar] [CrossRef]
- Busi, M.V.; Gomez-Casati, D.F. Exploring Frataxin Function. IUBMB Life 2012, 64, 56–63. [Google Scholar] [CrossRef]
- Han, T.H.L.L.; Camadro, J.M.; Santos, R.; Lesuisse, E.; Chahine, J.M.E.H.; Ha-Duong, N.T.; El Hage Chahine, J.M.; Ha-Duong, N.T. Mechanisms of Iron and Copper-Frataxin Interactions. Metallomics 2017, 9, 1073–1085. [Google Scholar] [CrossRef]
- Turowski, V.R.; Aknin, C.; Maliandi, M.V.; Buchensky, C.; Leaden, L.; Peralta, D.A.; Busi, M.V.; Araya, A.; Gomez-Casati, D.F. Frataxin Is Localized to Both the Chloroplast and Mitochondrion and Is Involved in Chloroplast Fe-S Protein Function in Arabidopsis. PLoS ONE 2015, 10, e0141443. [Google Scholar] [CrossRef] [Green Version]
- Zubimendi, J.P.; Martinatto, A.; Valacco, M.P.; Moreno, S.; Andreo, C.S.; Drincovich, F.; Tronconi, M.A. The Complex Allosteric and Redox Regulation of the Fumarate Hydratase and Malate Dehydratase Reactions of Arabidopsis thaliana Fumarase 1 and 2 Gives Clues for Understanding the Massive Accumulation of Fumarate. FEBS J. 2018, 285, 2205–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pracharoenwattana, I.; Zhou, W.; Keech, O.; Francisco, P.B.; Udomchalothorn, T.; Tschoep, H.; Stitt, M.; Gibon, Y.; Smith, S.M. Arabidopsis Has a Cytosolic Fumarase Required for the Massive Allocation of Photosynthate into Fumaric Acid and for Rapid Plant Growth on High Nitrogen. Plant J. 2010, 62, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Molina, A.; Marino, G.; Lehmann, M.; Leister, D. Systems Biology of Responses to Simultaneous Copper and Iron Deficiency in Arabidopsis. Plant J. 2020, 103, 2119–2138. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Lehmann, M.; Schneider, K.; Klingl, A.; Leister, D. Inactivation of Cytosolic FUMARASE2 Enhances Growth and Photosynthesis under Simultaneous Copper and Iron Deprivation in Arabidopsis. Plant J. 2021, 106, 766–784. [Google Scholar] [CrossRef] [PubMed]
- Yanqun, Z.; Yuan, L.; Schvartz, C.; Langlade, L.; Fan, L. Accumulation of Pb, Cd, Cu and Zn in Plants and Hyperaccumulator Choice in Lanping Lead—Zinc Mine Area, China. Environ. Int. 2004, 30, 567–576. [Google Scholar] [CrossRef]
- Leitenmaier, B.; Küpper, H. Compartmentation and Complexation of Metals in Hyperaccumulator Plants. Front. Plant Sci. 2013, 4, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Gurajala, H.K.; Wu, L.; Van Der Ent, A.; Qiu, R.; Baker, A.J.M.; Tang, Y.; Yang, X.; Shu, W. Hyperaccumulator Plants from China: A Synthesis of the Current State of Knowledge. Environ. Sci. Technol. 2018, 52, 11980–11994. [Google Scholar] [CrossRef] [Green Version]
- Angulo-Bajarano, P.I.; Puente-Rivera, J.; Cruz-Ortega, R. Metal and Metalloid Toxicity in Plants: An Overview on Molecular Aspects. Plants 2021, 10, 635. [Google Scholar] [CrossRef]
- Xu, W.; Xiang, P.; Liu, X.; Ma, L.Q. Closely-Related Species of Hyperaccumulating Plants and Their Ability in Accumulation of As, Cd, Cu, Mn, Ni, Pb and Zn. Chemosphere 2020, 251, 126334. [Google Scholar] [CrossRef]
- Reeves, R.D.; Baker, A.J.M.; Jaffré, T.; Erskine, P.D.; Echevarria, G.; van der Ent, A. A Global Database for Plants That Hyperaccumulate Metal and Metalloid Trace Elements. New Phytol. 2018, 218, 407–411. [Google Scholar] [CrossRef] [Green Version]
- van der Ent, A.; Baker, A.J.M.; Reeves, R.D.; Pollard, A.J.; Schat, H. Hyperaccumulators of Metal and Metalloid Trace Elements: Facts and Fiction. Plant Soil 2013, 362, 319–334. [Google Scholar] [CrossRef]
- Küpper, H.; Götz, B.; Mijovilovich, A.; Küpper, F.C.; Meyer-Klaucke, W. Complexation and Toxicity of Copper in Higher Plants. I. Characterization of Copper Accumulation, Speciation, and Toxicity in Crassula helmsii as a New Copper Accumulator. Plant Physiol. 2009, 151, 702–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, A.J.M.; Brooks, R.R. Terrestrial Higher Plants Which Hyper—Accumulate Metallic Elements—A Review of Their Distribution, Ecology and Phytochemistry. Biorecovery 2014, 1, 81–126. [Google Scholar]
- Jiang, L.Y.; Yang, X.E.; He, Z.L. Growth Response and Phytoextraction of Copper at Different Levels in Soils by Elsholtzia splendens. Chemosphere 2004, 55, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shan, X.-Q.; Wen, B.; Zhang, S.; Wang, Z. Responses of Antioxidative Enzymes to Accumulation of Copper in a Copper Hyperaccumulator of Commoelina communis. Arch. Environ. Contam. Toxicol. 2004, 192, 185–192. [Google Scholar] [CrossRef]
- Kobayashi, F.; Maki, T.; Nakamura, Y.; Ueda, K. Determination of Cu, Pb, Fe, and Zn in Plant Component Polymers of a Hyperaccumulating Plant. Anal. Sci. 2005, 21, 1553–1556. [Google Scholar] [CrossRef] [Green Version]
- Küpper, H.; Lombi, E.; Zhao, F.-J.; Wieshammer, G.; Mcgrath, S.P. Cellular Compartmentation of Nickel in the Hyperaccumulators Alyssum lesbiacum, Alyssum bertolonii and Thlaspi goesingense. J. Exp. Bot. 2001, 52, 2291–2300. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.G.; Zhao, F.J.; Mcgrath, S. Uptake and Transport of Zinc in the Hyperaccumulator Thiaspi caerulescens and the Non-Hyperaccumulator Thiaspi ochroleucum. Plant. Cell Environ. 1997, 20, 898–906. [Google Scholar] [CrossRef] [Green Version]
- van der Ent, A.; Malaisse, F.; Erskine, P.D.; Mesjasz-Przybyłowicz, J.; Przybyłowicz, W.J.; Barnabas, A.D.; Sosnicka, M.; Harris, H.H. Abnormal Concentrations of Cu–Co in Haumaniastrum katangense, Haumaniastrum robertii and Aeolanthus biformifolius: Contamination or Hyperaccumulation? Metallomics 2019, 11, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wang-müller, Q.; Witt, T.; Malaisse, F.; Küpper, H. Differences in Copper Accumulation and Copper Stress between Eight Populations of Haumaniastrum katangense. Environ. Exp. Bot. 2012, 79, 58–65. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wairich, A.; De Conti, L.; Lamb, T.I.; Keil, R.; Neves, L.O.; Brunetto, G.; Sperotto, R.A.; Ricachenevsky, F.K. Throwing Copper Around: How Plants Control Uptake, Distribution, and Accumulation of Copper. Agronomy 2022, 12, 994. https://doi.org/10.3390/agronomy12050994
Wairich A, De Conti L, Lamb TI, Keil R, Neves LO, Brunetto G, Sperotto RA, Ricachenevsky FK. Throwing Copper Around: How Plants Control Uptake, Distribution, and Accumulation of Copper. Agronomy. 2022; 12(5):994. https://doi.org/10.3390/agronomy12050994
Chicago/Turabian StyleWairich, Andriele, Lessandro De Conti, Thainá I. Lamb, Rosana Keil, Leonardo O. Neves, Gustavo Brunetto, Raul A. Sperotto, and Felipe K. Ricachenevsky. 2022. "Throwing Copper Around: How Plants Control Uptake, Distribution, and Accumulation of Copper" Agronomy 12, no. 5: 994. https://doi.org/10.3390/agronomy12050994
APA StyleWairich, A., De Conti, L., Lamb, T. I., Keil, R., Neves, L. O., Brunetto, G., Sperotto, R. A., & Ricachenevsky, F. K. (2022). Throwing Copper Around: How Plants Control Uptake, Distribution, and Accumulation of Copper. Agronomy, 12(5), 994. https://doi.org/10.3390/agronomy12050994






