Response of Nitrifier and Denitrifier Abundance to Salinity Gradients in Agricultural Soils at the Yellow River Estuary
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Background
2.2. DNA Extraction and PCR Amplification
2.3. Statistical Analysis
3. Results
3.1. Characteristics of the Soil Properties
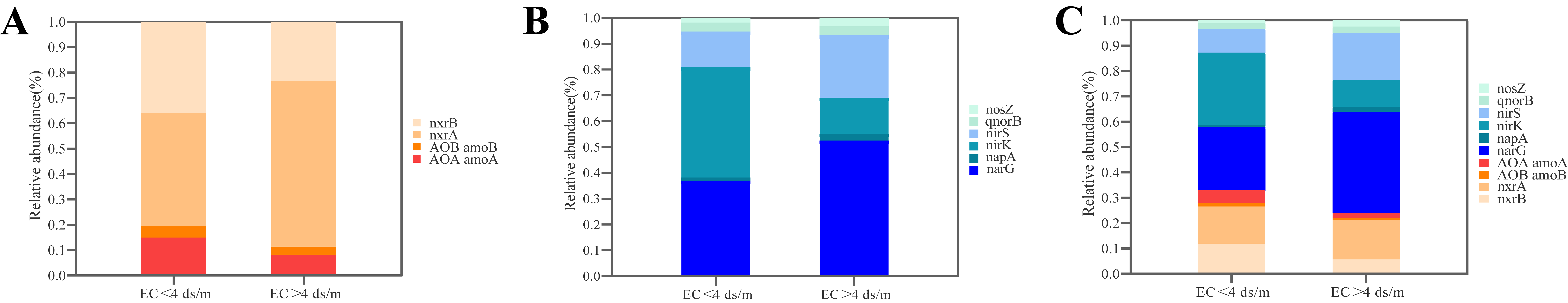
3.2. The Correlation between EC and N-Cycling Gene Abundance
3.3. The Effect of Salinity on Community Structure
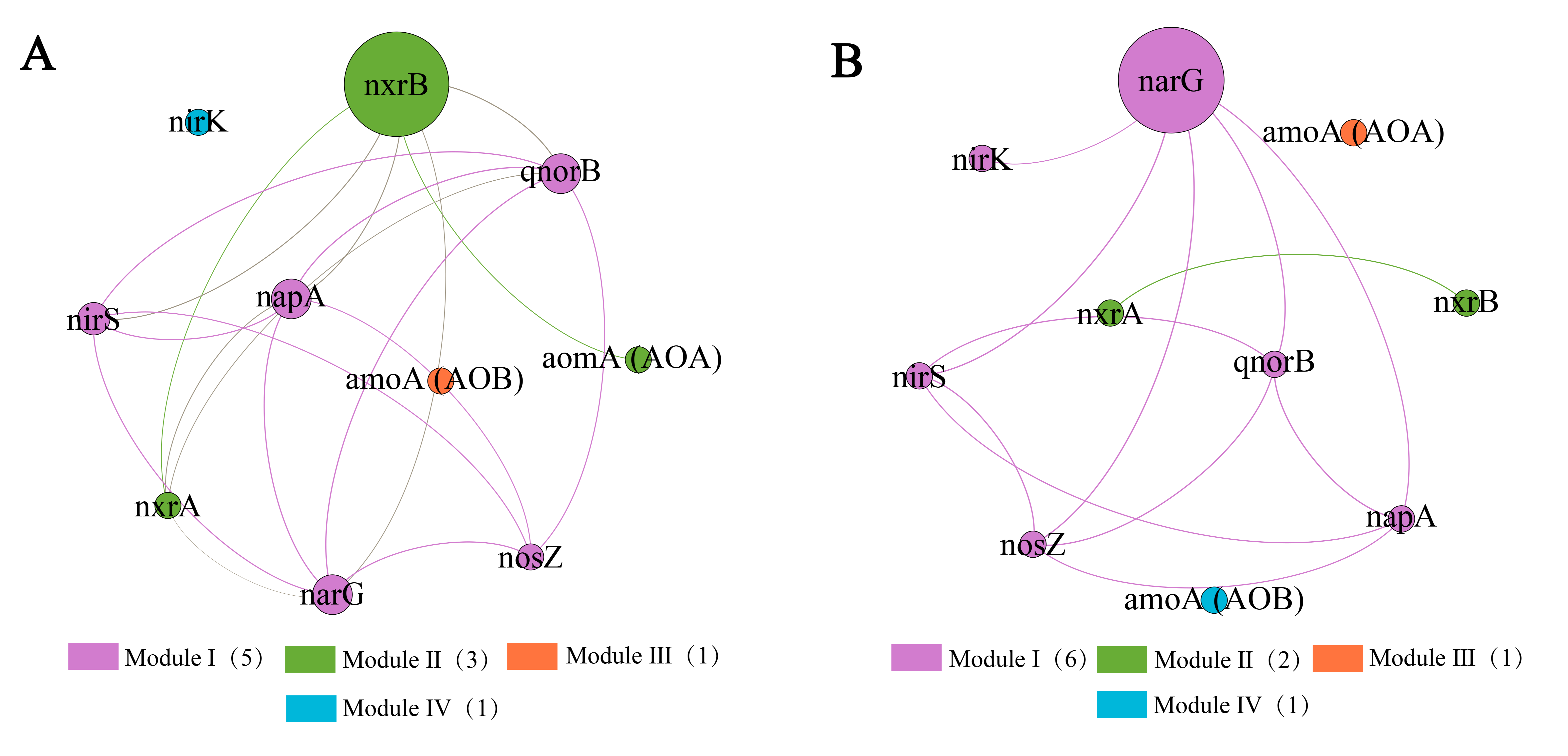
3.4. Co-Occurrence Network Analysis
4. Discussion
4.1. Responses of Gene Abundance to Salinity
4.2. Impact of Salinity on the Community Structure of Nitrifiers and Denitrifiers
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Devkota, K.P.; Devkota, M.; Rezaei, M.; Oosterbaan, R. Managing salinity for sustainable agricultural production in salt-affected soils of irrigated drylands. Agric. Syst. 2022, 198, 103390. [Google Scholar] [CrossRef]
- Rath, K.M.; Murphy, D.N.; Rousk, J. The microbial community size, structure, and process rates along natural gradients of soil salinity. Soil Biol. Biochem. 2019, 138, 107607. [Google Scholar] [CrossRef]
- Jesus, J.M.; Danko, A.S.; Fiúza, A.; Borges, M.-T. Phytoremediation of salt-affected soils: A review of processes, applicability, and the impact of climate change. Environ. Sci. Pollut. Res. 2015, 22, 6511–6525. [Google Scholar] [CrossRef]
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [Green Version]
- Ardón, M.; Morse, J.L.; Colman, B.P.; Bernhardt, E.S. Drought-induced saltwater incursion leads to increased wetland nitrogen export. Glob. Change Biol. 2013, 19, 2976–2985. [Google Scholar] [CrossRef]
- Dang, C.; Morrissey, E.M.; Neubauer, S.C.; Franklin, R.B. Novel microbial community composition and carbon biogeochemistry emerge over time following saltwater intrusion in wetlands. Glob. Change Biol. 2019, 25, 549–561. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, X.; Wang, S.; Yu, L.; Armanbek, G.; Yu, J.; Jiang, L.; Yuan, D.; Guo, Z.; Zhang, H.; et al. Towards a more labor-saving way in microbial ammonium oxidation: A review on complete ammonia oxidization (comammox). Sci. Total Environ. 2022, 829, 154590. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Abell, G.C.; Revill, A.T.; Smith, C.; Bissett, A.P.; Volkman, J.K.; Robert, S.S. Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 2010, 4, 286–300. [Google Scholar] [CrossRef]
- Zhai, S.; Ji, M.; Zhao, Y.; Su, X. Shift of bacterial community and denitrification functional genes in biofilm electrode reactor in response to high salinity. Environ. Res. 2020, 184, 109007. [Google Scholar] [CrossRef]
- Wang, H.; Gilbert, J.A.; Zhu, Y.; Yang, X. Salinity is a key factor driving the nitrogen cycling in the mangrove sediment. Sci. Total Environ. 2018, 631–632, 1342–1349. [Google Scholar] [CrossRef]
- Sheng, Q.; Wang, L.; Wu, J. Vegetation alters the effects of salinity on greenhouse gas emissions and carbon sequestration in a newly created wetland. Ecol. Eng. 2015, 84, 542–550. [Google Scholar] [CrossRef]
- Xie, W.; Zhang, C.; Zhou, X.; Wang, P. Salinity-dominated change in community structure and ecological function of Archaea from the lower Pearl River to coastal South China Sea. Appl. Microbiol. Biotechnol. 2014, 98, 7971–7982. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Dai, T.; Tian, J.; Wen, D. The influence of salinity on the abundance, transcriptional activity, and diversity of AOA and AOB in an estuarine sediment: A microcosm study. Appl. Microbiol. Biotechnol. 2015, 99, 9825–9833. [Google Scholar] [CrossRef]
- Guo, H.; Ma, L.; Liang, Y.; Hou, Z.; Min, W. Response of ammonia-oxidizing Bacteria and Archaea to long-term saline water irrigation in alluvial grey desert soils. Sci. Rep. 2020, 10, 489. [Google Scholar] [CrossRef] [Green Version]
- Cantera, J.J.L.; Jordan, F.L.; Stein, L.Y. Effects of irrigation sources on ammonia-oxidizing bacterial communities in a managed turf-covered aridisol. Biol. Fertil. Soils 2006, 43, 247–255. [Google Scholar] [CrossRef]
- Chandra, S.; Joshi, H.C.; Pathak, H.; Jain, M.C.; Kalra, N. Effect of potassium salts and distillery effluent on carbon mineralization in soil. Bioresour. Technol. 2002, 83, 255–257. [Google Scholar] [CrossRef]
- Zhou, M.; Butterbach-Bahl, K.; Vereecken, H.; Brüggemann, N. A meta-analysis of soil salinization effects on nitrogen pools, cycles and fluxes in coastal ecosystems. Glob. Change Biol. 2017, 23, 1338–1352. [Google Scholar] [CrossRef]
- Nejidat, A. Nitrification and occurrence of salt-tolerant nitrifying bacteria in the Negev desert soils. FEMS Microbiol. Ecol. 2005, 52, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Bernhard, A.E.; Landry, Z.C.; Blevins, A.; de la Torre, J.R.; Giblin, A.E.; Stahl, D.A. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl. Environ. Microbiol. 2010, 76, 1285–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Zhou, W.; Yuan, Y.; Zhong, H.; Zhong, C. Effects of salinity shock on simultaneous nitrification and denitrification by a membrane bioreactor: Performance, sludge activity, and functional microflora. Sci. Total Environ. 2021, 801, 149748. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Shao, T.; Long, X.; He, T.; Gao, X.; Zhou, Z.; Liu, Z.; Rengel, Z. Microbiome structure and function in rhizosphere of Jerusalem artichoke grown in saline land. Sci. Total Environ. 2020, 724, 138259. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wan, W.; Zheng, L.; Wang, A.; Luo, X.; Huang, Q.; Chen, W. Community assembly mechanisms and co-occurrence patterns of nitrite-oxidizing bacteria communities in saline soils. Sci. Total Environ. 2021, 772, 145472. [Google Scholar] [CrossRef]
- Gao, J.; Hou, L.; Zheng, Y.; Liu, M.; Yin, G.; Yu, C.; Gao, D. Shifts in the Community Dynamics and Activity of Ammonia-Oxidizing Prokaryotes Along the Yangtze Estuarine Salinity Gradient. J. Geophys. Res. Biogeosci. 2018, 123, 3458–3469. [Google Scholar] [CrossRef]
- Rysgaard, S.; Thastum, P.; Dalsgaard, T.; Sloth, C. Effects of Salinity on NH4+ Adsorption Capacity, Nitrification, and Denitrification in Danish Estuarine Sediments. Estuaries 1999, 22, 21–30. [Google Scholar] [CrossRef]
- Giblin, A.E.; Weston, N.B.; Banta, G.T.; Tucker, J.; Hopkinson, C.S. The Effects of Salinity on Nitrogen Losses from an Oligohaline Estuarine Sediment. Estuaries Coasts 2010, 33, 1054–1068. [Google Scholar] [CrossRef]
- Fear, J.M.; Thompson, S.P.; Gallo, T.E.; Paerl, H.W. Denitrification rates measured along a salinity gradient in the eutrophic Neuse River Estuary, North Carolina, USA. Estuaries 2005, 28, 608–619. [Google Scholar] [CrossRef]
- Gauthier, M.J.; Flatau, G.N.; Breittmayer, V.A. Protective Effect of Glycine Betaine on Survival of Escherichia coli Cells in Marine Environments. Water Sci. Technol. 1991, 24, 129–132. [Google Scholar] [CrossRef]
- Gao, J.; Hou, L.; Zheng, Y.; Liu, M.; Yin, G.; Li, X.; Lin, X.; Yu, C.; Wang, R.; Jiang, X.; et al. nirS-Encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Appl. Microbiol. Biotechnol. 2016, 100, 8573–8582. [Google Scholar] [CrossRef]
- Piao, Z.; Zhang, W.-W.; Ma, S.; Li, Y.-M.; Yin, S.-X. Succession of Denitrifying Community Composition in Coastal Wetland Soils Along a Salinity Gradient. Pedosphere 2012, 22, 367–374. [Google Scholar] [CrossRef]
- Yoshie, S.; Noda, N.; Tsuneda, S.; Hirata, A.; Inamori, Y. Salinity decreases nitrite reductase gene diversity in denitrifying bacteria of wastewater treatment systems. Appl. Environ. Microbiol. 2004, 70, 3152–3157. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wei, G.; Wang, N.; Gao, Z. Diversity and Distribution of nirK-Harboring Denitrifying Bacteria in the Water Column in the Yellow River Estuary. Microbes Environ. 2014, 29, 107–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Han, S.; Wan, W.; Zheng, L.; Chen, W.; Huang, Q. Manure fertilizes alter the nitrite oxidizer and comammox community composition and increase nitrification rates. Soil Tillage Res. 2020, 204, 104701. [Google Scholar] [CrossRef]
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef] [Green Version]
- Attard, E.; Poly, F.; Commeaux, C.; Laurent, F.; Terada, A.; Smets, B.F.; Recous, S.; Roux, X.L. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 2010, 12, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Pester, M.; Maixner, F.; Berry, D.; Rattei, T.; Koch, H.; Lücker, S.; Nowka, B.; Richter, A.; Spieck, E.; Lebedeva, E.; et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 2014, 16, 3055–3071. [Google Scholar] [CrossRef] [Green Version]
- Bru, D.; Sarr, A.; Philippot, L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 2007, 73, 5971–5974. [Google Scholar] [CrossRef] [Green Version]
- Hallin, S.; Lindgren, P.E. PCR detection of genes encoding nitrile reductase in denitrifying bacteria. Appl. Environ. Microbiol. 1999, 65, 1652–1657. [Google Scholar] [CrossRef] [Green Version]
- Throbäck, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef]
- Braker, G.; Tiedje, J.M. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 2003, 69, 3476–3483. [Google Scholar] [CrossRef] [Green Version]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Zhen, Y.; Mi, T.; Fu, L.; Yu, Z. Ammonia-Oxidizing Archaea and Bacteria Differentially Contribute to Ammonia Oxidation in Sediments from Adjacent Waters of Rushan Bay, China. Front. Microbiol. 2018, 9, 116. [Google Scholar] [CrossRef] [Green Version]
- Adair, K.L.; Schwartz, E. Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb. Ecol. 2008, 56, 420–426. [Google Scholar] [CrossRef]
- Fan, F.; Yang, Q.; Li, Z.; Wei, D.; Cui, X.A.; Liang, Y. Impacts of Organic and Inorganic Fertilizers on Nitrification in a Cold Climate Soil are Linked to the Bacterial Ammonia Oxidizer Community. Microb. Ecol. 2011, 62, 982–990. [Google Scholar] [CrossRef]
- Trias, R.; Ruiz-Rueda, O.; Garcia-Lledo, A.; Vilar-Sanz, A.; Lopez-Flores, R.; Quintana, X.D.; Hallin, S.; Baneras, L. Emergent Macrophytes Act Selectively on Ammonia-Oxidizing Bacteria and Archaea. Appl. Environ. Microbiol. 2012, 78, 6352–6356. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.H.; Wong, M.T.; Okabe, S.; Watanabe, Y. Dynamic response of nitrifying activated sludge batch culture to increased chloride concentration. Water Res. 2003, 37, 3125–3135. [Google Scholar] [CrossRef]
- Moussa, M.S.; Sumanasekera, D.U.; Ibrahim, S.H.; Lubberding, H.J.; Hooijmans, C.M.; Gijzen, H.J.; van Loosdrecht, M.C.M. Long term effects of salt on activity, population structure and floc characteristics in enriched bacterial cultures of nitrifiers. Water Res. 2006, 40, 1377–1388. [Google Scholar] [CrossRef]
- Mosier, A.C.; Francis, C.A. Denitrifier abundance and activity across the San Francisco Bay estuary. Environ. Microbiol. Rep. 2010, 2, 667–676. [Google Scholar] [CrossRef]
- Miao, Y.; Liao, R.; Zhang, X.-X.; Liu, B.; Li, Y.; Wu, B.; Li, A. Metagenomic insights into salinity effect on diversity and abundance of denitrifying bacteria and genes in an expanded granular sludge bed reactor treating high-nitrate wastewater. Chem. Eng. J. 2015, 277, 116–123. [Google Scholar] [CrossRef]
- Li, X.; Gao, D.; Hou, L.; Liu, M. Salinity stress changed the biogeochemical controls on CH4 and N2O emissions of estuarine and intertidal sediments. Sci. Total Environ. 2019, 652, 593–601. [Google Scholar] [CrossRef]
- Vera-Gargallo, B.; Chowdhury, T.R.; Brown, J.; Fansler, S.J.; Duran-Viseras, A.; Sanchez-Porro, C.; Bailey, V.L.; Jansson, J.K.; Ventosa, A. Spatial distribution of prokaryotic communities in hypersaline soils. Sci. Rep. 2019, 9, 1769. [Google Scholar] [CrossRef]
- Canfora, L.; Bacci, G.; Pinzari, F.; Lo Papa, G.; Dazzi, C.; Benedetti, A. Salinity and Bacterial Diversity: To What Extent Does the Concentration of Salt Affect the Bacterial Community in a Saline Soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Chi, Z.; Li, J.; Wu, H.; Yan, B. Bacterial community structure and function in soils from tidal freshwater wetlands in a Chinese delta: Potential impacts of salinity and nutrient. Sci. Total Environ. 2019, 696, 411–420. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, Y.; Cui, X.; Yue, P.; Li, K.; Liu, X.; Tripathi, B.M.; Chu, H. Salinity Is a Key Determinant for Soil Microbial Communities in a Desert Ecosystem. Msystems 2019, 4, e00225-18. [Google Scholar] [CrossRef] [Green Version]
- Rath, K.M.; Rousk, J. Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: A review. Soil Biol. Biochem. 2015, 81, 108–123. [Google Scholar] [CrossRef]
- Rietz, D.N.; Haynes, R.J. Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 2003, 35, 845–854. [Google Scholar] [CrossRef]
- Rath, K.M.; Maheshwari, A.; Rousk, J. Linking Microbial Community Structure to Trait Distributions and Functions Using Salinity as an Environmental Filter. mBio 2019, 10, e01607-19. [Google Scholar] [CrossRef] [Green Version]
- Piccini, C.; Garcia-Alonso, J. Bacterial diversity patterns of the intertidal biofilm in urban beaches of Rio de la Plata. Mar. Pollut. Bull. 2015, 91, 476–482. [Google Scholar] [CrossRef]
- Guo, X.P.; Niu, Z.S.; Lu, D.P.; Feng, J.N.; Chen, Y.R.; Tou, F.Y.; Liu, M.; Yang, Y. Bacterial community structure in the intertidal biofilm along the Yangtze Estuary, China. Mar. Pollut. Bull. 2017, 124, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]



| Genes | Primer Name | Amplicon Size (bp) | Reference |
|---|---|---|---|
| for nitrifiers | |||
| amoA (AOA) | amoA23F/amoA616R | 593 | [35] |
| amoA (AOB) | amoA-1F/amoA-2R | 491 | [36] |
| nxrA | F1norA/R2norA | 282 | [37] |
| nxrB | nxrB169F/nxrB638R | 469 | [38] |
| for denitrifiers | |||
| narG | narGf/narGr | 173 | [39] |
| napA | V17m/napA4r | 152 | [39] |
| nirK | F1aCu/R3Cu | 473 | [40] |
| nirS | cd3aF/R3cd | 425 | [41] |
| qnorB | qnorB2F/qnorB5R | 262 | [42] |
| nosZ | nosZ1F/nosZ1R | 259 | [43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, D.; Li, X.; Luo, X. Response of Nitrifier and Denitrifier Abundance to Salinity Gradients in Agricultural Soils at the Yellow River Estuary. Agronomy 2022, 12, 1642. https://doi.org/10.3390/agronomy12071642
Huang D, Li X, Luo X. Response of Nitrifier and Denitrifier Abundance to Salinity Gradients in Agricultural Soils at the Yellow River Estuary. Agronomy. 2022; 12(7):1642. https://doi.org/10.3390/agronomy12071642
Chicago/Turabian StyleHuang, Daqing, Xiang Li, and Xuesong Luo. 2022. "Response of Nitrifier and Denitrifier Abundance to Salinity Gradients in Agricultural Soils at the Yellow River Estuary" Agronomy 12, no. 7: 1642. https://doi.org/10.3390/agronomy12071642





