In-Field LAMP Detection of Flavescence Dorée Phytoplasma in Crude Extracts of the Scaphoideus titanus Vector
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Plant Cultivation
2.2. DNA Extraction
2.3. Isothermal Amplification Assays
2.4. Analytic Sensitivity of the Selected Assay
2.5. Development of a Crude Extract Preparation Method for LAMP
2.6. LAMP and Real-Time PCR from Field-Captured Insects
2.6.1. Laboratory Assay
2.6.2. In-Field LAMP Assay
3. Results
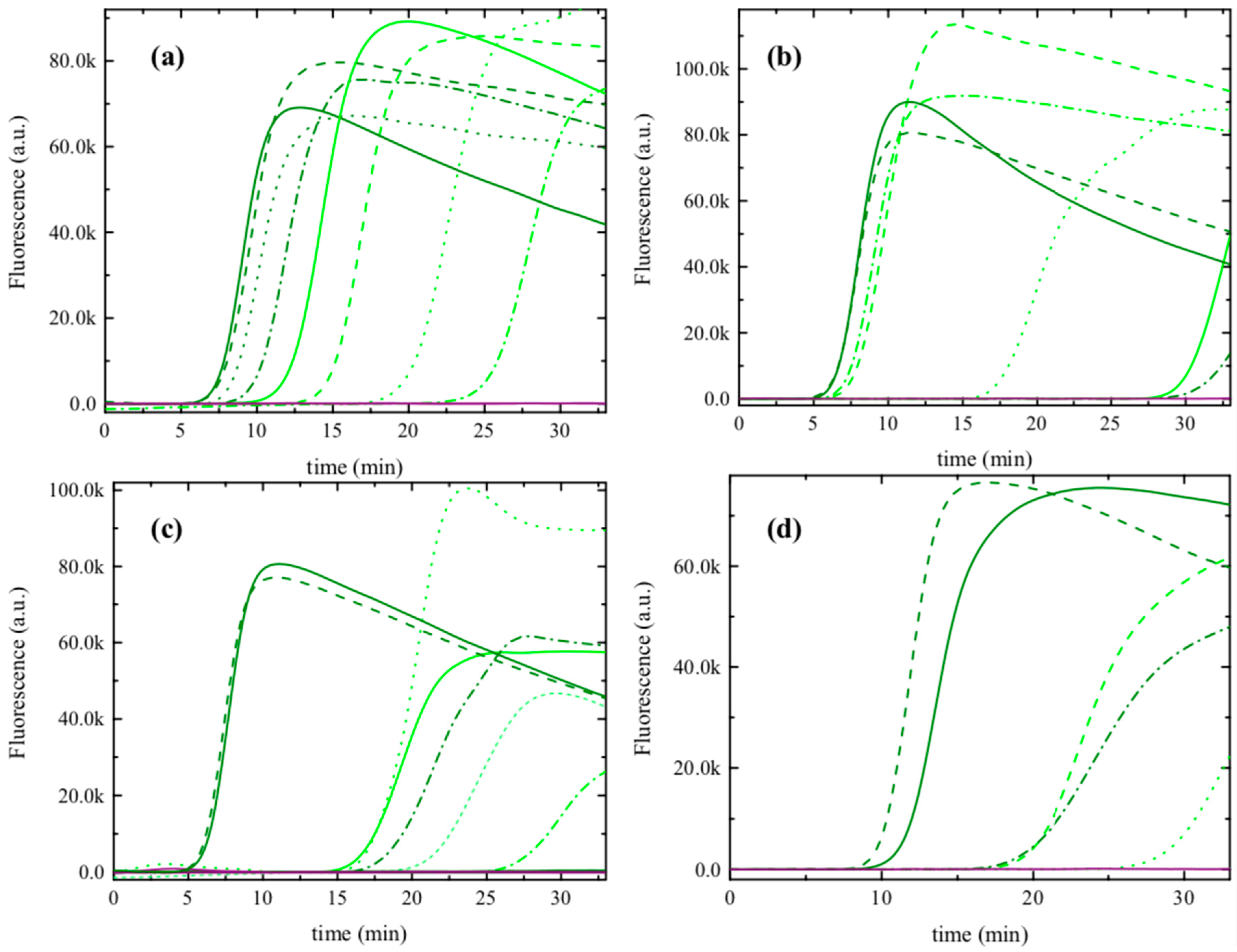
3.1. Selection of the Best Isothermal Amplification Protocol for FDp Detection in S. titanus
3.2. Analytical Sensitivity of the LAMP 1 Protocol
3.3. Selection of the Best Method for Obtaining Crude S. titanus Extracts for LAMP
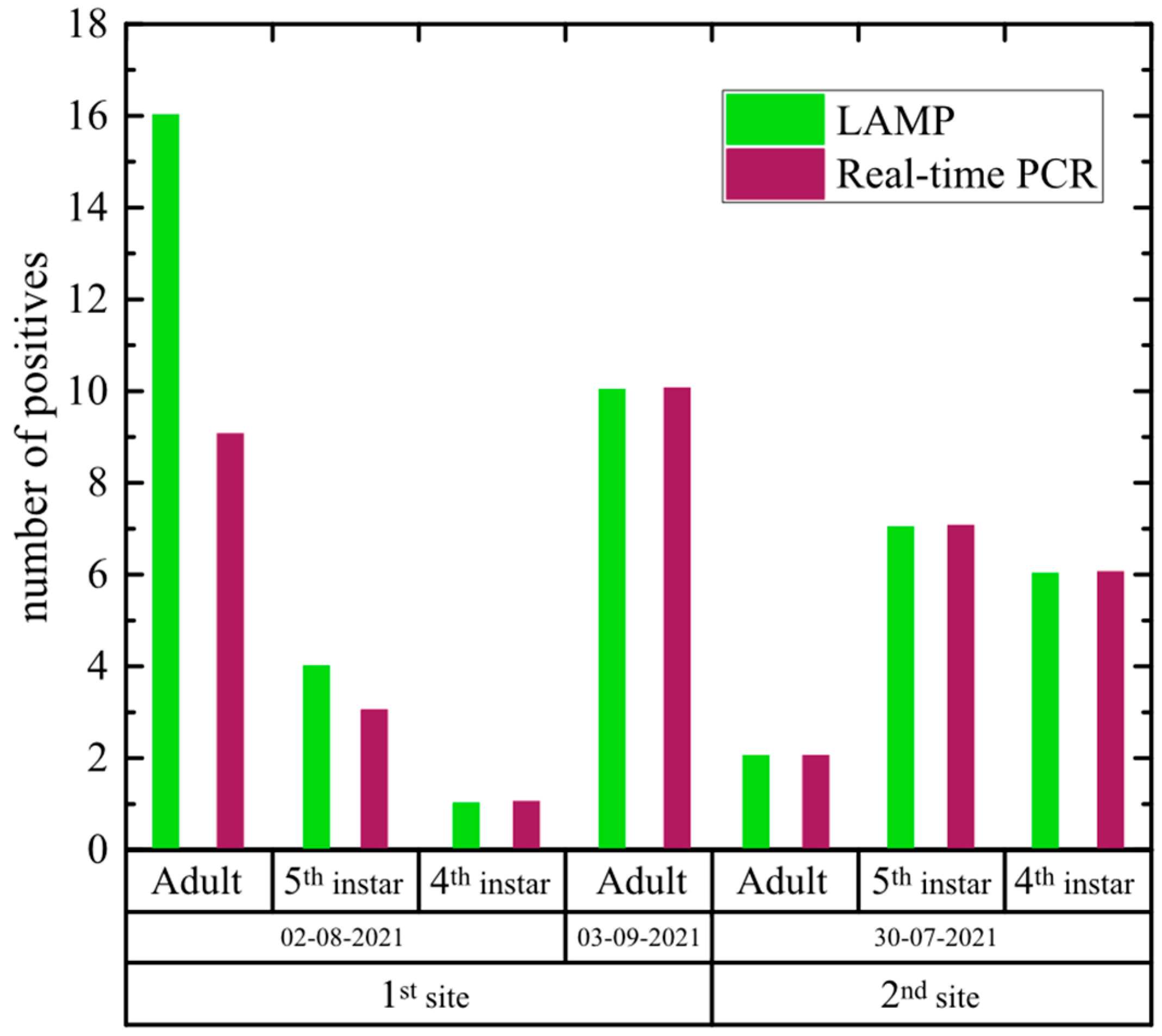
3.4. Evaluation of S. titanus Infectivity in the Field
3.5. Application of the LAMP Assay in the Field
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loebenstein, G.; Katis, N.I. Advances in Virus Research. Control of Plant Virus Diseases: Vegetatively-Propagated Crops; Elsevier: Waltham, MA, USA, 2015; Volume 91. [Google Scholar]
- Malembic-Maher, S.; Desqué, D.; Khalil, D.; Salar, P.; Bergey, B.; Danet, J.-L.; Duret, S.; Dubrana-Ourabah, M.-P.; Beven, L.; Ember, I.; et al. When a Palearctic bacterium meets a Nearctic insect vector: Genetic and ecological insights into the emergence of the grapevine Flavescence dorée epidemics in Europe. PLOS Pathog. 2020, 16, e1007967. [Google Scholar] [CrossRef] [Green Version]
- Morone, C.; Boveri, M.; Giosuè, S.; Gotta, P.; Rossi, V.; Scapin, I.; Marzachì, C. Epidemiology of flavescence dorée in vineyards in northwestern Italy. Phytopathology 2007, 97, 1422–1427. [Google Scholar] [CrossRef] [Green Version]
- Duduk, B.; Botti, S.; Ivanović, M.; Krstić, B.; Dukić, N.; Bertaccini, A. Identification of Phytoplasmas Associated with Grapevine Yellows in Serbia. J. Phytopathol. 2004, 152, 575–579. [Google Scholar] [CrossRef]
- Belli, G.; Bianco, P.A.; Conti, M. Grapevine yellows in Italy: Past, present and future. J. Plant Pathol. 2010, 92, 303–326. [Google Scholar]
- Mehle, N.; Kogovšek, P.; Constable, F.; De Jonghe, K.; Loiseau, M.; Veratti, F.; Marzachi, C.; Ferretti, L.; Angelini, E.; Filippin, L.; et al. Test performance study of isothermal amplification tests for the detection of Grapevine flavescence dorée phytoplasma and ‘Candidatus Phytoplasma solani’. Bull. OEPP 2017, 47, 18–23. [Google Scholar] [CrossRef]
- Foissac, X.; Maixner, M. Spread of grapevine phytoplasma diseases in Europe. Phytopathogenic Mollicutes 2013, 3, 47–50. [Google Scholar] [CrossRef]
- Plavec, J.; Križanac, I.; Budinščak, Ž.; Škorić, D.; Musić, M.Š. Distribution and epidemiology of Flavescence doree phytoplasma in Croatia. Glas. Biljn. Zaštite 2013, 13, 385–390. [Google Scholar]
- Oliveira, M.J.R.A.; Castro, S.; Paltrinieri, S.; Bertaccini, A.; Sottomayor, M.; Santos, C.S.; Vasconcelos, M.W.; Carvalho, S.M.P. “Flavescence dorée” impacts growth, productivity and ultrastructure of Vitis vinifera plants in Portuguese “Vinhos Verdes” region. Sci. Hortic. 2020, 261, 108742. [Google Scholar] [CrossRef]
- Mori, N.; Bressan, A.; Martini, M.; Guadagnini, M.; Girolami, V.; Bertaccini, A. Experimental transmission by Scaphoideus titanus Ball of two Flavescence doree-type phytoplasmas. Vitis 2002, 41, 99–102. [Google Scholar]
- Chuche, J.; Thiéry, D. Biology and ecology of the Flavescence dorée vector Scaphoideus titanus: A review. Agron. Sustain. Dev. 2014, 34, 381–403. [Google Scholar] [CrossRef] [Green Version]
- Alma, A.; Tedeschi, R.; Lessio, F.; Picciau, L.; Gonella, E.; Ferracini, C. Insect vectors of plant pathogenic Mollicutes in the Euro-Mediterranean region. Phytopathogenic Mollicutes 2015, 5, 53. [Google Scholar] [CrossRef]
- Belli, G.; Fortusini, A.; Osler, R.; Amici, A. Presenza di una malattia del tipo “Flavescence dorée” in vigneti dell’Oltrepò pavese. Riv. Patol. Veg. 1973, 9, 50–56. [Google Scholar]
- European Commission. Commission Implementing Regulation (EU) 2019/2072 of 28 November 2019 Establishing Uniform Conditions for the Implementation of Regulation (EU) 2016/2031 of the European Parliament and the Council, as Regards Protective Measures against Pests of Plants, and Repealing Commission Regulation (EC) No 690/2008 and Amending Commission Implementing Regulation (EU) 2018/2019. Off. J. Eur. Union 2019, 1, 1–279. [Google Scholar]
- OEPP/EPPO. PM 1/2 (30), EPPO A1 and A2 Lists of Pests Recommended for Regulation as Quarantine Pests. Available online: https://gd.eppo.int/standards/PM1/ (accessed on 25 March 2022).
- OEPP/EPPO. Hot water treatment of grapevine to control Grapevine flavescence doree phytoplasma. Bull. OEPP 2012, 42, 490–492. [Google Scholar]
- OEPP/EPPO. PM 7/079 (2) Grapevine flavescence dorée phytoplasma. Bull. OEPP 2016, 46, 78–93. [Google Scholar]
- Deng, S.; Hiruki, C. Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Schneider, B.; Cousin, M.T.; Klinkong, S.; Seemüller, E. Taxonomic relatedness and phylogenetic positions of phytoplasmas associated with diseases of faba bean, sunnhemp, sesame, soybean, and eggplant. Z. Pflanzenkrankh. Pflanzenschutz 1995, 102, 225–232. [Google Scholar]
- Daire, X.; Clair, D.; Reinert, W.; Boudon-Padieu, E. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. Eur. J. Plant Pathol. 1997, 103, 507–514. [Google Scholar] [CrossRef]
- Lee, I.-M.; Gundersen, D.E.; Hammond, R.W.; Davis, R.E. Use of mycoplasmalike organism (MLO) group-specific oligonucleotide primers for nested-PCR assays to detect mixed-MLO infections in a single host plant. Phytopathology 1994, 84, 559–566. [Google Scholar] [CrossRef]
- Clair, D.; Larrue, J.; Aubert, G.; Gillet, J.; Cloquemin, G.; Boudon-Padieu, E. A multiplex nested-PCR assay for sensitive and simultaneous detection and direct identification of phytoplasma in the Elm yellows group and Stolbur group and its use in survey of grapevine yellows in France. Vitis 2003, 42, 151–157. [Google Scholar]
- Hren, M.; Boben, J.; Rotter, A.; Kralj, P.; Gruden, K.; Ravnikar, M. Real-time PCR detection systems for Flavescence dorée and Bois noir phytoplasmas in grapevine: Comparison with conventional PCR detection and application in diagnostics. Plant Pathol. 2007, 56, 785–796. [Google Scholar] [CrossRef]
- Pelletier, C.; Salar, P.; Gillet, J.; Cloquemin, G.; Very, P.; Foissac, X.; Malembic-Maher, S. Triplex real-time PCR assay for sensitive and simultaneous detection of grapevine phytoplasmas of the 16SrV and 16SrXII-A groups with an endogenous analytical control. Vitis 2009, 48, 87–95. [Google Scholar]
- Kogovšek, P.; Hodgetts, J.; Hall, J.; Prezelj, N.; Nikolić, P.; Mehle, N.; Lenarčič, R.; Rotter, A.; Dickinson, M.; Boonham, N.; et al. LAMP assay and rapid sample preparation method for on-site detection of flavescence dorée phytoplasma in grapevine. Plant Pathol. 2015, 64, 286–296. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Gallmetzer, A.; Innerebner, G.; Roschatt, C.; Reyes-Dominguez, Y. Dynamics of Scaphoideus titanus population in southern South Tyrol (Italy) and detection of Grapevine Flavescence Dorée phytoplasma in the insect with a multiplex real-time PCR. Vitis 2020, 59, 85–89. [Google Scholar]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Panno, S.; Matić, S.; Tiberini, A.; Caruso, A.G.; Bella, P.; Torta, L.; Stassi, R.; Davino, S. Loop Mediated Isothermal Amplification: Principles and Applications in Plant Virology. Plants 2020, 9, 461. [Google Scholar] [CrossRef] [Green Version]
- Ortega, S.F.; Matic, S.; Spadaro, D. Impiego di nuove tecniche molecolari per la diagnosi delle malattie. Prot. Colt. 2018, 2, 9–14. [Google Scholar]
- Bertacca, S.; Caruso, A.G.; Trippa, D.; Marchese, A.; Giovino, A.; Matic, S.; Noris, E.; Ambrosio, M.I.F.S.; Alfaro, A.; Panno, S.; et al. Development of a Real-Time Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Olea Europaea Geminivirus. Plants 2022, 11, 660. [Google Scholar] [CrossRef]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.-J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Tomlinson, J.A.; Ostoja-Starzewska, S.; Webb, K.; Cole, J.A.; Barnes, A.V.; Dickinson, M.; Boonham, N. A loop-mediated isothermal amplification-based method for confirmation of Guignardia citricarpa in citrus black spot lesions. Eur. J. Plant Pathol. 2013, 136, 217–224. [Google Scholar] [CrossRef]
- Constable, F.E.; Gibb, K.S.; Symons, R.H. Seasonal distribution of phytoplasmas in Australian grapevines. Plant Pathol. 2003, 52, 267–276. [Google Scholar] [CrossRef]
- Kogovšek, P.; Mehle, N.; Pugelj, A.; Jakomin, T.; Schroers, H.-J.; Ravnikar, M.; Dermastia, M. Rapid loop-mediated isothermal amplification assays for grapevine yellows phytoplasmas on crude leaf-vein homogenate has the same performance as qPCR. Eur. J. Plant Pathol. 2017, 148, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, D.D.S.; Cassimiro, A.P.A.; de Oliveira, C.D.; Rodrigues, N.B.; Moreira, L.A. Wolbachia detection in insects through LAMP: Loop mediated isothermal amplification. Parasites Vectors 2014, 7, 228. [Google Scholar] [CrossRef] [Green Version]
- Sasaya, T. Detection Methods for Rice Viruses by a Reverse-Transcription Loop-Mediated Isothermal Amplification (RT-LAMP). In Plant Virology Protocols; Methods in Molecular Biology (Methods and, Protocols); Uyeda, I., Masuta, C., Eds.; Humana Press: New York, NY, USA, 2015. [Google Scholar]
- Yaseen, T.; Drago, S.; Valentini, F.; Elbeaino, T.; Stampone, G.; Digiaro, M.; D’Onghia, A.M. On-site detection of Xylella fastidiosa in host plants and in “spy insects” using the real-time loop-mediated isothermal amplification method. Phytopathol. Mediterr. 2015, 54, 488–496. [Google Scholar]
- Lu, H.; Wilson, B.A.L.; Ash, G.J.; Woruba, S.B.; Fletcher, M.J.; You, M.; Yang, G.; Gurr, G.M. Determining putative vectors of the Bogia Coconut Syndrome phytoplasma using loop-mediated isothermal amplification of single-insect feeding media. Sci. Rep. 2016, 6, 35801. [Google Scholar] [CrossRef] [Green Version]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Babu, B.; Ochoa-Corona, F.M.; Paret, M.L. Recombinase polymerase amplification applied to plant virus detection and potential implications. Anal. Biochem. 2018, 546, 72–77. [Google Scholar] [CrossRef]
- Bertin, S.; Bosco, D. Molecular identification of phytoplasma vector species. In Phytoplasma Methods and Protocols; Dickinson, M., Hodgetts, J., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 87–108. [Google Scholar]
- Hibbert, D.B.; Gooding, J. Data Analysis for Chemistry: An Introductory Guide for Students and Laboratory Scientists; Oxford University Press: Oxford, UK, 2005. [Google Scholar]
- Arseneau, J.-R.; Steeves, R.; Laflamme, M. Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 2016, 17, 686–693. [Google Scholar] [CrossRef]
- Inglis, P.W.; Pappas, M.D.C.R.; Resende, L.V.; Grattapaglia, D. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS ONE 2018, 13, e0206085. [Google Scholar] [CrossRef]
- Romero, J.L.R.; Carver, G.D.; Johnson, P.A.; Perry, K.L.; Thompson, J.R. A rapid, sensitive and inexpensive method for detection of grapevine red blotch virus without tissue extraction using loop-mediated isothermal amplification. Arch. Virol. 2019, 164, 1453–1457. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- John, M.E. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acids Res. 1992, 20, 2381. [Google Scholar] [CrossRef] [Green Version]
- Fang, G.; Hammar, S.; Grumet, R. A quick and inexpensive method for removing polysaccharides from plant genomic DNA. BioTechniques 1992, 13, 52–54. [Google Scholar]
- Maixner, M.; Reinert, W.; Darimont, H. Transmission of grapevine yellows by Oncopsis alni (Schrank) (Auchenorrhyncha: Macropsinae). Vitis 2000, 39, 83–84. [Google Scholar]
- Lessio, F.; Picciau, L.; Gonella, E.; Tota, F.; Mandrioli, M.; Alma, A. The mosaic leafhopper Orientus ishidae: Host plants, spatial distribution, infectivity, and transmission of 16SrV phytoplasmas to vines. Bull. Insectology 2016, 69, 277–289. [Google Scholar]
- Belgeri, E.; Rizzoli, A.; Jermini, M.; Angelini, E.; Filippin, L.; Rigamonti, I.E. First report of Flavescence dorée phytoplasma identification and characterization in three species of leafhoppers. J. Plant Pathol. 2022, 104, 375–379. [Google Scholar] [CrossRef]
- Krstic, O.; Cvrković, T.; Marinković, S.; Jakovljević, M.; Mitrović, M.; Toševski, I.; Jović, J. Genetic Diversity of Flavescence Doree Phytoplasmas in Vineyards of Serbia: From the Widespread Occurrence of Autochthonous Map-M51 to the Emergence of Endemic Map-FD2 (Vectotype II) and New Map-FD3 (Vectotype III) Epidemic Genotypes. Agronomy 2022, 12, 448. [Google Scholar] [CrossRef]


| Host/Pathogen | No. of samples | LAMP 1 | LAMP 2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA | DNA | ||||||||||||
| 4 ng/µL * | 0.4 ng/µL | 0.04 ng/µL | 4 ng/µL | 0.4 ng/µL | 0.04 ng/µL | ||||||||
| Scaphoideus titanus insects infected with FDp-D | 5 | 7.3 ± 0.8 | (85.10 ± 0.10) | 8.8 ± 1.0 | (85.05 ± 0.21) | 8.1 ± 2.0 | (84.90 ± 0.00) | 5.8 ± 1.3 | (84.90 ± 0.10) | 6.8 ± 1.8 | (85.00 ± 0.14) | 13 ± 3 | (84.95 ± 0.07) |
| S. titanus insects infected with FDp-C | 5 | 8.4 ± 1.4 | (85.20 ± 0.14) | 8.87 ± 0.23 | (85.10 ± 0.00) | 9.3 ± 2.0 | (85.00 ± 0.14) | 9.2 ± 0.8 | (85.15 ± 0.07) | 7.1 ± 0.3 | (85.83 ± 0.29) | 18.4 ± 5.1 | (85.15 ± 0.07) |
| Grapevine plants infected with FDp-D | 5 | 9.6 ± 0.8 | (84.50 ± 0.00) | 9.0 ± 0.9 | (85.0 ± 0.10) | 10.5 ± 0.4 | 85.20 ± 0.14 | 8.3 ± 1.3 | (84.05 ± 0.11) | 12.1 ± 1.0 | (84.70 ± 0.14) | 22 ± 5 | (85.0 ± 1.3) |
| Grapevine plants infected with FDp-C | 5 | 9.6 ± 1.4 | (85.15 ± 0.07) | 8.0 ± 1.1 | (84.4 ± 0.4) | 12.4 ± 1.4 | 84.80 ± 0.28 | 8.3 ± 2.1 | (84.80 ± 0.25) | 9.9 ± 1.7 | (85.2 ± 0.4) | 18.4 ± 1.4 | (85.4 ± 0.8) |
| Hyalesthes obsoletus insects infected with 16Sr-XII-A (‘Ca. P. solani’) | 3 | nd ** | (nd) | nd | (nd) | nd | (nd) | 18.2 ± 2.5 | (85.50 ± 0.25) | 25 ± 3 | (82.4 ± 0.5) | 32.2 ± 0.5 | (83.30 ± 0.00) |
| Cacopsylla melanoneura insects infected with 16Sr-X-A (‘Ca. P. mali’) | 3 | nd | (nd) | nd | (nd) | nd | (nd) | 26.8 ± 1.6 | (86.00 ± 0.00) | 29 ± 3 | (85.8 ± 0.4) | 37.4 ± 2.4 | (85.8 ± 0.4) |
| Healthy S. titanus | 5 | nd | (nd) | nd | (nd) | nd | (nd) | 20 ± 4 | (84.0 ± 0.8) | 22 ± 3 | (81.2 ± 1.2) | 34 ± 4 | (85.6 ± 0.4) |
| Healthy grapevine | 5 | nd | (nd) | nd | (nd) | nd | (nd) | 18 ± 4 | (81.45 ± 0.21) | 27 ± 4 | (84.8 ± 0.4) | 33 ± 3 | (84 ± 3) |
| Insect DNA Dilution | Estimated Copy Number of FDp DNA | LAMP Tp (min ± SD) * | Real-Time PCR Ct ± SD ** |
|---|---|---|---|
| 1 × 10 | 126,013,887.61 | 7.6 ± 0.5 | 21.8 ± 0.4 |
| 1 × 10−1 | 2,473,463.12 | 8.6 ± 0.8 | 24.27 ± 0.15 |
| 1 × 10−2 | 56,098.85 | 9.4 ± 0.9 | 29.8 ± 0.8 |
| 1 × 10−3 | 65.76 | 11.7 ± 0.4 | 35.38 ± 0.23 |
| 1 × 10−4 | 2.33 | 26.3 ± 1.3 | nd *** |
| 1 × 10−5 | nd | nd | nd |
| Host/Pathogen | Time/ Temperature | TET Buffer | ELISA Extraction Buffer | OptiGene Lysis Buffer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Undiluted | 1:5 | 1:7.5 | 1:10 | Undiluted | 1:5 | 1:7.5 | 1:10 | Undiluted | 1:5 | 1:7.5 | 1:10 | ||
| Scaphoideus titanus insects infected by FDp-D | Tp (min ± SD) * | nd ** | 6.2 ± 1.1 | 11.8 ± 2.4 | 18 ± 3 | nd | 8 ± 3 | 14.6 ± 1.9 | 21 ± 8 | nd | 11 ± 3 | 21.4 ± 1.8 | 30 ± 7 |
| Tmelt ± SD *** | nd | 84.60 ± 0.07 | 85.0 ± 0.5 | 85.5 ± 0.4 | nd | 85.05 ± 0.07 | 85.1 ± 0.8 | 85.0 ± 0.5 | nd | 85.2 ± 0.4 | 84.8 ± 0.8 | 84 ± 4 | |
| S. titanus insects infected by FDp-C | Tp (min ± SD) | nd | 6.6 ± 1.5 | 14.1 ± 1.9 | 22 ± 3 | nd | 8.1 ± 2.0 | 20 ± 6 | 22 ± 6 | nd | 10 ± 4 | 24 ± 3 | 32 ± 6 |
| Tmelt ± SD | nd | 84.70 ± 0.14 | 85.30 ± 0.27 | 84.4 ± 1.9 | nd | 84.90 ± 0.14 | 84.7 ± 0.4 | 85.6 ± 0.4 | nd | 85.8 ± 0.5 | 85.1 ± 0.4 | 84 ± 3 | |
| Healthy S. titanus insects | Tp (min ± SD) | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Tmelt ± SD | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| Isolate | LAMP | |
|---|---|---|
| Tp (Min) * | Tmelt (Min) ** | |
| St_F1 | nd *** | nd |
| St_F2 | 22.55 | 84.34 |
| St_F3 | 15.41 | 84.54 |
| St_F4 | nd | nd |
| St_F5 | nd | nd |
| St_F6 | 13.49 | 84.54 |
| St_F7 | 14.27 | 84.71 |
| St_F8 | nd | nd |
| St_F9 | 16.36 | 84.38 |
| St_F10 | nd | nd |
| St_F11 | 18.01 | 84.35 |
| St_F12 | 26.55 | 84.24 |
| St_F13 | nd | nd |
| St_F14 | 12.26 | 84.39 |
| St_F15 | 11.33 | 84.56 |
| St_F16 | nd | nd |
| St_F17 | nd | nd |
| St_F18 | nd | nd |
| St_F19 | nd | nd |
| St_F20 | nd | nd |
| St_F21 | 10.18 | 85.04 |
| St_F22 | 9.26 | 84.72 |
| Mean value ± SD | 15.48 | 84.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matić, S.; Candian, V.; D’Errico, C.; Pierro, R.; Panno, S.; Davino, S.; Noris, E.; Tedeschi, R. In-Field LAMP Detection of Flavescence Dorée Phytoplasma in Crude Extracts of the Scaphoideus titanus Vector. Agronomy 2022, 12, 1645. https://doi.org/10.3390/agronomy12071645
Matić S, Candian V, D’Errico C, Pierro R, Panno S, Davino S, Noris E, Tedeschi R. In-Field LAMP Detection of Flavescence Dorée Phytoplasma in Crude Extracts of the Scaphoideus titanus Vector. Agronomy. 2022; 12(7):1645. https://doi.org/10.3390/agronomy12071645
Chicago/Turabian StyleMatić, Slavica, Valentina Candian, Chiara D’Errico, Roberto Pierro, Stefano Panno, Salvatore Davino, Emanuela Noris, and Rosemarie Tedeschi. 2022. "In-Field LAMP Detection of Flavescence Dorée Phytoplasma in Crude Extracts of the Scaphoideus titanus Vector" Agronomy 12, no. 7: 1645. https://doi.org/10.3390/agronomy12071645
APA StyleMatić, S., Candian, V., D’Errico, C., Pierro, R., Panno, S., Davino, S., Noris, E., & Tedeschi, R. (2022). In-Field LAMP Detection of Flavescence Dorée Phytoplasma in Crude Extracts of the Scaphoideus titanus Vector. Agronomy, 12(7), 1645. https://doi.org/10.3390/agronomy12071645











