Effects of Soil Properties and Microbiome on Highbush Blueberry (Vaccinium corymbosum) Growth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing
2.3. Measurement Methods
2.3.1. Soil pH and EC
2.3.2. Determination of Soil Enzymes
2.3.3. DNA Extraction and Next-Generation Sequencing of Soil Microorganisms
2.3.4. Nutrient Elements in Roots, Stems and Leaves
2.3.5. Root Metabolome
2.4. Data Analysis
3. Results
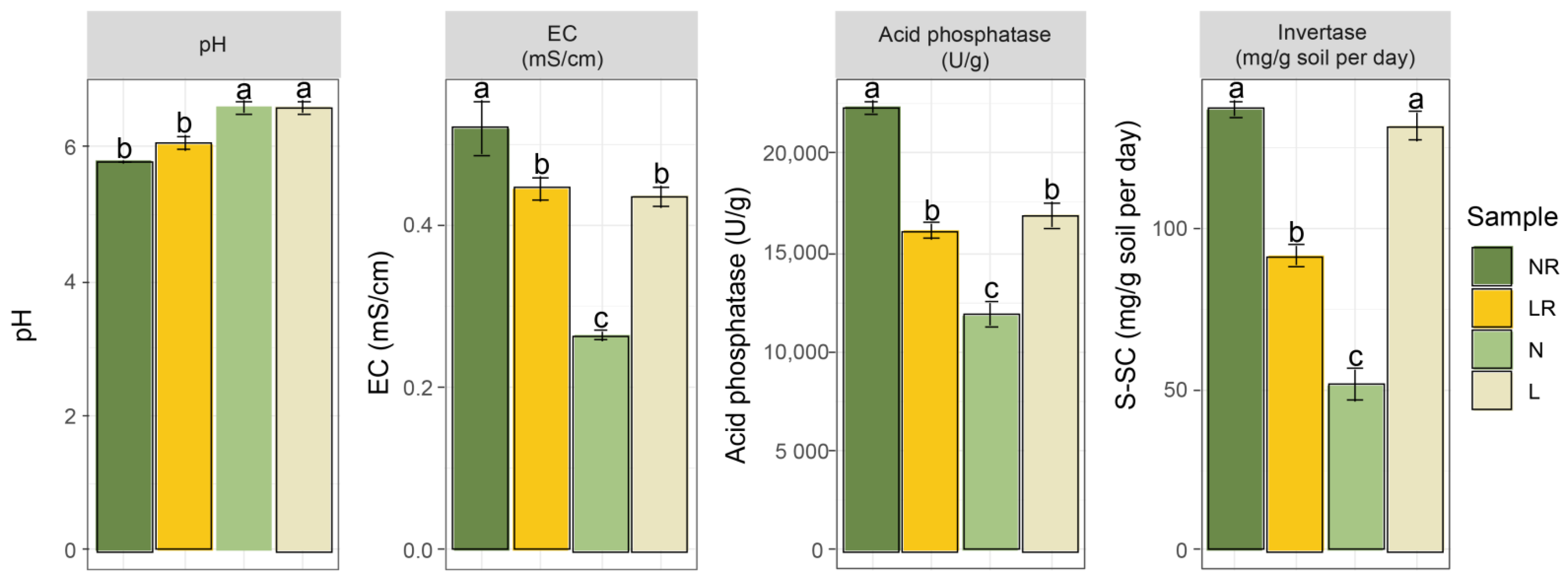
3.1. Soil Chemical Properties
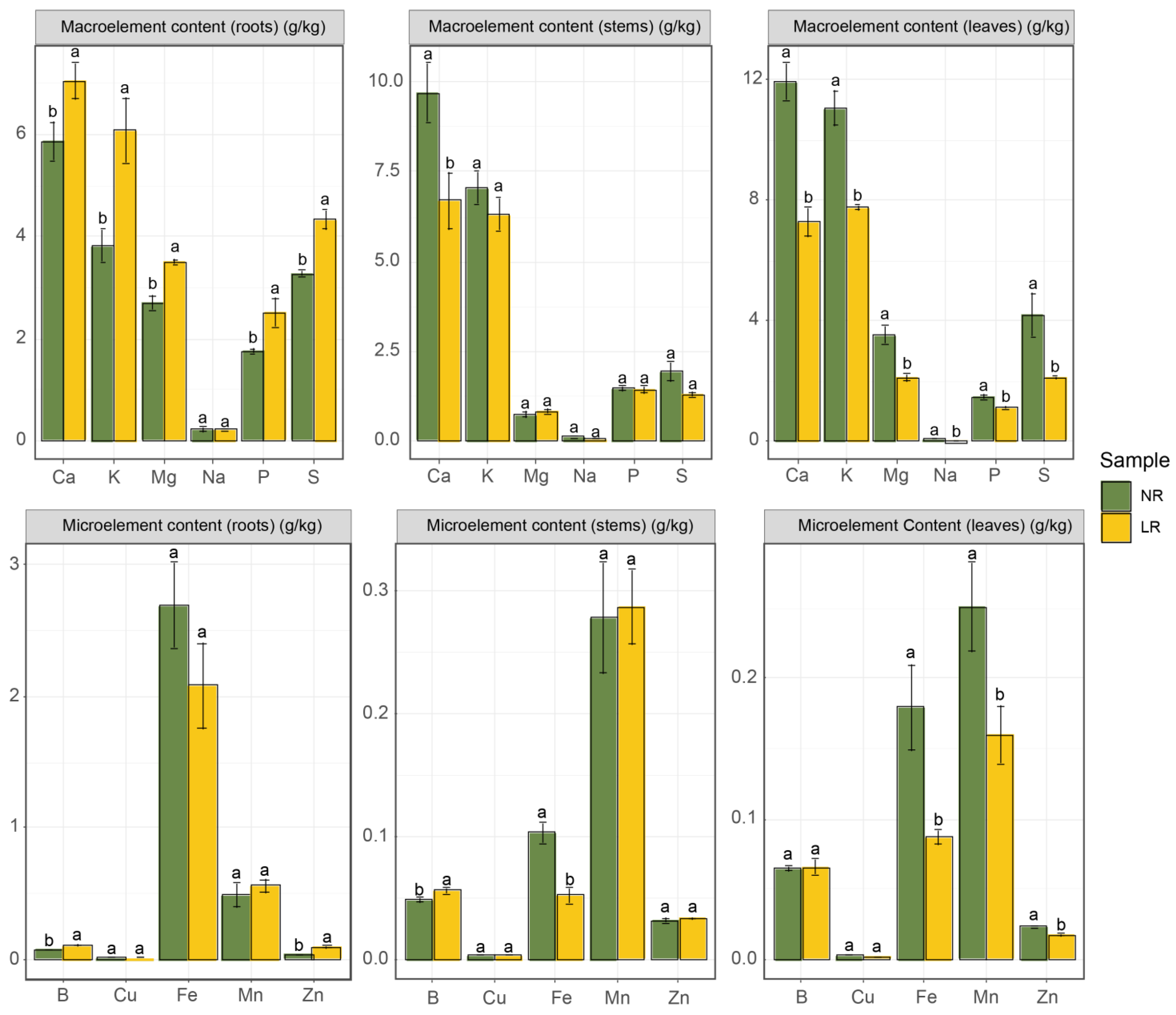
3.2. Plant Tissue Nutrient Content
3.3. Soil Microorganisms
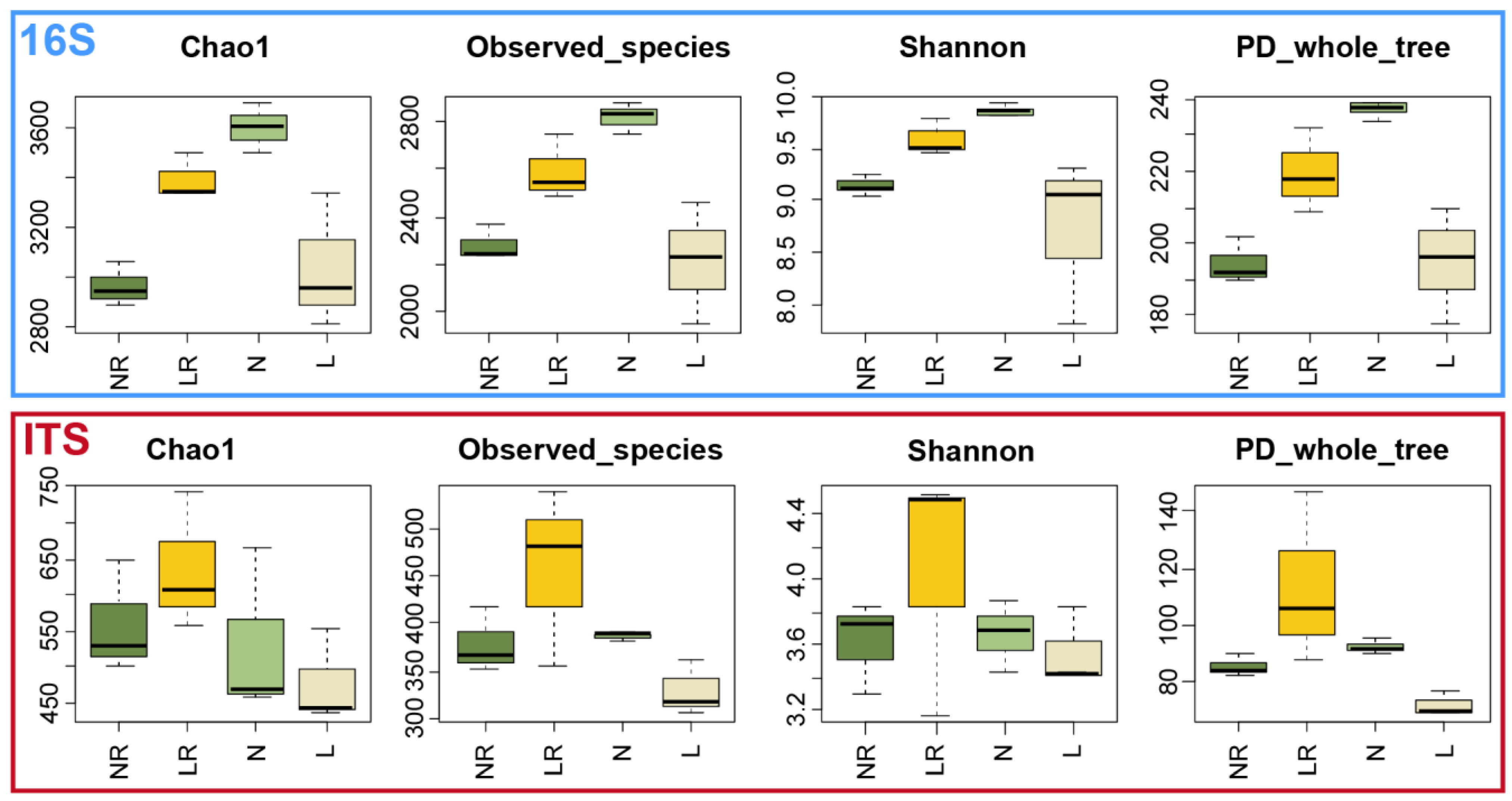
3.3.1. Alpha Diversity Analysis
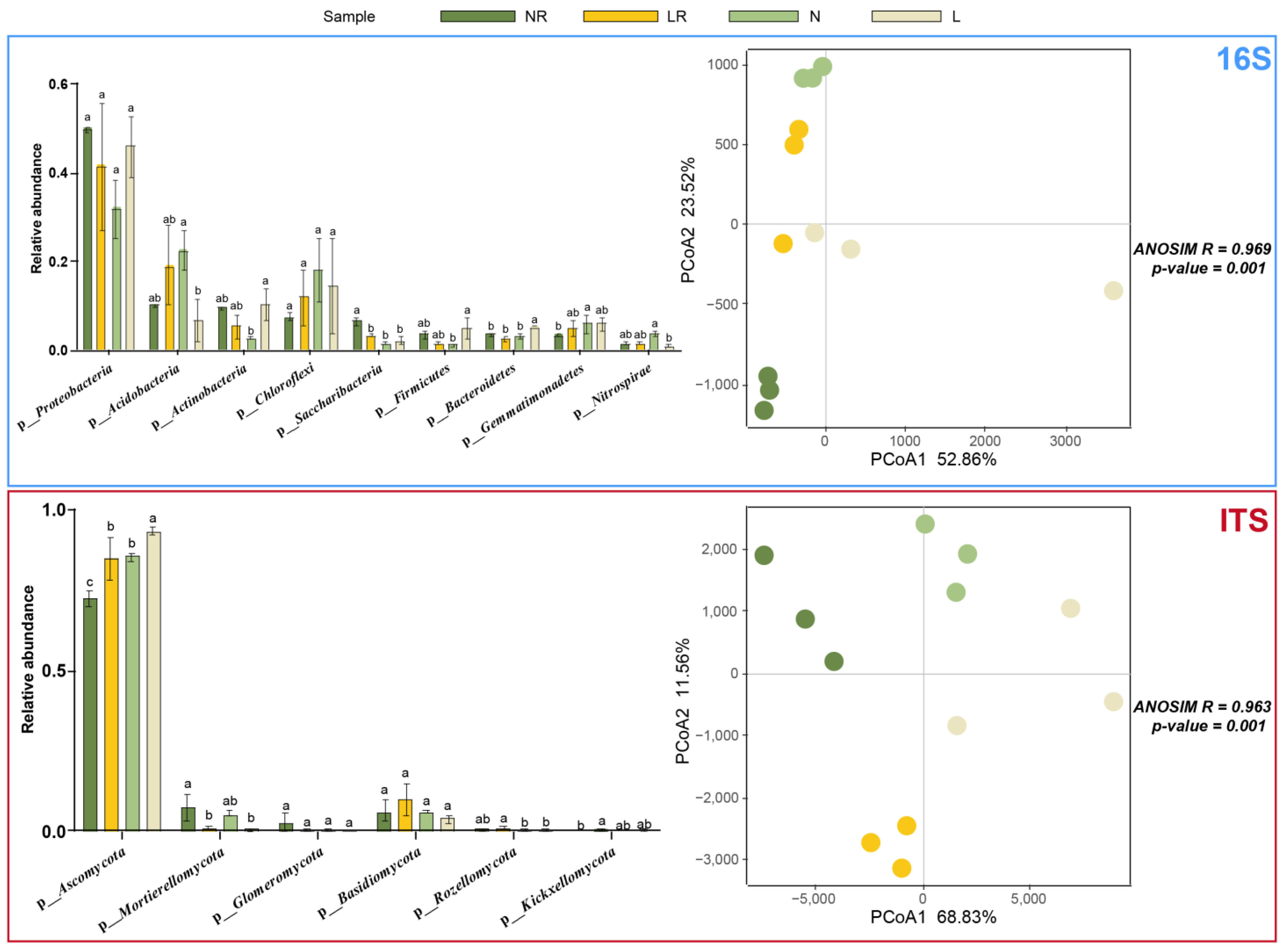
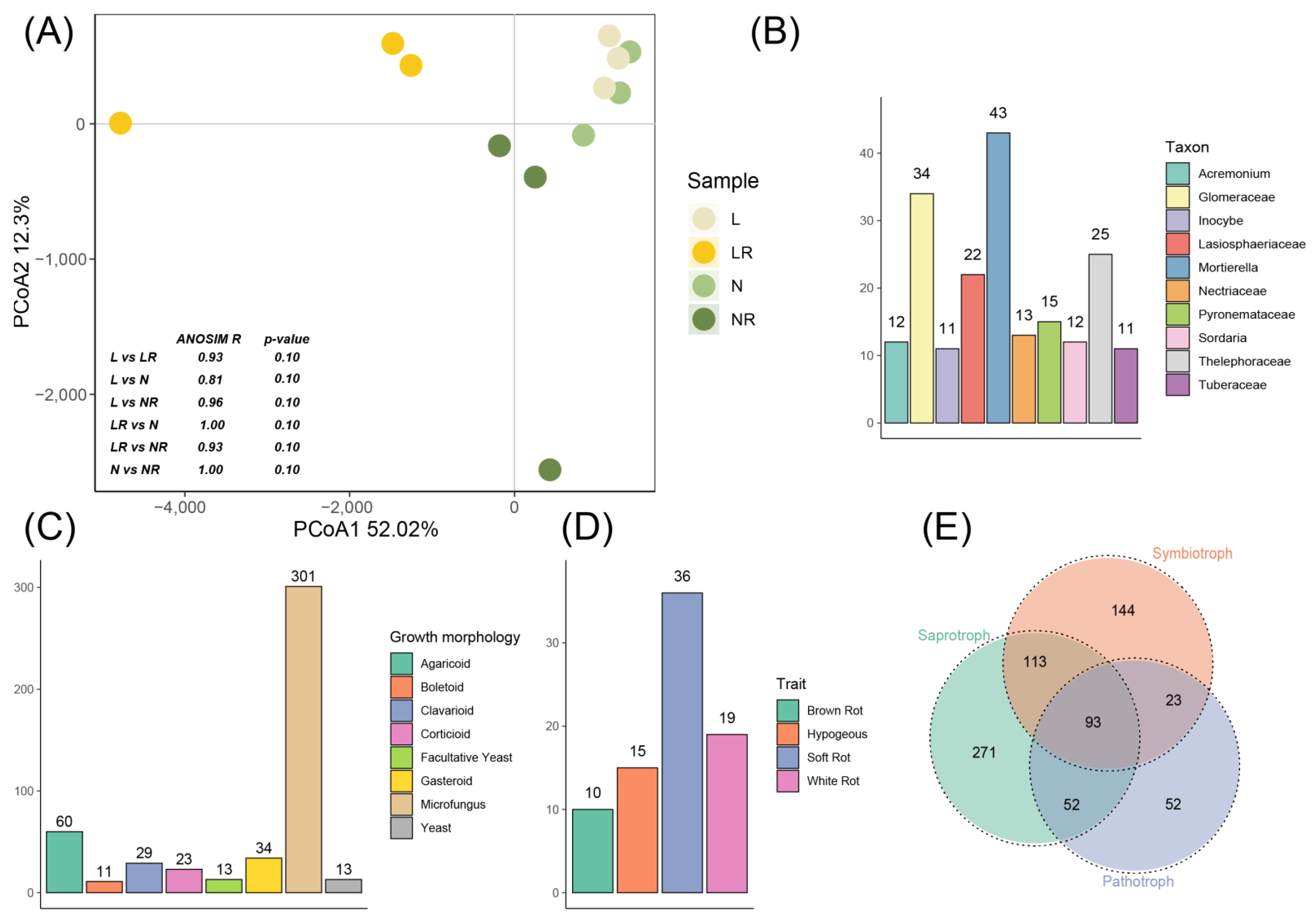
3.3.2. Analysis of Species Composition
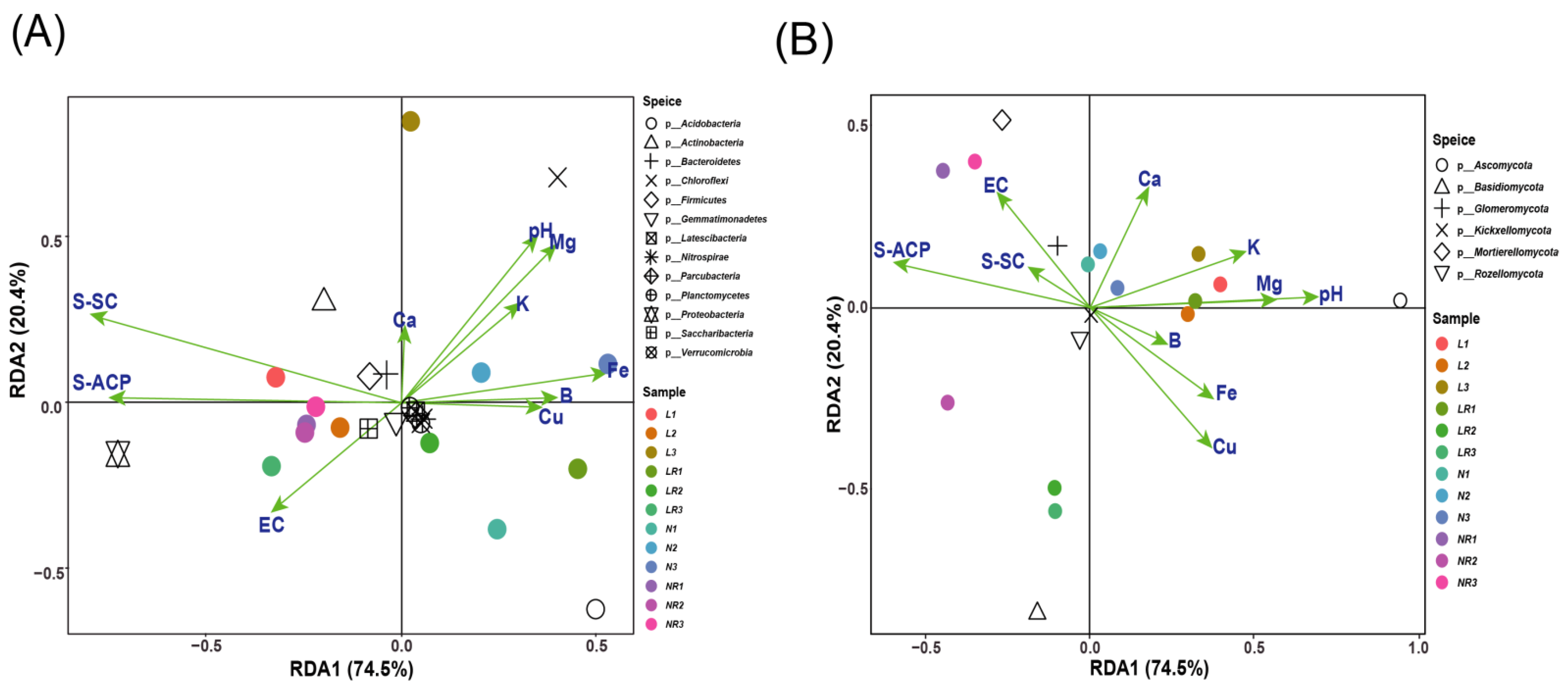
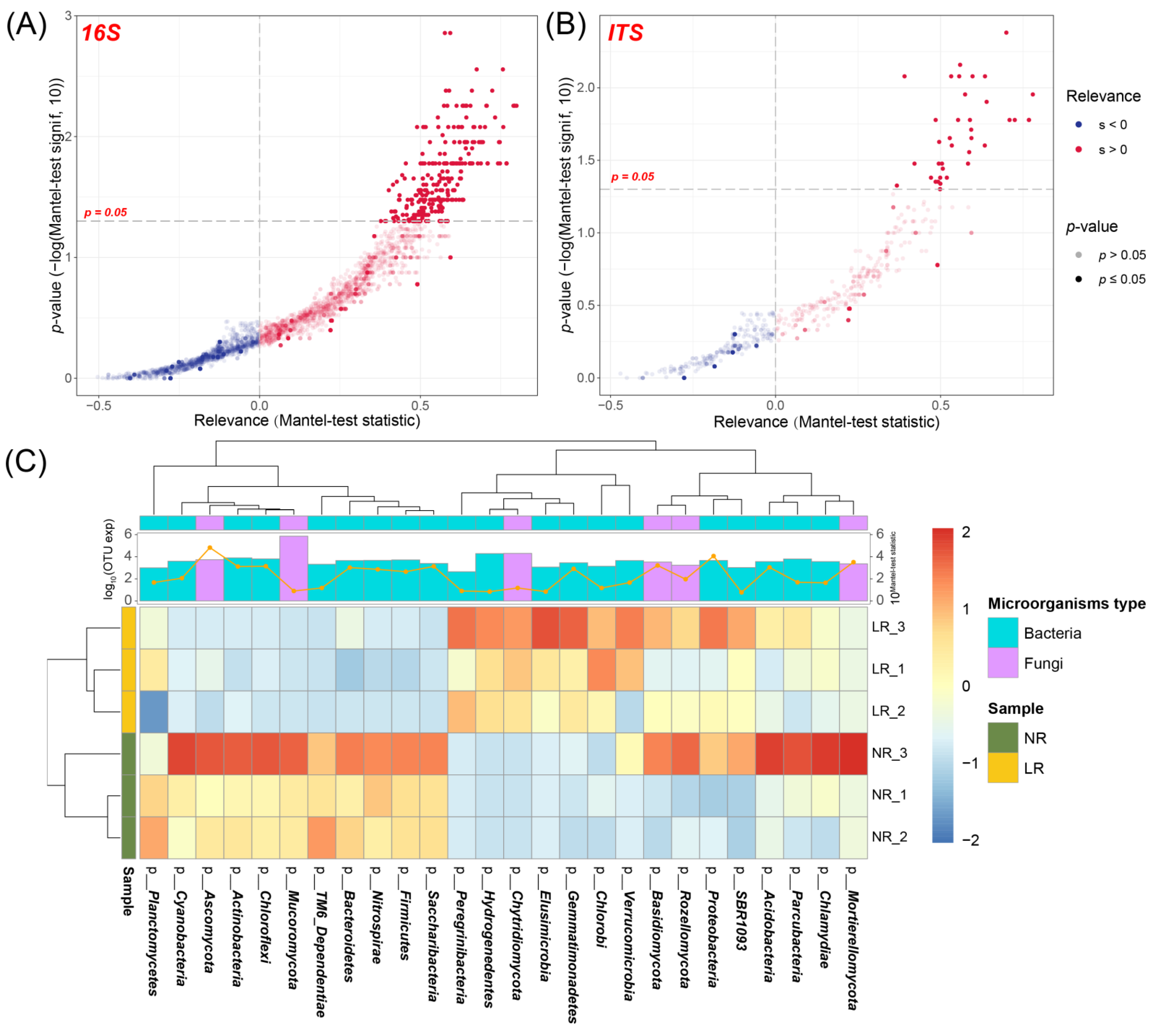
3.3.3. RDA Analysis
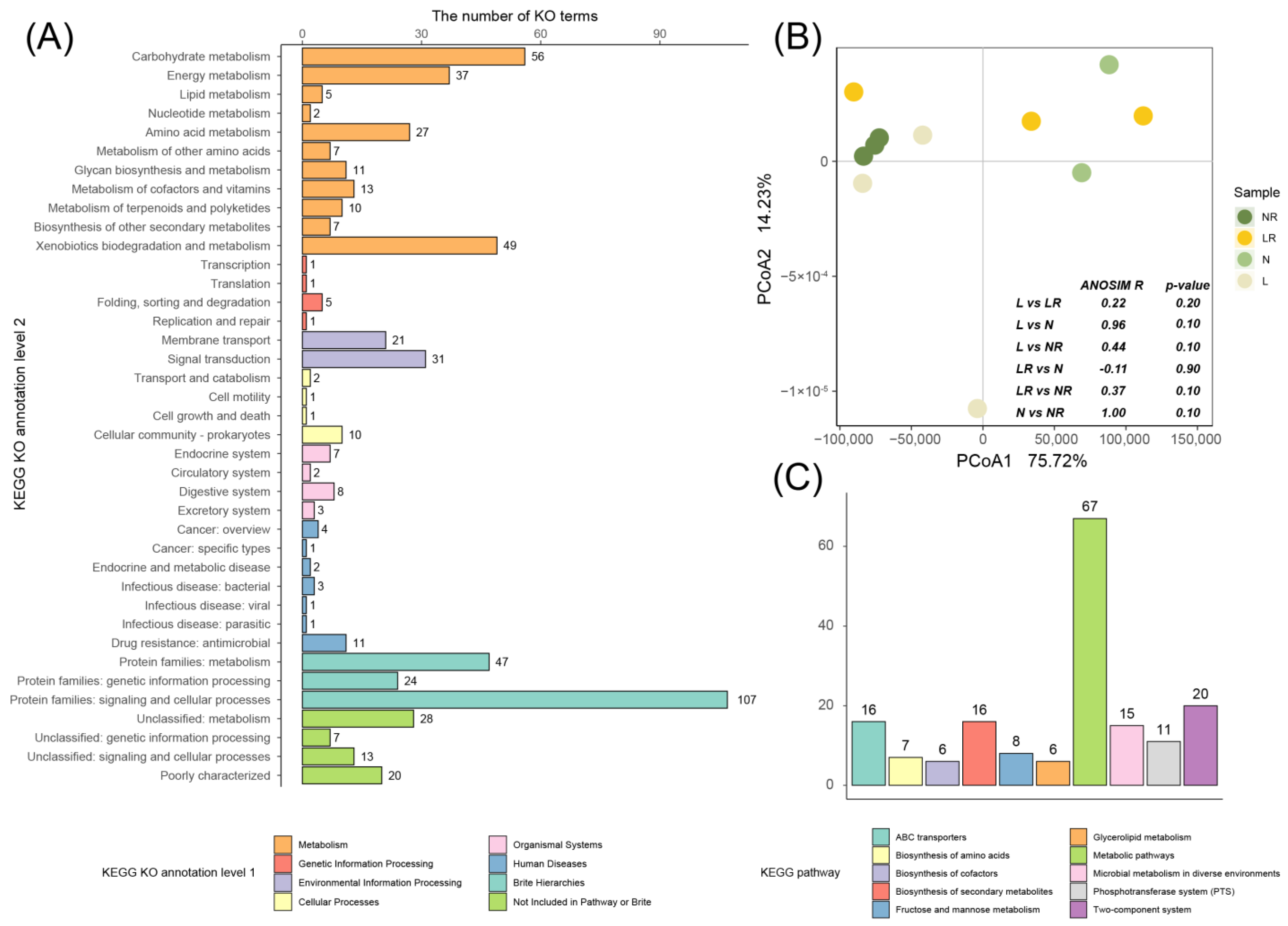
3.3.4. Prediction of Microbial Function
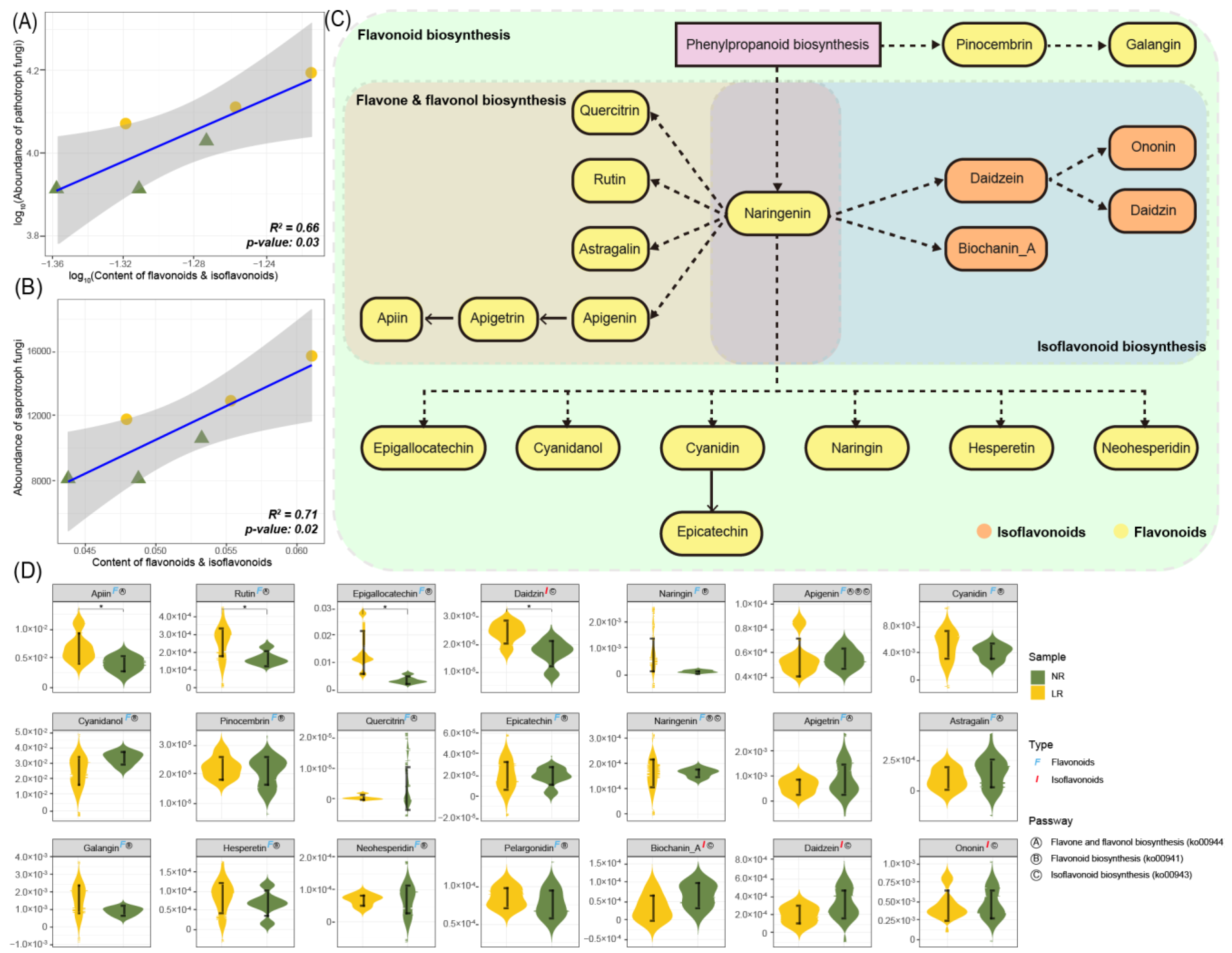
3.4. Root Metabolome
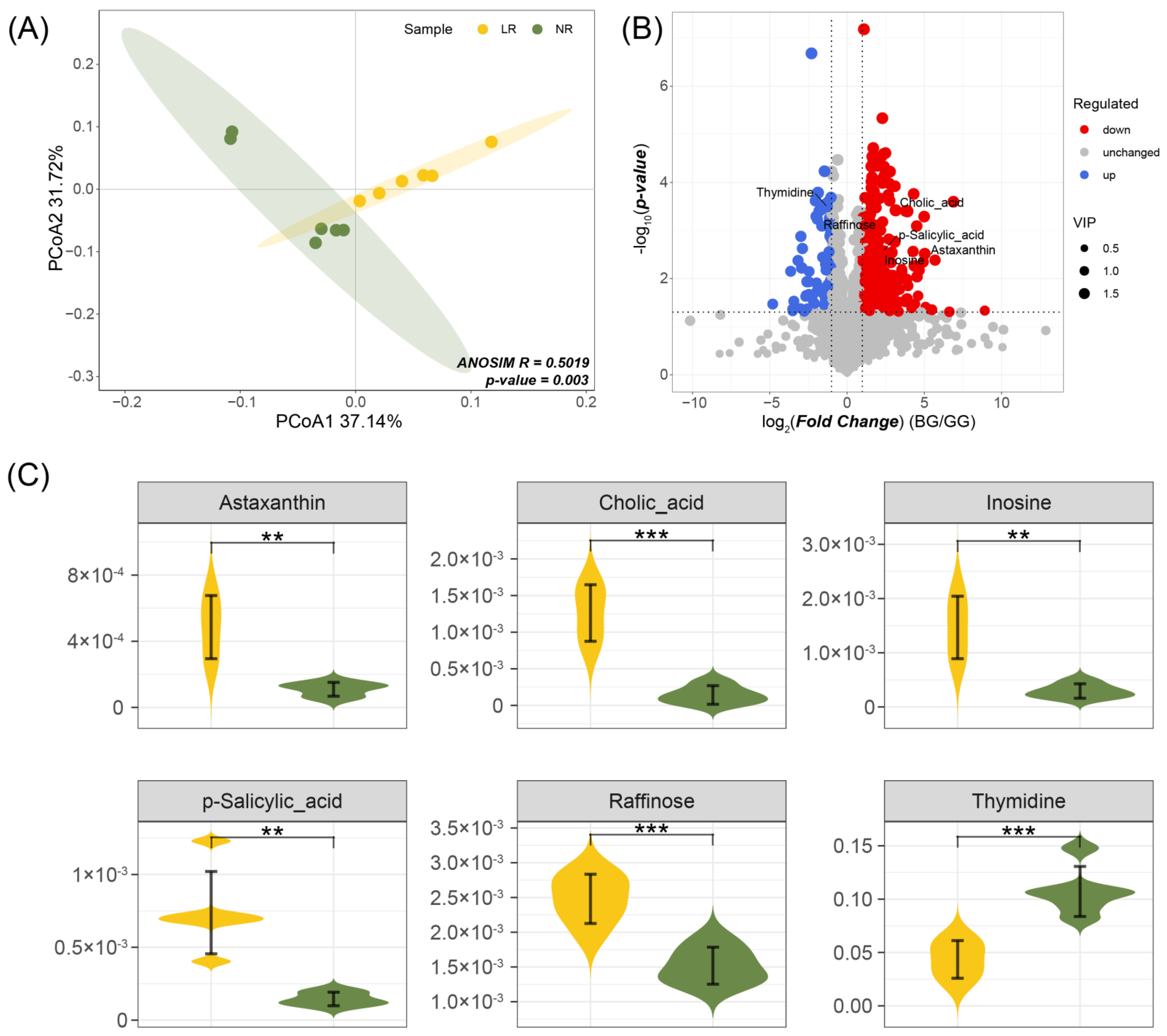
PCoA Analysis and Differential Metabolite Screening
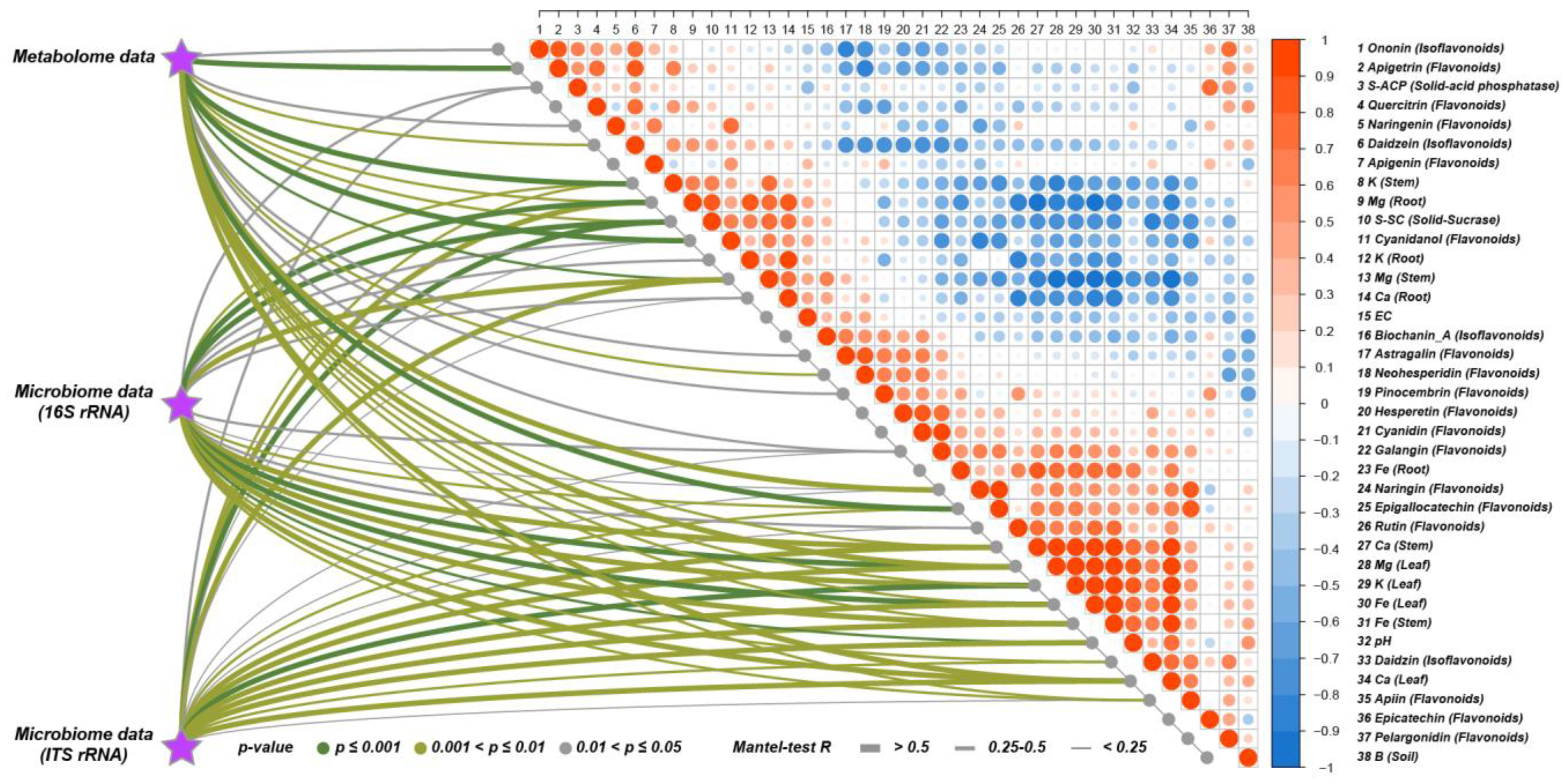
3.5. Joint Analysis of Root Metabolome and Soil Microorganisms
3.5.1. Screening of Microorganisms Related to Root Metabolome
3.5.2. Analysis of Flavonoids in Roots
3.6. Correlation Analysis between Environmental Factors and Root Metabolites
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lo Vasco, V.R.; Leopizzi, M.; Di Maio, V.; Di Raimo, T.; Cesa, S.; Masci, A.; Rocca, C.D. LPS, Oleuropein and blueberry extracts affect the survival, morphology and phosphoinositide signalling in stimulated human endothelial cells. J. Cell Commun. Signal. 2017, 11, 317–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Li, B.; Ma, Y.; Sun, X.; Lin, Y.; Meng, X. Polyphenols, anthocyanins, and flavonoids contents and the antioxidant capacity of various cultivars of highbush and half-high blueberries. J. Food Compos. Anal. 2017, 62, 84–93. [Google Scholar] [CrossRef]
- Holzapfel, E.A.; Hepp, R.F.; Mariño, M.A. Effect of irrigation on fruit production in blueberry. Agric. Water Manag. 2004, 67, 173–184. [Google Scholar] [CrossRef]
- Paal, T.; Starast, M.; Noormets-Šanski, M.; Vool, E.; Tasa, T.; Karp, K. Influence of liming and fertilization on lowbush blueberry in harvested peat field condition. Sci. Hortic. 2011, 130, 157–163. [Google Scholar] [CrossRef]
- Chang, C.J.; Donaldson, R.; Brannen, P.; Krewer, G.; Boland, R. Bacterial Leaf Scorch, a new blueberry disease caused by Xylella fastidiosa. HortScience 2009, 44, 413–417. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Bryla, D.R.; Vargas, O. Effects of salinity induced by ammonium sulfate fertilizer on root and shoot growth of highbush blueberry. Acta Hortic. 2014, 1017, 407–414. [Google Scholar] [CrossRef] [Green Version]
- Spiers, J.M.; Braswell, J.H. Soil-applied sulfur affects elemental leaf content and growth of ‘tifblue’ rabbiteye blueberry. J. Am. Soc. Hortic. Sci. 1992, 117, 230–233. [Google Scholar] [CrossRef] [Green Version]
- Tamir, G.; Zilkah, S.; Dai, N.; Shawahna, R.; Cohen, S.; Bar Tal, A. Combined effects of CaCO3 and the proportion of N-NH4+ among the total applied inorganic N on the growth and mineral uptake of rabbiteye blueberry. J. Soil Sci. Plant Nutr. 2020, 21, 35–48. [Google Scholar] [CrossRef]
- Anna, G.; Karolina, G.; Jarosław, G.; Magdalena, F.; Jerzy, K. Microbial community diversity and the interaction of soil under maize growth in different cultivation techniques. Plant Soil Environ. 2017, 63, 264–270. [Google Scholar] [CrossRef] [Green Version]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Salloum, M.S.; Menduni, M.F.; Benavides, M.P.; Larrauri, M.; Luna, C.M.; Silvente, S. Polyamines and flavonoids: Key compounds in mycorrhizal colonization of improved and unimproved soybean genotypes. Symbiosis 2018, 76, 265–275. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, L.J.; Jothibasu, K.; Reese, R.N.; Brözel, V.S.; Subramanian, S. Spatio temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Mol. Plant-Microbe Interact. 2015, 28, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhu, Y.; Shao, T.; Long, X.; Gao, X.; Zhou, Z. Relationship between rhizosphere soil properties and disease severity in highbush blueberry (Vaccinium corymbosum). Appl. Soil Ecol. 2019, 137, 187–194. [Google Scholar] [CrossRef]
- Zhu, T.; Shao, T.; Liu, J.; Li, N.; Long, X.; Gao, X.; Rengel, Z. Improvement of physico-chemical properties and microbiome in different salinity soils by incorporating Jerusalem artichoke residues. Appl. Soil Ecol. 2021, 158, 103791. [Google Scholar] [CrossRef]
- Shao, T.; Long, X.; Liu, Y.; Gao, X.; Liu, M.; Rengel, Z. Effect of industrial crop Jerusalem artichoke on the micro-ecological rhizosphere environment in saline soil. Appl. Soil Ecol. 2021, 166, 104080. [Google Scholar] [CrossRef]
- Shao, T.; Gu, X.; Zhu, T.; Pan, X.; Zhu, Y.; Long, X.; Shao, H.; Liu, M.; Rengel, Z. Industrial crop Jerusalem artichoke restored coastal saline soil quality by reducing salt and increasing diversity of bacterial community. Appl. Soil Ecol. 2019, 138, 195–206. [Google Scholar] [CrossRef]
- Li, N.; Shao, T.; Zhou, Y.; Cao, Y.; Hu, H.; Sun, Q.; Long, X.; Yue, Y.; Gao, X.; Rengel, Z. Effects of planting Melia azedarach L. on soil properties and microbial community in saline-alkali soil. Land Degrad. Dev. 2021, 32, 2951–2961. [Google Scholar] [CrossRef]
- Souza, S.O.; Costa, S.S.L.; Brum, B.C.T.; Santos, S.H.; Garcia, C.A.B.; Araujo, R.G.O. Determination of nutrients in sugarcane juice using slurry sampling and detection by ICP OES. Food Chem. 2019, 273, 57–63. [Google Scholar] [CrossRef]
- Finn, C.E.; Luby, J.J.; Rosen, C.J.; Ascher, P.D. Blueberry germplasm screening at several soil pH regimes. I. Plant Survival and Growth. J. Am. Soc. Hortic. Sci. 1993, 118, 377–382. [Google Scholar] [CrossRef] [Green Version]
- Haynes, R.J.; Swift, R.S. Effects of liming on the extractability of Fe, Mn, Zn and Cu from a peat medium and the growth and micronutrient uptake of highbush blueberry plants. Plant Soil 1985, 84, 213–223. [Google Scholar] [CrossRef]
- Tiffin, L.O. Mineral Nutrition of Plants: Principles and Perspectives. Soil Sci. Soc. Am. J. 1972, 36, 78. [Google Scholar] [CrossRef]
- Schmidt, S.B.; Husted, S. The Biochemical Properties of Manganese in Plants. Plants 2019, 8, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiel, R.S. Book review: Soil organic matter and biological activity. Outlook Agric. 1987, 16, 203. [Google Scholar] [CrossRef]
- Schomburg, I.; Chang, A.; Placzek, S.; Söhngen, C.; Rother, M.; Lang, M.; Munaretto, C.; Ulas, S.; Stelzer, M.; Grote, A.; et al. BRENDA in 2013: Integrated reactions, kinetic data, enzyme function data, improved disease classification: New options and contents in BRENDA. Nucleic Acids Res. 2012, 41, D764–D772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, B.L.; Cade-Menun, B.J.; Condron, L.M.; Newman, S. Extraction of soil organic phosphorus. Talanta 2005, 66, 294–306. [Google Scholar] [CrossRef]
- Lee, J.J.; Park, R.D.; Kim, Y.W.; Shim, J.H.; Chae, D.H.; Rim, Y.S.; Sohn, B.K.; Kim, T.H.; Kim, K.Y. Effect of food waste compost on microbial population, soil enzyme activity and lettuce growth. Bioresour. Technol. 2004, 93, 21–28. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16s rRNA and 16s rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [Green Version]
- Kersters, K.; De Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. Introduction to the proteobacteria. In The Prokaryotes: A Handbook on the Biology of Bacteri; Springer: Berlin/Heidelberg, Germany, 2006; pp. 3–37. [Google Scholar] [CrossRef]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X.; et al. Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Change Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef]
- Roley, S.S.; Xue, C.; Hamilton, S.K.; Tiedje, J.M.; Robertson, G.P. Isotopic evidence for episodic nitrogen fixation in switchgrass (Panicum virgatum L.). Soil Biol. Biochem. 2019, 129, 90–98. [Google Scholar] [CrossRef]
- Szymańska, S.; Borruso, L.; Brusetti, L.; Hulisz, P.; Furtado, B.; Hrynkiewicz, K. Bacterial microbiome of root-associated endophytes of Salicornia europaea in correspondence to different levels of salinity. Environ. Sci. Pollut. Res. 2018, 25, 25420–25431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smit, E.; Leeflang, P.; Gommans, S.; Broek, J.; van den Mil, S.; van Wernars, K. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Ka, J.O.; Cho, J.C. Members of the phylum acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindaichi, T.; Yamaoka, S.; Uehara, R.; Ozaki, N.; Ohashi, A.; Albertsen, M.; Nielsen, P.H.; Nielsen, J.L. Phylogenetic diversity and ecophysiology of Candidate phylum Saccharibacteria in activated sludge. FEMS Microbiol. Ecol. 2016, 92, fiw078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starr, E.P.; Shi, S.; Blazewicz, S.J.; Probst, A.J.; Herman, D.J.; Firestone, M.A.; Banfield, J.F. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially processed plant derived carbon. Microbiome 2018, 6, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, L.V.; Porras-Alfaro, A.; Kuske, C.R. Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genom. 2019, 20, 976. [Google Scholar] [CrossRef] [Green Version]
- Berbee, M.L. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant Pathol. 2001, 59, 165–187. [Google Scholar] [CrossRef] [Green Version]
- Kito, K.; Yamane, K.; Yamamori, T.; Matsuhira, H.; Tanaka, Y.; Takabe, T. Isolation, functional characterization and stress responses of raffinose synthase genes in sugar beet. J. Plant Biochem. Biotechnol. 2018, 27, 36–45. [Google Scholar] [CrossRef]
- Chu, Q.; Sha, Z.; Maruyama, H.; Yang, L.; Pan, G.; Xue, L.; Watanabe, T. Metabolic reprogramming in nodules, roots, and leaves of symbiotic soybean in response to iron deficiency. Plant Cell Environ. 2019, 42, 3027–3043. [Google Scholar] [CrossRef]
- George, S.; Manoharan, D.; Li, J.; Britton, M.; Parida, A. Drought and salt stress in macrotyloma uniflorum leads to common and specific transcriptomic responses and reveals importance of raffinose family oligosaccharides in stress tolerance. Gene Rep. 2018, 10, 7–16. [Google Scholar] [CrossRef]
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol. 2019, 180, 1725–1739. [Google Scholar] [CrossRef] [PubMed]
- Torun, H. Time-course analysis of salicylic acid effects on ROS regulation and antioxidant defense in roots of hulled and hulless barley under combined stress of drought, heat and salinity. Physiol. Plant. 2019, 165, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Kamran, M.; Imran, M.; Al-Harrasi, A.; Al-Rawahi, A.; Al-Amri, I.; Lee, I.-J.; Khan, A.L. Silicon and salicylic acid confer high-pH stress tolerance in tomato seedlings. Sci. Rep. 2019, 9, 19788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, S.; Wang, B.; Song, Y.; Xie, Z.; Li, C.; Li, S.; Huang, Y.; Jiang, M. Astaxanthin and its gold nanoparticles mitigate cadmium toxicity in rice by inhibiting cadmium translocation and uptake. Sci. Total Environ. 2021, 786, 147496. [Google Scholar] [CrossRef]
- Pietta, P.G. Flavonoids as Antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef]
- Fini, A.; Brunetti, C.; Ferdinando, M.D.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef]
- Gharibi, S.; Sayed Tabatabaei, B.E.; Saeidi, G.; Talebi, M.; Matkowski, A. The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech.f. Phytochemistry 2019, 162, 90–98. [Google Scholar] [CrossRef]
- Fejes, S.; Lemberkovics, E.; Balázs, A.; Apáti, P.; Kristó, T.S.; Szöke, E.; Kery, A.; Blázovics, A. Antioxidant activity of different compounds from Anthriscus cerefolium L. (Hoffm.). Acta Hortic. 2003, 597, 191–198. [Google Scholar] [CrossRef]
- Pradas del Real, A.E.; Silvan, J.M.; de Pascual-Teresa, S.; Guerrero, A.; García-Gonzalo, P.; Lobo, M.C.; Pérez-Sanz, A. Role of the polycarboxylic compounds in the response of Silene vulgaris to chromium. Environ. Sci. Pollut. Res. 2017, 24, 5746–5756. [Google Scholar] [CrossRef]
- De Matos, Y.M.L.S.; Vasconcelos, D.L.M.; Barreto, A.C.H.; Rocha, J.E.; Neto, J.B.; Campina, F.F.; de A. M. Guedes, T.T.; Sousa, A.K.; Teixeira, R.N.P.; Alvarez-Pizarro, J.C.; et al. Reduction of the phytotoxic effect of mercury chloride by rutin and evaluation of interactions by vibrational spectroscopy (Raman and FTIR). Vib. Spectrosc. 2020, 109, 103084. [Google Scholar] [CrossRef]
- Yang, L.-L.; Yang, L.; Yang, X.; Zhang, T.; Lan, Y.; Zhao, Y.; Han, M.; Yang, L.-M. Drought stress induces biosynthesis of flavonoids in leaves and saikosaponins in roots of Bupleurum chinense DC. Phytochemistry 2020, 177, 112434. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.J.; Jeong, H.Y.; Gil, C.S.; Kwon, S.J.; Na, J.K.; Lee, C.; Eom, S.H. Isoflavone accumulation and the metabolic gene expression in response to persistent UV-B irradiation in soybean sprouts. Food Chem. 2020, 303, 125376. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.J.; Lyu, J.I.; Kwon, S.J.; Eom, S.H. Effects of UV-A radiation on organ-specific accumulation and gene expression of isoflavones and flavonols in soybean sprout. Food Chem. 2021, 339, 128080. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liu, Y.; Zhang, X.; Gao, X.; Shao, T.; Long, X.; Rengel, Z. Effects of Soil Properties and Microbiome on Highbush Blueberry (Vaccinium corymbosum) Growth. Agronomy 2022, 12, 1263. https://doi.org/10.3390/agronomy12061263
Zhou Y, Liu Y, Zhang X, Gao X, Shao T, Long X, Rengel Z. Effects of Soil Properties and Microbiome on Highbush Blueberry (Vaccinium corymbosum) Growth. Agronomy. 2022; 12(6):1263. https://doi.org/10.3390/agronomy12061263
Chicago/Turabian StyleZhou, Yujie, Yuqing Liu, Xun Zhang, Xiumei Gao, Tianyun Shao, Xiaohua Long, and Zed Rengel. 2022. "Effects of Soil Properties and Microbiome on Highbush Blueberry (Vaccinium corymbosum) Growth" Agronomy 12, no. 6: 1263. https://doi.org/10.3390/agronomy12061263
APA StyleZhou, Y., Liu, Y., Zhang, X., Gao, X., Shao, T., Long, X., & Rengel, Z. (2022). Effects of Soil Properties and Microbiome on Highbush Blueberry (Vaccinium corymbosum) Growth. Agronomy, 12(6), 1263. https://doi.org/10.3390/agronomy12061263





