Apple Fruit Size QTLs on Chromosomes 8 and 16 Characterized in ‘Honeycrisp’-Derived Germplasm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Germplasm
2.2. Phenotypic Data
2.3. Genotypic Data
2.4. QTL Detection
2.5. Haplotype Characterization of QTL Alleles
3. Results
3.1. Phenotypic Data
3.2. QTL Detection
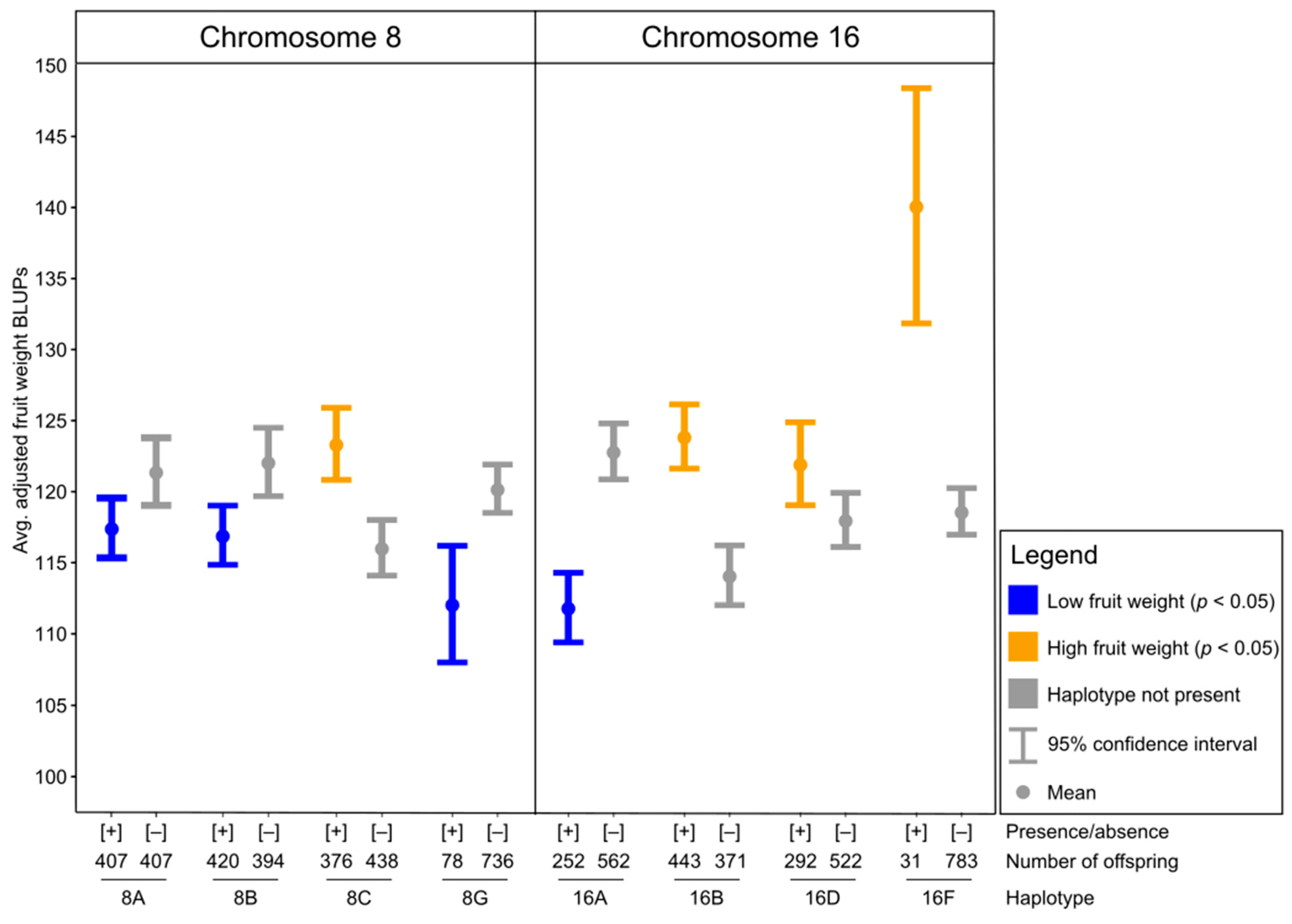
3.3. Chromosome 8 QTL Haplotype Analysis
3.4. Chromosome 16 QTL Haplotype Analysis
3.5. Interactions at and among the Chromosomes 8 and 16 QTLs
4. Discussion
4.1. Quantitative Variation for Fruit Weight Observed within Germplasm Set
4.2. Identities of Chromosomes 8 and 16 QTLs
4.3. Chromosome 16 QTL Colocalized with Other Important QTLs
4.4. ‘Minneiska’ Was Heterozygous for Fruit Weight QTL on Chromosome 8
4.5. Unique Low Fruit Weight Haplotype at the Chromosome 16 QTL in ‘Honeycrisp’
4.6. Putative Large Effect Fruit Weight Haplotype at the Chromosome 16 QTL in ‘Wildung’
4.7. Tracing of ‘MonArk’ Fruit Weight Haplotype Sources Hampered by Incomplete Pedigree
4.8. ‘Frostbite’, a Grandparent of ‘Honeycrisp’, Had Three Low Fruit Weight Haplotypes
4.9. Non-Additive Interactions at and among QTLs
4.10. Putative QTLs on Chromosomes 2, 4, 6, 10, and 14
4.11. Study Limitations
4.12. Breeding Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Musacchi, S.; Serra, S. Apple fruit quality: Overview on pre-harvest factors. Sci. Hortic. 2018, 234, 409–430. [Google Scholar] [CrossRef]
- Lee, J.; Mattheis, J.P.; Rudell, D.R. Fruit size affects physiological attributes and storage disorders in cold-stored ‘Royal Gala’ apples. HortScience 2013, 48, 1518–1520. [Google Scholar] [CrossRef] [Green Version]
- Robinson, T.L.; Watkins, C.B. Cropload of Honeycrisp affects not only fruit size but many quality attributes. N. Y. Fruit Q. 2003, 11, 7–10. [Google Scholar]
- Lakso, A.N.; Goffinet, M.C. Apple fruit growth. N. Y. Fruit Q. 2013, 21, 11–14. [Google Scholar]
- Autio, W.; Robinson, T.; Blatt, S.; Cochran, D.; Francescato, P.; Hoover, E.; Kushad, M.; Lang, G.; Lordan, J.; Miller, D.; et al. Budagovsky, Geneva, Pillnitz, and Malling apple rootstocks affect ‘Honeycrisp’ performance over eight years in the 2010 NC-140 ‘Honeycrisp’ apple rootstock trial. J. Am. Pomol. 2020, 74, 182–195. [Google Scholar]
- Cline, J.A.; Autio, W.; Clements, J.; Cowgill, W.; Crassweller, R.; Einhorn, T.; Fallahi, E.; Francescatto, P.; Hoover, E.; Lang, G.; et al. Early performance of ‘Honeycrisp’ apple trees on several size-controlling rootstocks in the 2014 NC-140 rootstock trial. J. Am. Pomol. 2021, 75, 189–202. [Google Scholar]
- Serra, S.; Leisso, R.; Giordani, L.; Kalcsits, L.; Musacchi, S. Crop load influences fruit quality, nutritional balance, and return bloom in ‘Honeycrisp’ apple. HortScience 2016, 51, 236–244. [Google Scholar] [CrossRef]
- Chang, Y.; Sun, R.; Sun, H.; Zhao, Y.; Han, Y.; Chen, D.; Wang, Y.; Zhang, X.; Han, Z. Mapping of quantitative trait loci corroborates independent genetic control of apple size and shape. Sci. Hortic. 2014, 174, 126–132. [Google Scholar] [CrossRef]
- Devoghalaere, F.; Doucen, T.; Guitton, B.; Keeling, J.; Payne, W.; Ling, T.J.; Ross, J.J.; Hallett, I.C.; Gunaseelan, K.; Dayatilake, G.A.; et al. A genomics approach to understanding the role of auxin in apple (Malus × domestica) fruit size. BMC Plant Biol. 2012, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Kenis, K.; Keulemans, J.; Davey, M.W. Identification and stability of QTLs for fruit traits in apple. Tree Genet. Genomes 2008, 4, 647–661. [Google Scholar] [CrossRef]
- King, G.J.; Lynn, J.R.; Dover, C.J.; Evans, K.M.; Seymour, G.B. Resolution of quantitative trait loci for mechanical measures accounting for genetic variation in fruit texture of apple (Malus pumila Mill.). Theor. Appl. Genet. 2001, 102, 1227–1235. [Google Scholar] [CrossRef]
- Potts, S.M.; Khan, M.A.; Han, Y.; Kushad, M.M.; Korban, S.S. Identification of quantitative trait loci (QTLs) for fruit quality traits in apple. Plant Mol. Biol. Rep. 2014, 32, 109–116. [Google Scholar] [CrossRef]
- Fresnedo-Ramírez, J.; Frett, J.T.; Sandefur, P.J.; Salgado-Rojas, A.; Clark, J.R.; Gasic, K.; Peace, C.P.; Anderson, J.; Hartmann, T.P.; Byrne, D.d.H.; et al. QTL mapping and breeding value estimation through pedigree-based analysis of fruit size and weight in four diverse peach breeding programs. Tree Genet. Genomes 2016, 12, 25. [Google Scholar] [CrossRef]
- Zhang, G.; Sebolt, A.M.; Sooriyapathirana, S.S.; Wang, D.; Bink, M.C.A.M.; Olmstead, J.W.; Iezzoni, A.F. Fruit size QTL analysis of an F1 population derived from a cross between a domesticated sweet cherry cultivar and a wild forest sweet cherry. Tree Genet. Genomes 2010, 6, 25–36. [Google Scholar] [CrossRef]
- Rosyara, U.R.; Bink, M.C.A.M.; van de Weg, E.; Zhang, G.; Wang, D.; Sebolt, A.; Dirlewanger, E.; Quero-Garcia, J.; Schuster, M.; Iezzoni, A.F. Fruit size QTL identification and the prediction of parental QTL genotypes and breeding values in multiple pedigreed populations of sweet cherry. Mol. Breed. 2013, 32, 875–887. [Google Scholar] [CrossRef]
- Grandillo, S.; Ku, H.M.; Tanksley, S.D. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 1999, 99, 978–987. [Google Scholar] [CrossRef]
- Prudent, M.; Causse, M.; Génard, M.; Tripodi, P.; Grandillo, S.; Bertin, N. Genetic and physiological analysis of tomato fruit weight and composition: Influence of carbon availability on QTL detection. J. Exp. Bot. 2009, 60, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Liebhard, R.; Kellerhals, M.; Pfammatter, W.; Jertmini, M.; Gessler, C. Mapping quantitative physiological traits in apple (Malus × domestica Borkh.). Plant Mol. Biol. 2003, 52, 511–526. [Google Scholar] [CrossRef]
- Luby, J.J.; Bedford, D.S. Honeycrisp Apple. Minnesota Report 225-1992. 1992. Available online: https://conservancy.umn.edu/bitstream/handle/11299/141481/MR225.pdf?sequence=1&isAllowed=y (accessed on 3 March 2022).
- Evans, E.M.; Barritt, B.H.; Konishi, B.S.; Brutcher, L.J.; Ross, C.F. ‘WA 38’ apple. HortScience 2012, 47, 1177–1179. [Google Scholar] [CrossRef] [Green Version]
- Changé, D.; Crowhurst, R.N.; Troggio, M.; Davey, M.W.; Gilmore, B.; Lawley, C.; Vanderzande, S.; Hellens, R.P.; Kumar, S.; Cestaro, A.; et al. Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS ONE 2012, 7, e31745. [Google Scholar] [CrossRef] [Green Version]
- Bianco, L.; Cestaro, A.; Sargent, D.J.; Banchi, E.; Derdak, S.; Di Guardo, M.; Salvi, S.; Jansen, J.; Viola, R.; Gut, I.; et al. Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS ONE 2014, 9, e110377. [Google Scholar] [CrossRef] [Green Version]
- Vanderzande, S.; Howard, N.P.; Cai, L.; Da Silva Linge, C.; Antanaviciute, L.; Bink, M.C.A.M.; Kruisselbrink, J.W.; Bassil, N.; Iezzoni, A.; van de Weg, E.; et al. High-quality, genome-wide SNP genotypic data for pedigreed germplasm of the diploid outcrossing species apple, peach, and sweet cherry through a common workflow. PLoS ONE 2019, 14, e0210928. [Google Scholar] [CrossRef] [Green Version]
- Bink, M.C.A.M.; Jansen, J.; Madduri, M.; Voorrips, R.E.; Durel, C.-E.; Kouassi, A.B.; Laurens, F.; Mathis, F.; Gessler, C.; Gobbin, D.; et al. Bayesian QTL analyses using pedigreed families of an outcrossing species, with application to fruit firmness in apple. Theor. Appl. Genet. 2014, 127, 1073–1090. [Google Scholar] [CrossRef] [PubMed]
- Bink, M.C.A.M.; Uimari, P.; Sillanpää, M.; Janss, L.; Jansen, R. Multiple QTL mapping in related plant populations via a pedigree-analysis approach. Theor. Appl. Genet. 2012, 104, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Bink, M.C.A.M.; Boer, M.P.; ter Braak, C.J.F.; Jansen, J.; Voorrips, R.E.; van de Weg, W.E. Bayesian analysis of complex traits in pedigreed plant populations. Euphytica 2008, 161, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Bink, M.C.A.M.; Totir, L.R.; ter Braak, C.J.F.; Winkler, C.R.; Boer, M.P.; Smith, O.S. QTL linkage analysis of connected populations using ancestral marker and pedigree information. Theor. Appl. Genet. 2012, 124, 1097–1113. [Google Scholar] [CrossRef] [Green Version]
- Peace, C.P.; Luby, J.J.; van de Weg, W.E.; Bink, M.C.A.M.; Iezzoni, A.F. A strategy for developing representative germplasm sets for systematic QTL validation, demonstrated for apple, peach, and sweet cherry. Tree Genet. Genomes 2014, 10, 1679–1694. [Google Scholar] [CrossRef]
- Blanpied, G.D.; Silsby, K.J. Predicting harvest date windows for apples. Cornell Coop. Ext. 1992, 221. Available online: https://ecommons.cornell.edu/handle/1813/3299 (accessed on 3 April 2022).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Amyotte, B.; Bowen, A.J.; Banks, T.; Rajcan, I.; Somers, D.J. Mapping the sensory perception of apple using descriptive sensory evaluation in a genome wide association study. PLoS ONE 2012, 12, e0171710. [Google Scholar] [CrossRef]
- Kostick, S.A.; Teh, S.L.; Norelli, J.L.; Vanderzande, S.; Peace, C.; Evans, K.M. Fire blight QTL analysis in a multi-family apple population identifies a reduced-susceptibility allele in ‘Honeycrisp’. Hortic. Res. 2021, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Howard, N.P.; van de Weg, E.; Bedford, D.S.; Peace, C.P.; Vanderzande, S.; Clark, M.D.; Teh, S.; Cai, L.; Luby, J.J. Elucidation of the ‘Honeycrisp’ pedigree through haplotype analysis with a multi-family integrated SNP linkage map and a large apple (Malus × domestica) pedigree-connected SNP data set. Hortic. Res. 2017, 4, 17003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, A.; Bink, M.C.A.M.; Martinez, S.; Kelner, J.-J.; Legave, J.-M.; di Guardo, M.; Di Pierro, E.A.; Laurens, F.; van de Weg, E.W.; Costes, E. Detecting QTLs and putative candidate genes involved in budbreak and flowering time in an apple multiparental population. J. Exp. Bot. 2016, 67, 2875–2888. [Google Scholar] [CrossRef] [Green Version]
- Di Guardo, M.; Bink, M.C.A.M.; Guerra, W.; Letschka, T.; Lozano, L.; Busatto, N.; Poles, L.; Tadiello, A.; Bianco, L.; Visser, R.G.F.; et al. Deciphering the genetic control of fruit texture in apple by multiple family-based analysis and genome-wide association. J. Exp. Bot. 2017, 68, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Peace, C.; Rudell, D.; Verma, S.; Evans, K. QTLs detected for individual sugars and soluble solids content in apple. Mol. Breed. 2015, 35, 135. [Google Scholar] [CrossRef]
- Howard, N.P.; van de Weg, E.; Tillman, J.; Tong, C.B.S.; Silverstein, K.A.T.; Luby, J.J. Two QTL characterized for soft scald and soggy breakdown in apple (Malus × domestica) through pedigree-based analysis of a large population of interconnected families. Tree Genet. Genomes 2018, 14, 2. [Google Scholar] [CrossRef] [Green Version]
- Howard, N.P.; Tillman, J.; Vanderzande, S.; Luby, J.J. Genetics of zonal leaf chlorosis and genetic linkage to a major gene regulating skin anthocyanin production (MdMYB1) in the apple (Malus × domestica) cultivar Honeycrisp. PLoS ONE 2019, 14, e0210611. [Google Scholar] [CrossRef] [Green Version]
- Van de Weg, E.; Di Guardo, M.; Jänsch, M.; Socquet-Juglard, D.; Costa, F.; Baumgartner, I.; Broggini, G.A.L.; Kellerhals, M.; Troggio, M.; Laurens, F.; et al. Epistatic fire blight resistance QTL alleles in the apple cultivar ‘Enterprise’ and selection X-6398 discovered and characterized through pedigree-informed analysis. Mol. Breed. 2018, 38, 5. [Google Scholar] [CrossRef]
- Verma, S.; Evans, K.; Guan, Y.; Luby, J.J.; Rosyara, U.R.; Howard, N.P.; Bassil, N.; Bink, M.C.A.M.; van de Weg, W.E.; Peace, C.P. Two large-effect QTLs, Ma and Ma3, determine genetic potential for acidity in apple fruit: Breeding insights from a multi-family study. Tree Genet. Genomes 2019, 15, 18. [Google Scholar] [CrossRef] [Green Version]
- Kass, R.E.; Raftery, A.E. Bayes factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Luby, J.J.; Howard, N.P.; Tillman, J.R.; Bedford, D.S. Extended pedigrees of apple cultivars from the University of Minnesota breeding program elucidated using SNP array markers. HortScience 2022, 57, 472–477. [Google Scholar] [CrossRef]
- Howard, N.P.; Albach, D.C.; Luby, J.J. The identification of apple pedigree information on a large diverse set of apple germplasm and its application in apple breeding using new genetic tools. In Proceedings of the 18th International Conference on Organic Fruit Growing, Hohenheim, Germany, 19–21 February 2018; pp. 88–91. [Google Scholar]
- Kassambara, A. Ggpubr: ‘Ggplot2’ Based Publication Read Plots. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 24 April 2022).
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. Available online: http://tarwi.lamolina.edu.pe/~fmendiburu (accessed on 24 April 2022).
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Garrick, D.J.; Bink, M.C.; Whitworth, C.; Changé, D.; Volz, R.K. Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genom. 2013, 14, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buti, M.; Poles, L.; Caset, D.; Magnago, P.; Fernandez-Fernandez, F.; Colgan, R.J.; Velasco, R.; Sargent, D.J. Identification and validation of a QTL influencing bitter pit symptoms in apple (Malus × domestica). Mol. Breed. 2015, 35, 29. [Google Scholar] [CrossRef]
- Ferguson, I.B.; Triggs, C.M. Sampling factors affecting the use of mineral analysis of apple fruit for the prediction of bitter pit. N. Z. J. Crop Hortic. Sci. 1990, 18, 147–152. [Google Scholar] [CrossRef]
- Cheng, L.; Sazo, M.M. Why is ‘Honeycrisp’ so susceptible to bitter pit? N. Y. State Hortic. Soc. Fruit Q. 2018, 26, 19–23. [Google Scholar]
- Muranty, H.; Denancé, C.; Feugey, L.; Crépin, J.-L.; Barbier, Y.; Tartarini, S.; Ordidge, M.; Troggio, M.; Lateur, M.; Nybom, H.; et al. Using whole-genome SNP data to reconstruct a large multi-generational pedigree in apple germplasm. BMC Plant Biol. 2020, 20, 2. [Google Scholar] [CrossRef] [Green Version]
- University of Minnesota—Minnesota Agricultural Experiment Station. FrostbiteTM. Available online: https://mnhardy.umn.edu/varieties/fruit/apples/frostbite#:~:text=The%20Frostbite%E2%84%A2%20Apple%20(MN,a%20distinctive%20late%20season%20option (accessed on 3 March 2022).
- Evans, K.; Peace, C. Advances in marker-assisted breeding of apples. In Achieving Sustainable Cultivation of Apples; Evans, K., Ed.; Burleigh Dodds Science Publishing: Cambridge, UK, 2017; pp. 165–192. [Google Scholar]
- McClure, K.A.; Gardner, K.M.; Douglas, G.M.; Song, J.; Forney, C.F.; DeLong, J.; Fan, L.; Du, L.; Toivonen, P.M.A.; Somers, D.J.; et al. A genome-wide association study of apple quality and scab resistance. Plant Genome 2018, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Minamikawa, M.F.; Kunihisa, M.; Noshita, K.; Moriya, S.; Abe, K.; Hayashi, T.; Katayose, Y.; Matsumoto, T.; Nishitani, C.; Terakami, S.; et al. Tracing founder haplotypes of Japanese apple variaties: Application in genomic prediction and genome-wide association study. Hortic. Res. 2021, 8, 49. [Google Scholar] [CrossRef]
- Muranty, H.; Troggio, M.; Sadok, I.B.; Al Rifaı, M.; Auwerkerken, A.; Banchi, E.; Velasco, R.; Stevanato, P.; van de Weg, W.E.; Di Guardo, M.; et al. Accuracy and responses of genomic selection on key traits in apple. Hortic. Res. 2015, 2, 15060. [Google Scholar] [CrossRef] [Green Version]

| Family | Parents | No. of Offspring | |
|---|---|---|---|
| Maternal | Paternal | ||
| 1 | Dayton | Minnewashta | 23 |
| 2 | Honeycrisp | Jonafree | 53 |
| Jonafree | Honeycrisp | ||
| 3 | Honeycrisp | MN1702 | 52 |
| MN1702 | Honeycrisp | ||
| 4 | Honeycrisp | MN1836 | 49 |
| 5 | Honeycrisp | MN1915 | 30 |
| 6 | Honeycrisp | MonArk | 75 |
| 7 | Honeycrisp | Pitmaston Pine Apple | 51 |
| 8 | Honeycrisp | WA2 | 48 |
| 9 | Honeycrisp | Minnewashta | 149 |
| 10 | Minneiska | MN1965 | 49 |
| 11 | Minneiska | MN55 | 124 |
| 12 | Minneiska | Wildung | 70 |
| 13 | MN1702 | Minneiska | 41 |
| Total | 814 | ||
| Chr a | Replicate Run b | BF(1 vs. 0) c | BF(2 vs. 1) d | QTL Interval (cM) e | Mode (cM) f | Approx. Physical Position (Mbp) g | PVE (%) h |
|---|---|---|---|---|---|---|---|
| 2 | 1 | 8.0 | −2.8 | 47–53 | 49 | 16.7–23.7 | - |
| 2 | 8.5 | −2.7 | 47–53 | 49 | 16.7–23.7 | ||
| 4 | 1 | 6.8 | 6.2 | 31–37 | 33 | 20.8–23.5 | - |
| 2 | 5.6 | 3.4 | 29–37 | 34 | 20.5–23.5 | ||
| 6 | 1 | 6.0 | −1.2 | 52–60 | 54 | 32.1–32.9 | - |
| 2 | 6.6 | −0.8 | 52–58 | 55 | 32.1–32.8 | ||
| 8 | 1 | 15.6 | 2.4 | 0–4 | 1 | 0.3–1.9 | 8.0 |
| 2 | 15.8 | 1.7 | 0–8 | 1 | 0.3–2.7 | ||
| 10 | 1 | 8.4 | −3.5 | 72–80 | 75 | 39.0–40.8 | - |
| 2 | 8.9 | −3.0 | 68–78 | 70 | 37.6–40.5 | ||
| 14 | 1 | 5.2 | −2.2 | 3–11 | 4 | 1.6–3.8 | - |
| 2 | 6.0 | −0.8 | 3–13 | 4 | 1.6–3.8 | ||
| 16 | 1 | 30.0 | −0.4 | 9–13 | 10 | 3.1–4.2 | 7.0 |
| 2 | 29.9 | 0.5 | 9–13 | 10 | 3.1–4.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostick, S.A.; Luby, J.J. Apple Fruit Size QTLs on Chromosomes 8 and 16 Characterized in ‘Honeycrisp’-Derived Germplasm. Agronomy 2022, 12, 1279. https://doi.org/10.3390/agronomy12061279
Kostick SA, Luby JJ. Apple Fruit Size QTLs on Chromosomes 8 and 16 Characterized in ‘Honeycrisp’-Derived Germplasm. Agronomy. 2022; 12(6):1279. https://doi.org/10.3390/agronomy12061279
Chicago/Turabian StyleKostick, Sarah A., and James J. Luby. 2022. "Apple Fruit Size QTLs on Chromosomes 8 and 16 Characterized in ‘Honeycrisp’-Derived Germplasm" Agronomy 12, no. 6: 1279. https://doi.org/10.3390/agronomy12061279





