Large-Effect QTLs for Titratable Acidity and Soluble Solids Content Validated in ‘Honeycrisp’-Derived Apple Germplasm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotypic Data
2.3. Genotypic Data
2.4. QTL Mapping and Haplotype Analyses
2.4.1. QTL Analyses
2.4.2. Haplotype Analyses
3. Results
3.1. Phenotypic Data
3.2. QTL Detection
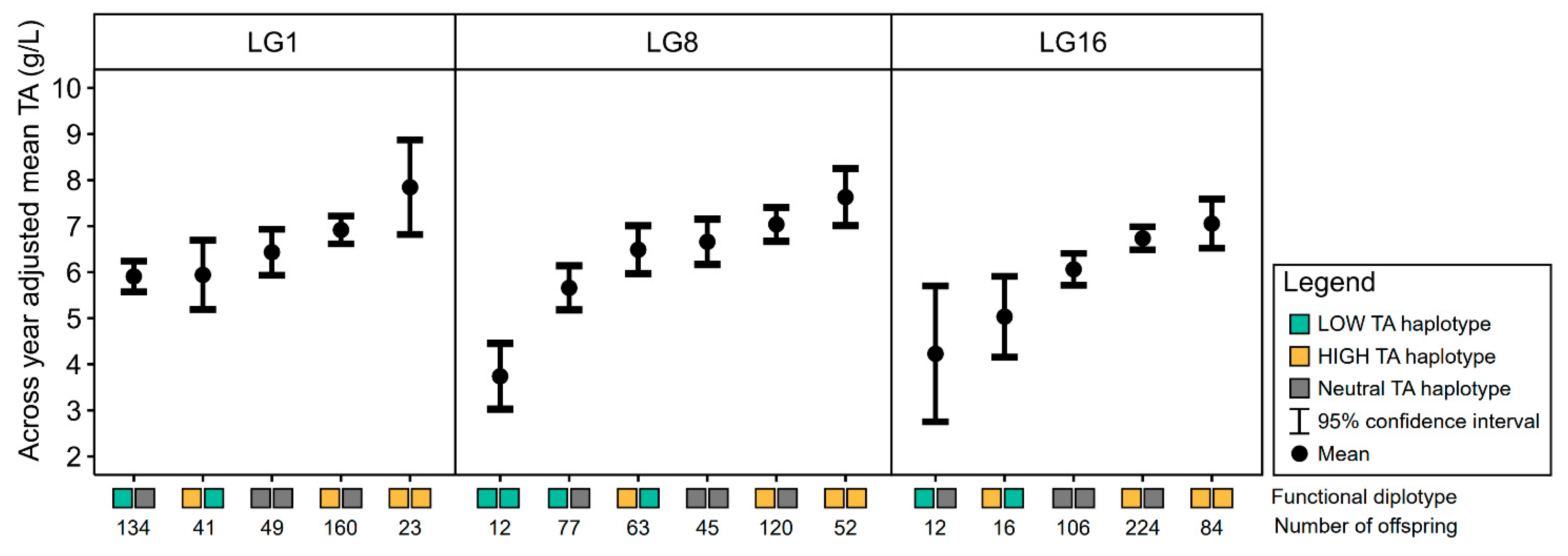
3.3. Haplotype Analyses for Titratable Acidity QTLs
3.3.1. LG1 Titratable Acidity QTL
3.3.2. LG8 Titratable Acidity QTL
3.3.3. LG16 Titratable Acidity QTL
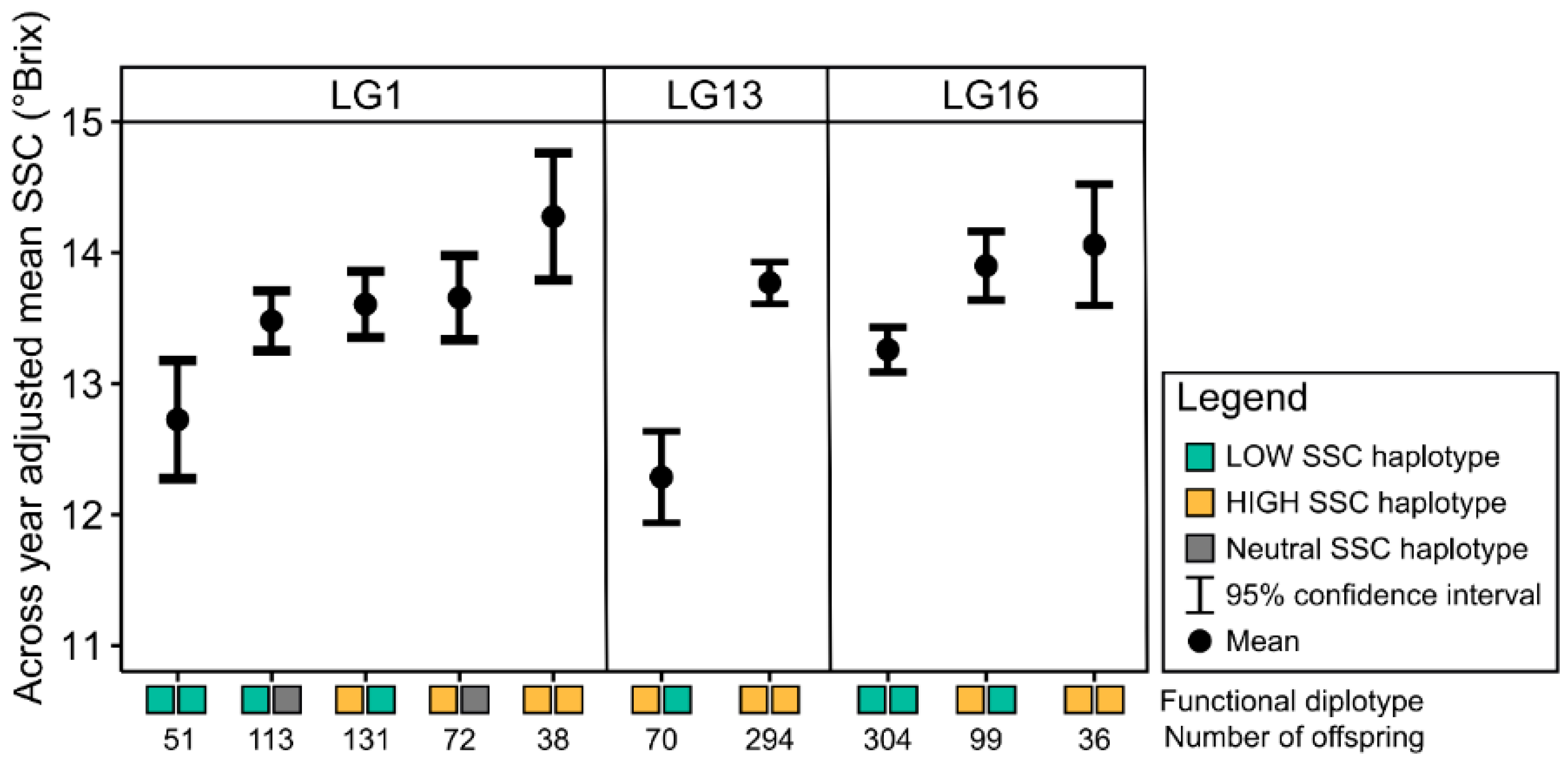
3.4. Haplotype Analyses for Soluble Solids Content QTLs
3.4.1. LG1 Soluble Solids Content QTL
3.4.2. LG13 Soluble Solids Content QTL
3.4.3. LG16 Soluble Solids Content QTL
3.5. Interactions at and among QTLs
3.5.1. Titratable Acidity QTL Interactions
3.5.2. Soluble Solids Content QTL Interactions
4. Discussion
4.1. QTL Identities
4.2. Breeding Parents Heterozygous for the Characterized QTLs
4.3. LG1 QTL Haplotype Putatively Associated with Low Titratable Acidity and High Soluble Solids Content
4.4. LG8 and LG16 Titratable Acidity QTL Haplotype Effects Validated
4.5. Most LG16 QTL Haplotypes Putatively Associated with High Titratable Acidity and Low Soluble Solids Content
4.6. Interactions at and among Detected QTLs
4.7. Study Limitations
4.8. Breeding Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoehn, E.; Gasser, F.; Guggenbuhl, B.; Kunsch, U. Efficacy of instrumental measurements for determination of minimum requirements of firmness, soluble solids, and acidity of several apple varieties in comparison to consumer expectations. Postharvest Biol. Technol. 2003, 27, 27–37. [Google Scholar] [CrossRef]
- Rymenants, M.; Van de Weg, E.; Auwerkerken, A.; De Wit, I.; Czech, A.; Nijland, B.; Heuven, H.; De Storme, N.; Keulemans, W. Detection of QTL for apple fruit acidity and sweetness using sensorial evaluation in multiple pedigreed full-sib families. Tree Genet. Genomes 2020, 16, 71. [Google Scholar] [CrossRef]
- Verma, S.; Evans, K.; Guan, Y.; Luby, J.J.; Rosyara, U.R.; Howard, N.P.; Bassil, N.; Bink, M.C.A.M.; van de Weg, W.E.; Peace, C.P. Two large-effect QTLs, Ma and Ma3, determine genetic potential for acidity in apple fruit: Breeding insights from a multi-family study. Tree Genet. Genomes 2019, 15, 18. [Google Scholar] [CrossRef] [Green Version]
- Maliepaard, C.; Alston, F.H.; van Arkel, G.; Brown, L.M.; Chevreau, E.; Dunemann, F.; Evans, K.M.; Gardiner, S.; Guilford, P.; van Heusden, A.W.; et al. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor. Appl. Genet. 1998, 97, 60–73. [Google Scholar] [CrossRef]
- Guan, Y.; Peace, C.; Rudell, D.; Verma, S.; Evans, K. QTLs detected for individual sugars and soluble solids content in apple. Mol. Breed. 2015, 35, 135. [Google Scholar] [CrossRef]
- Liebhard, R.; Kellerhals, M.; Pfammatter, W.; Jertmini, M.; Gessler, C. Mapping quantitative physiological traits in apple (Malus × domestica Borkh.). Plant Mol. Biol. 2003, 52, 511–526. [Google Scholar] [CrossRef]
- Kenis, K.; Keulemans, J.; Davey, M.W. Identification and stability of QTLs for fruit quality traits in apple. Tree Genet. Genomes 2008, 4, 647–661. [Google Scholar] [CrossRef]
- Sun, R.; Chang, Y.; Yang, F.; Wang, Y.; Li, H.; Zhao, Y.; Chen, D.; Wu, T.; Zhang, X.; Han, Z. A dense SNP genetic map constructed using restriction site-associated DNA sequencing enables detection of QTLs controlling apple fruit quality. BMC Genom. 2015, 16, 747. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Wang, A.; Brown, S. Genetic characterization of the Ma locus with pH and titratable acidity in apple. Mol. Breed. 2012, 30, 899–912. [Google Scholar] [CrossRef]
- Liu, Z.; Bao, D.; Liu, D.; Zhang, Y.; Ashraf, M.A.; Chen, X. Construction of a Genetic Linkage Map and QTL Analysis of Fruit-related Traits in an F1 Red Fuji x Hongrou Apple Hybrid. Open Life Sci. 2016, 11, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Jia, D.; Shen, F.; Wang, Y.; Wu, T.; Xu, X.; Zhang, X.; Han, Z. Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH, and MdALMTII. Plant J. 2018, 95, 427–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, S.; Xu, K. Identification of two QTLs associated with high fruit acidity in apple using pooled genome sequencing analysis. Hortic. Res. 2020, 7, 171. [Google Scholar] [CrossRef]
- Zhen, Q.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 2018, 5, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chagné, D.; Crowhurst, R.N.; Troggio, M.; Davey, M.W.; Gilmore, B.; Lawley, C.; Vanderzande, S.; Hellens, R.P.; Kumar, S.; Cestaro, A.; et al. Genome-Wide SNP Detection, Validation, and Development of an 8K SNP Array for Apple. PLoS ONE 2012, 7, e31745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianco, L.; Cestaro, A.; Sargent, D.J.; Banchi, E.; Derdak, S.; Di Guardo, M.; Salvi, S.; Jansen, J.; Viola, R.; Gut, I.; et al. Development and Validation of a 20K Single Nucleotide Polymorphism (SNP) Whole Genome Genotyping Array for Apple (Malus × domestica Borkh). PLoS ONE 2014, 9, e110377. [Google Scholar] [CrossRef] [Green Version]
- Bink, M.C.A.M.; Uimari, P.; Sillanpaa, M.J.; Jans, L.L.G.; Jansen, R.C. Multiple QTL mapping in related plant populations via a pedigree-analysis approach. Theor. Appl. Genet. 2002, 104, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Bink, M.C.A.M.; Radu Totir, L.; ter Braak, C.J.F.; Winkler, C.R.; Boer, M.P.; Smith, O.S. QTL linkage analysis of connected populations using ancestral marker and pedigree information. Theor. Appl. Genet. 2012, 124, 1097–1113. [Google Scholar] [CrossRef] [Green Version]
- Evans, K.; Guan, Y.; Luby, J.; Clark, M.; Schmitz, C.; Brown, S.; Orcheski, B.; Peace, C.; Van de Weg, E.; Iezzoni, A. Large-scale standardized phenotyping of apple in RosBREED. Acta Hortic. 2012, 945, 233–238. [Google Scholar] [CrossRef]
- Blanpied, G.D.; Silsby, K.J. Predicting Harvest Date Windows for Apples; Information Bulletin 221; Cornell Cooperative Extension: Ithaca, NY, USA, 1992; Available online: https://hdl.handle.net/1813/3299 (accessed on 29 October 2021).
- Kostick, S.A.; Teh, S.L.; Norelli, J.L.; Vanderzande, S.; Peace, C.; Evans, K.M. Fire blight QTL analysis in a multi-family apple population identifies a reduced-susceptibility allele in ‘Honeycrisp’. Hortic. Res. 2021, 8, 28. [Google Scholar] [CrossRef]
- Kostick, S.A.; Luby, J.J. Apple Fruit Size QTLs on Chromosomes 8 and 16 Characterized in ‘Honeycrisp’-Derived Germplasm. Agronomy 2022, 12, 1279. [Google Scholar] [CrossRef]
- Vanderzande, S.; Howard, N.P.; Cai, L.; Da Silva Linge, C.; Antanaviciute, L.; Bink, M.C.A.M.; Kruisselbrink, J.W.; Bassil, N.; Gasic, K.; Iezzoni, A.; et al. High-quality, genome-wide SNP genotypic data for pedigreed germplasm of the diploid outbreeding species apple, peach, and sweet cherry through a common workflow. PLoS ONE 2019, 14, e0210928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, N.; Van de Weg, E.; Bedford, D.S.; Peace, C.P.; Vanderzande, S.; Clark, M.D.; Teh, S.L.; Cai, L.; Luby, J.J. Elucidation of the ‘Honeycrisp’ pedigree through haplotype analysis with a multi-family integrated SNP linkage map and a large apple (Malus × domestica) pedigree-connected SNP data set. Hortic. Res. 2017, 4, 17003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kass, R.E.; Raftery, A.E. Bayes factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Howard, N.P.; Van de Weg, E.; Tillman, J.; Tong, C.B.S.; Silverstein, K.A.T.; Luby, J.J. Two QTL characterized for soft scald and soggy breakdown in apple (Malus × domestica) through pedigree-based analysis of a large population of interconnected families. Tree Genet. Genomes 2018, 14, 2. [Google Scholar] [CrossRef] [Green Version]
- Luby, J.J.; Howard, N.P.; Tillman, J.R.; Bedford, D.S. Extended pedigrees of apple cultivars from the University of Minnesota breeding program elucidated using SNP array markers. HortScience 2022, 57, 3. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Zhang, B.; Han, Y. Genomics of Fruit Acidity and Sugar Content in Apple. In The Apple Genome; Korban, S.S., Ed.; Springer: Cham, Switzerland, 2021; pp. 297–309. [Google Scholar] [CrossRef]
- Ma, B.; Zhao, S.; Wu, B.; Wang, D.; Peng, Q.; Owiti, A.; Fang, T.; Liao, L.; Ogutu, C.; Korban, S.S.; et al. Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet. Genomes 2016, 12, 1. [Google Scholar] [CrossRef]
- Kumar, S.; Garrick, D.; Bink, M.C.A.M.; Whitworth, C.; Chagné, D.; Volz, R.K. Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genom. 2013, 14, 393. [Google Scholar] [CrossRef] [Green Version]
- Visser, T.; Verhaegh, J.J. Inheritance and selection of some fruit characters of apple. I. Inheritance of low and high acidity. Euphytica 1978, 27, 753–760. [Google Scholar] [CrossRef]
- Bai, Y.; Dougherty, L.; Li, M.; Fazio, G.; Cheng, L.; Xu, K. A natural mutation-led truncation in one of the two aluminum-activated malate transporter-like genes at the Ma locus is associated with low fruit acidity in apple. Mol. Genet. Genom. 2012, 287, 663–678. [Google Scholar] [CrossRef] [Green Version]
- Chagné, D.; Dayatilake, D.; Diack, R.; Oliver, M.; Ireland, H.; Watson, A.; Gardiner, S.E.; Johnston, J.W.; Schaffer, R.J.; Tustin, S. Genetic and environmental control of fruit maturation, dry matter and firmness in apple (Malus × domestica Borkh.). Hortic. Res. 2014, 1, 14046. [Google Scholar] [CrossRef] [Green Version]
- Kunihisa, M.; Takita, Y.U.; Yamaguchi, N.; Okada, H.; Sato, M.; Komori, S.; Nishitani, C.; Terakami, S.; Yamamoto, T. The use of a fertile doubled haploid apple line for QTL analysis of fruit traits. Breed. Sci. 2019, 69, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, T.; Ogawa, H.; Fukuda, N.; Moriguchi, T. Changes in the taste and textural attributes of apples in response to climate change. Sci. Rep. 2013, 3, 2418. [Google Scholar] [CrossRef] [Green Version]
- Iwanami, H. Breeding for fruit quality in apple. In Breeding for Fruit Quality; Jenks, M.A., Bebeli, P.J., Eds.; John Wiley & Sons: West Sussex, UK, 2011; pp. 175–200. [Google Scholar] [CrossRef]
- Chagné, D.; Vanderzande, S.; Kirk, C.; Profitt, N.; Weskett, R.; Gardiner, S.E.; Peace, C.P.; Volz, R.K.; Bassil, N.V. Validation of SNP markers for fruit quality and disease resistance loci in apple (Malus × domestica Borkh.) using the OpenArray® platform. Hortic. Res. 2019, 6, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Chagné, D.; Bink, M.C.A.M.; Volz, R.K.; Whitworth, C.; Carlisle, C. Genomic Selection for Fruit Quality Traits in Apple (Malus × domestica Borkh.). PLoS ONE 2012, 7, e36674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, M.; Keller, B.; Roth, M.; Aranzana, M.J.; Auwerkerken, A.; Guerra, W.; Al-Rifaï, M.; Lewandowski, M.; Sanin, N.; Rymemnants, M.; et al. Genetic architecture and genomic predictive ability of apple quantitative traits across environments. Hortic. Res. 2022, 9, uhac028. [Google Scholar] [CrossRef]
| Trait z | Year y | LG x | BF w | QTL Region (cM) v | QTL Mode (cM) u | Position (Mbp) t |
|---|---|---|---|---|---|---|
| TA | 2017 | 1 | 6.6 | 57–63 | 62 | 31.1–32.5 |
| 2015 | 11.4 | 31–41 | 39 | 23.6–24.2 | ||
| 2018 | 31.1 | 49–53 | 51 | 28.4–30.0 | ||
| 2014–2018 | 31.1 | 47–53 | 47 | 27.7–30.0 | ||
| 2014 | 31.3 | 45–55 | 53 | 26.9–30.9 | ||
| 2014 | 8 | 8.2 | 3–9 | 5 | 1.3–2.7 | |
| 2014–2018 | 30 | 23–33 | 29 | 7.4–12.0 | ||
| 2018 | 30.8 | 27–33 | 29 | 9.4–12.0 | ||
| 2017 | 31 | 31–35 | 33 | 11.0–12.6 | ||
| 2014–2018 | 10 | 7.9 | 19–25 | 23 | 9.4–18.9 | |
| 2017 | 16 | 9.2 | 3–13 | 13 | 2.0–4.2 | |
| 2014 | 10.6 | 7–11 | 9 | 3.0–3.6 | ||
| 2015 | 29.9 | 11–13 | 11 | 3.6–4.2 | ||
| 2018 | 30.8 | 7–11 | 9 | 3.0–3.6 | ||
| 2014–2018 | 30.8 | 9–11 | 9 | 3.2–3.6 | ||
| SSC | 2014–2018 | 1 | 7.6 | 47–53 | 51 | 27.7–30.0 |
| 2014 | 9 | 5.2 | 23–25 | 25 | 8.0–8.5 | |
| 2017 | 12 | 5.5 | 25–27 | 27 | 12.9–17.2 | |
| 2016 | 13 | 7 | 43–47 | 43 | 12.2–14.1 | |
| 2018 | 15.1 | 65–69 | 67 | 23.4–26.1 | ||
| 2014–2018 | 15.1 | 63–67 | 65 | 22.1–25.7 | ||
| 2014–2018 | 15 | 7.5 | 35–43 | 37 | 9.8–12.2 | |
| 2017 | 16 | 7.1 | 3–9 | 7 | 2.0–3.2 | |
| 2014–2018 | 6.9 | 1–9 | 7 | 0.9–3.2 | ||
| 2017 | 17 | 5.8 | 33–39 | 35 | 11.8–13.9 |
| LG | Haplotype Name | Haplotype Sequence z | Mean (g/L) | Variance | No. Offspring | Relative Effect | Reported Effect y |
|---|---|---|---|---|---|---|---|
| 1 | 1A | ABBABA | 6.9 | 5.0 | 255 | High | - x |
| 1B | ABBAAA | 7.2 | 5.6 | 39 | Neutral | - | |
| 1C | BABABB | 6.5 | 4.8 | 152 | Neutral | - | |
| 1D | BBAABA | 6.3 | 3.3 | 214 | Neutral | - | |
| 1E | BBABAB | 6.0 | 4.6 | 154 | Low | - | |
| 1F | BBABAA | 5.8 | 5.8 | 33 | Low | - | |
| 8 | 8A | BBABBBBAABAAAB | 7.4 | 5.7 | 52 | High | Unknown w |
| 8B | AAAABAAABAAAAA | 7.1 | 6.5 | 51 | High | Unknown | |
| 8C | BABBBBBBABAABB | 7.0 | 4.5 | 212 | High | qv | |
| 8D | BABAABBBABABAB | 6.9 | 4.4 | 20 | Neutral | Unknown | |
| 8E | BAABBAAABAAAAA | 6.6 | 2.3 | 73 | Neutral | Unknown | |
| 8F | AABBABBBABABAA | 6.4 | 4.4 | 161 | Neutral | q | |
| 8G | BAABBBBAABABAB | 6.3 | 6.5 | 41 | Neutral | Unknown | |
| 8H | BAABBBAAAAAAAB | 6.4 | 8.2 | 24 | Neutral | q | |
| 8I | BAABBBAAABBABA | 5.7 | 4.8 | 184 | Low | q | |
| 16 | 16A | BABAAB | 6.8 | 5.0 | 205 | High | Qu |
| 16B | ABBBAA | 6.8 | 5.4 | 207 | High | Q | |
| 16C | BBBBBB | 6.6 | 3.8 | 143 | Neutral | Q | |
| 16D | BBAAAB | 6.5 | 4.6 | 141 | Neutral | Q | |
| 16E | ABBBBA | 6.0 | 2.7 | 43 | Neutral | q | |
| 16F | BBBBAA | 6.0 | 4.4 | 104 | Neutral | Unknown | |
| 16G | BBAAAB | 5.9 | 4.8 | 23 | Neutral | Q | |
| 16H | ABBBAA | 4.8 | 4.1 | 29 | Low | q |
| LG | Haplotype Name | Haplotype Sequence z | Mean (°Brix) | Variance | No. Offspring | Relative Effect |
|---|---|---|---|---|---|---|
| 1 | 1A | ABBABA | 13.7 | 2.3 | 253 | High |
| 1B | ABBAAA | 13.7 | 1.2 | 39 | Neutral | |
| 1C | BABABB | 13.5 | 1.9 | 153 | Neutral | |
| 1D | BBAABA | 13.3 | 2.3 | 212 | Low | |
| 1E | BBABAB | 13.3 | 2.2 | 152 | Low | |
| 1F | BBABAA | 14.1 | 1.5 | 33 | High | |
| 13 | 13A | BABABABABBBAAABBAB | 14.1 | 1.0 | 36 | High |
| 13B | AAABBAABAAAABBBAAB | 14.1 | 2.3 | 24 | High | |
| 13C | BABABABABAABABAABB | 14.0 | 1.2 | 31 | High | |
| 13D | ABBBABBABBABABAABA | 13.9 | 1.8 | 50 | High | |
| 13E | AAAABBBABBABABAABA | 13.7 | 2.0 | 176 | High | |
| 13F | ABBBBABABAAABBBABA | 13.5 | 1.8 | 25 | High | |
| 13G | AAABBABABAAABBBABA | 13.5 | 2.2 | 263 | High | |
| 13H | AAABBABBAAAAAABBBB | 13.3 | 0.9 | 22 | High | |
| 13I | ABBBABBABAABABAABB | 13.3 | 2.4 | 89 | High | |
| 13J | ABBBABBBAAAAABAABA | 12.3 | 2.0 | 77 | Low | |
| 16 | 16A | BABAAB | 13.4 | 2.4 | 201 | Low |
| 16B | ABBBAA | 13.2 | 2.6 | 204 | Low | |
| 16C | BBBBBB | 13.3 | 2.3 | 145 | Low | |
| 16D | BBAAAB | 13.1 | 2.4 | 136 | Low | |
| 16E | ABBBBA | 14.0 | 2.1 | 45 | High | |
| 16F | BBBBAA | 13.9 | 1.8 | 103 | High | |
| 16G | BBAAAB | 13.9 | 1.2 | 24 | Low | |
| 16H | ABBBAA | 14.1 | 1.3 | 28 | High |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, B.A.; Kostick, S.A.; Luby, J.J. Large-Effect QTLs for Titratable Acidity and Soluble Solids Content Validated in ‘Honeycrisp’-Derived Apple Germplasm. Agronomy 2022, 12, 1703. https://doi.org/10.3390/agronomy12071703
Miller BA, Kostick SA, Luby JJ. Large-Effect QTLs for Titratable Acidity and Soluble Solids Content Validated in ‘Honeycrisp’-Derived Apple Germplasm. Agronomy. 2022; 12(7):1703. https://doi.org/10.3390/agronomy12071703
Chicago/Turabian StyleMiller, Baylee A., Sarah A. Kostick, and James J. Luby. 2022. "Large-Effect QTLs for Titratable Acidity and Soluble Solids Content Validated in ‘Honeycrisp’-Derived Apple Germplasm" Agronomy 12, no. 7: 1703. https://doi.org/10.3390/agronomy12071703
APA StyleMiller, B. A., Kostick, S. A., & Luby, J. J. (2022). Large-Effect QTLs for Titratable Acidity and Soluble Solids Content Validated in ‘Honeycrisp’-Derived Apple Germplasm. Agronomy, 12(7), 1703. https://doi.org/10.3390/agronomy12071703






