Choosing Feature Selection Methods for Spatial Modeling of Soil Fertility Properties at the Field Scale
Abstract
:1. Introduction
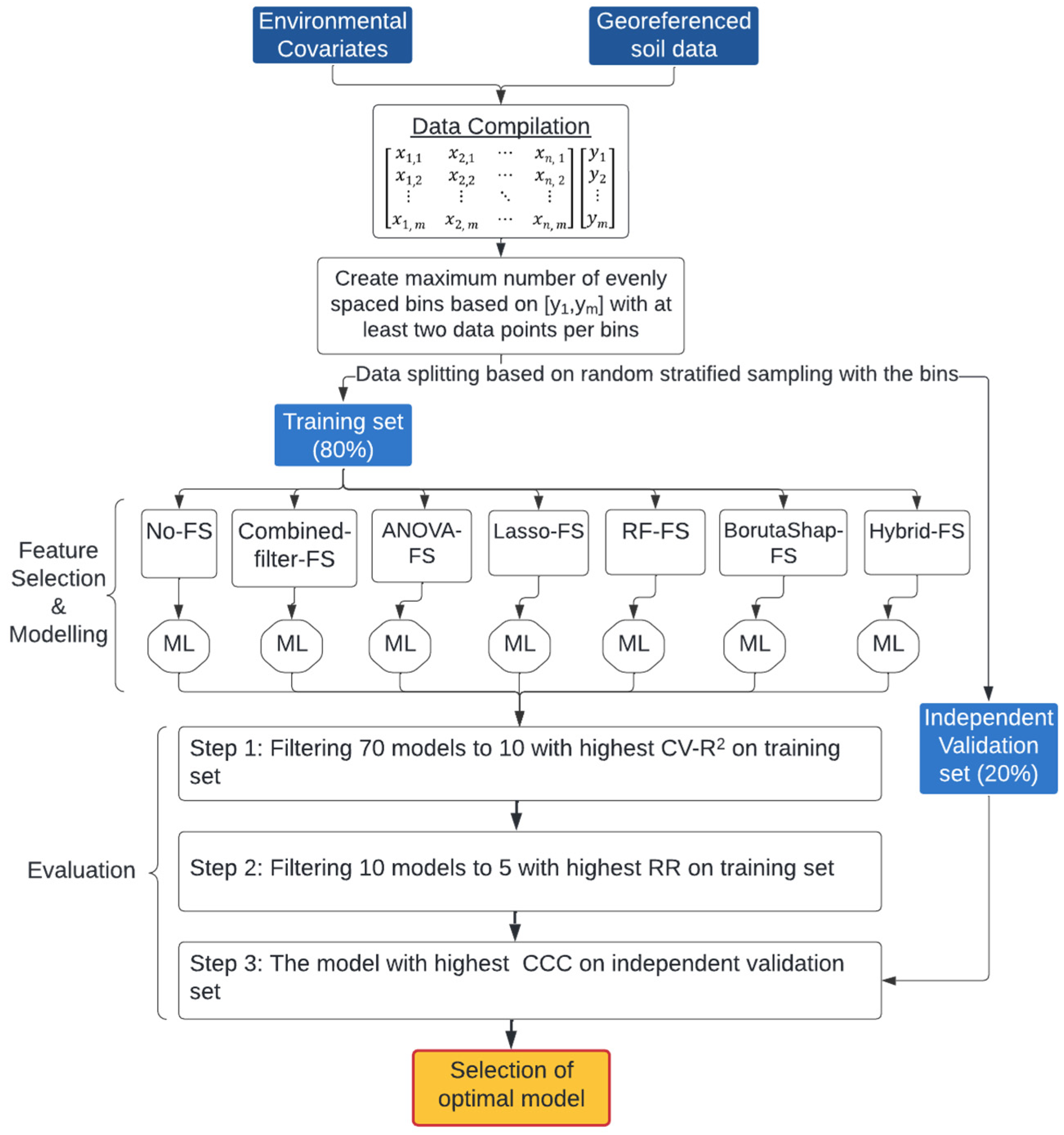
2. Materials and Methods
2.1. Study Fields and Soil Sampling
2.2. Environmental Covariates
2.3. Feature Selection
2.4. Machine Learning
2.5. Model Evaluation
2.5.1. Cross-Validation
2.5.2. Robustness Ratio
2.5.3. Independent Validation
3. Results
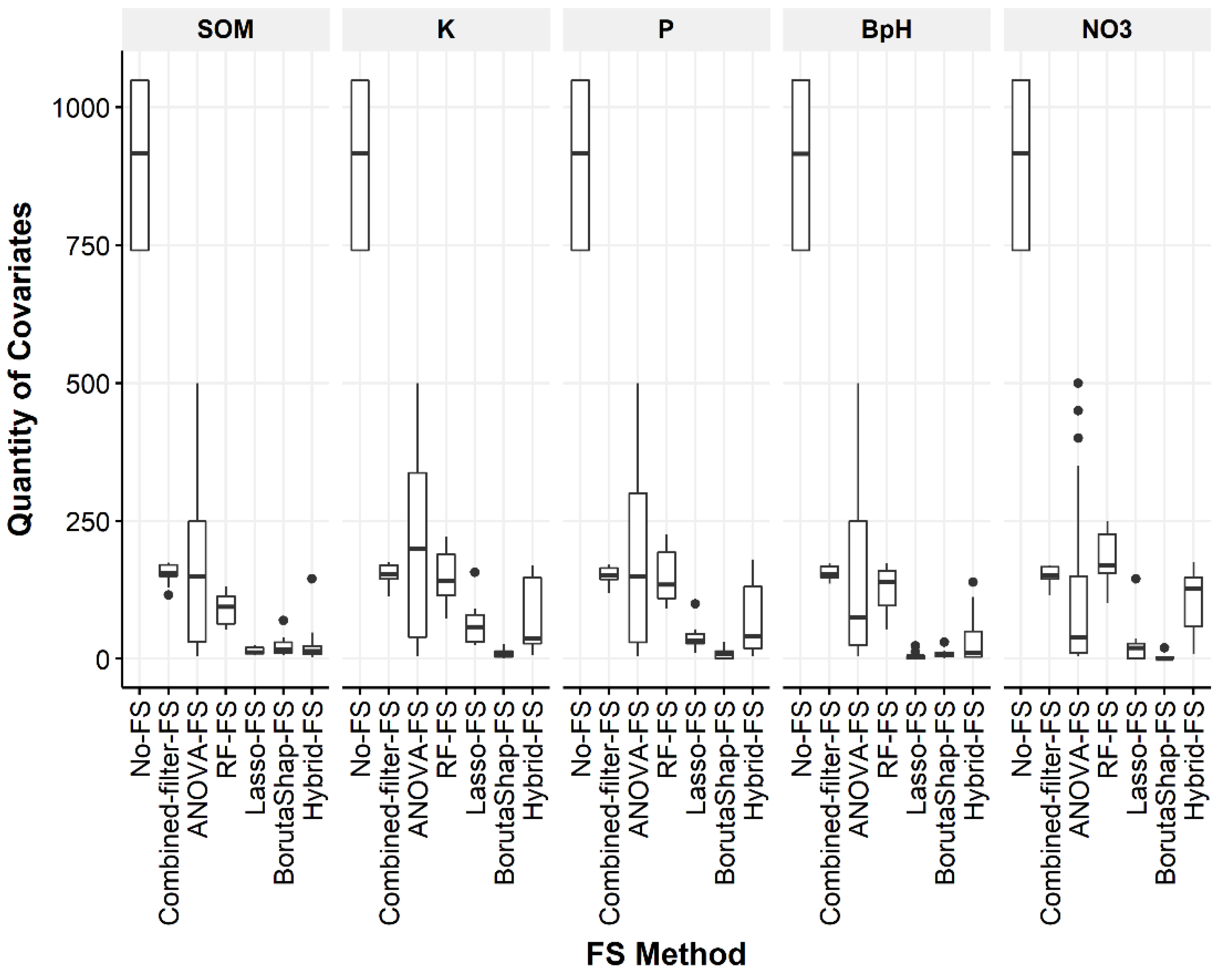
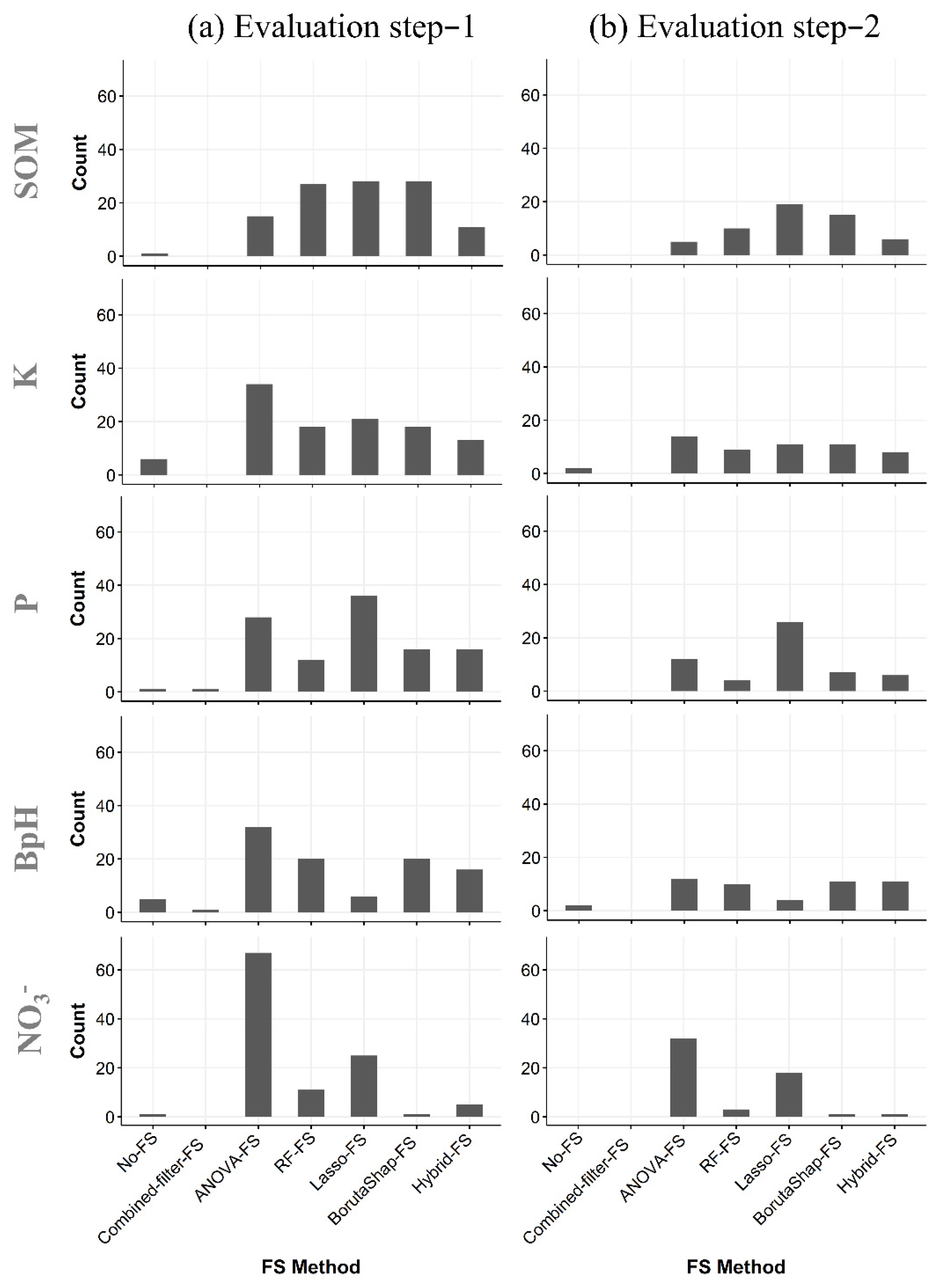
3.1. Quantity of Covariates Selected
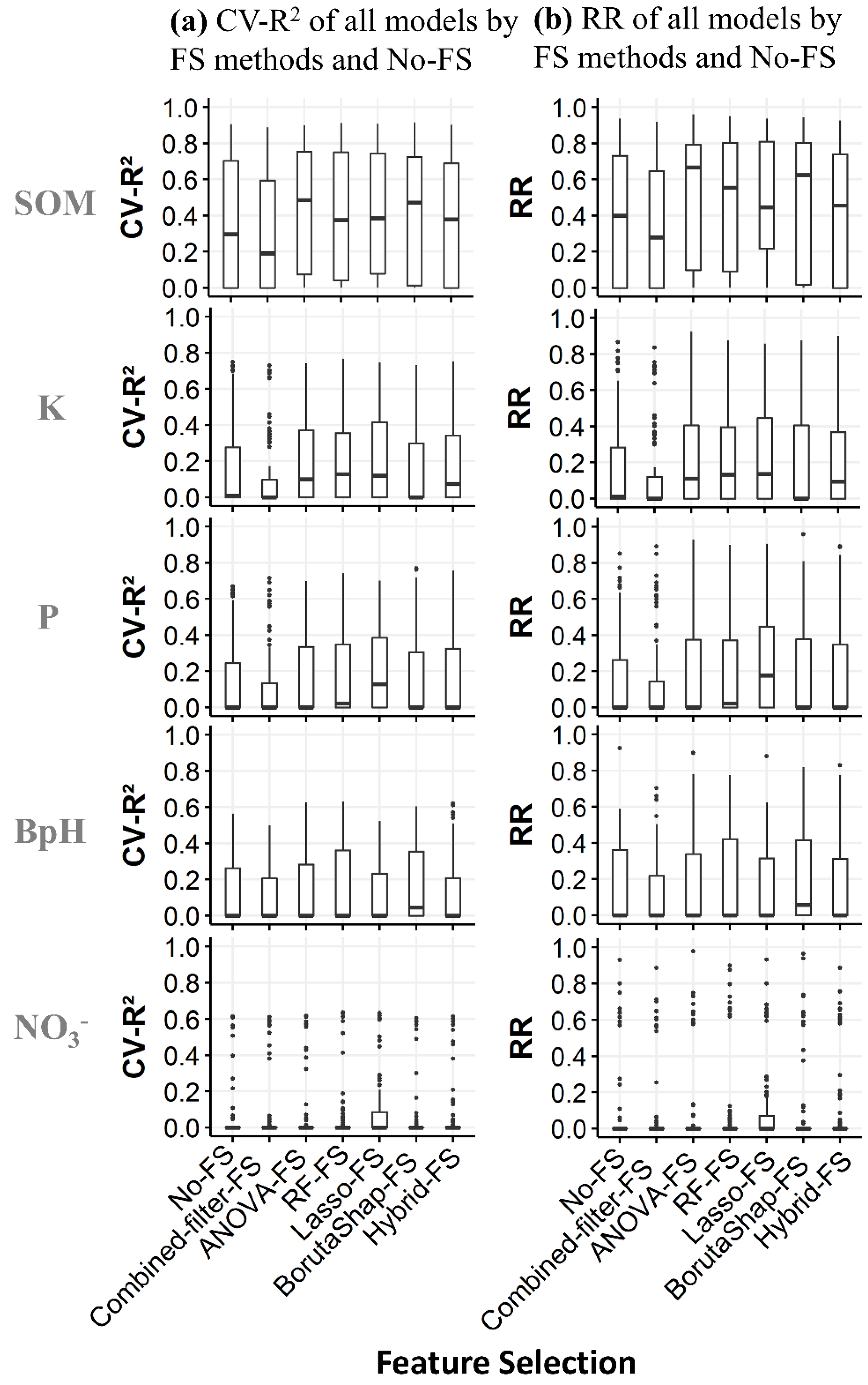
3.2. Cross-Validation
3.3. Robustness
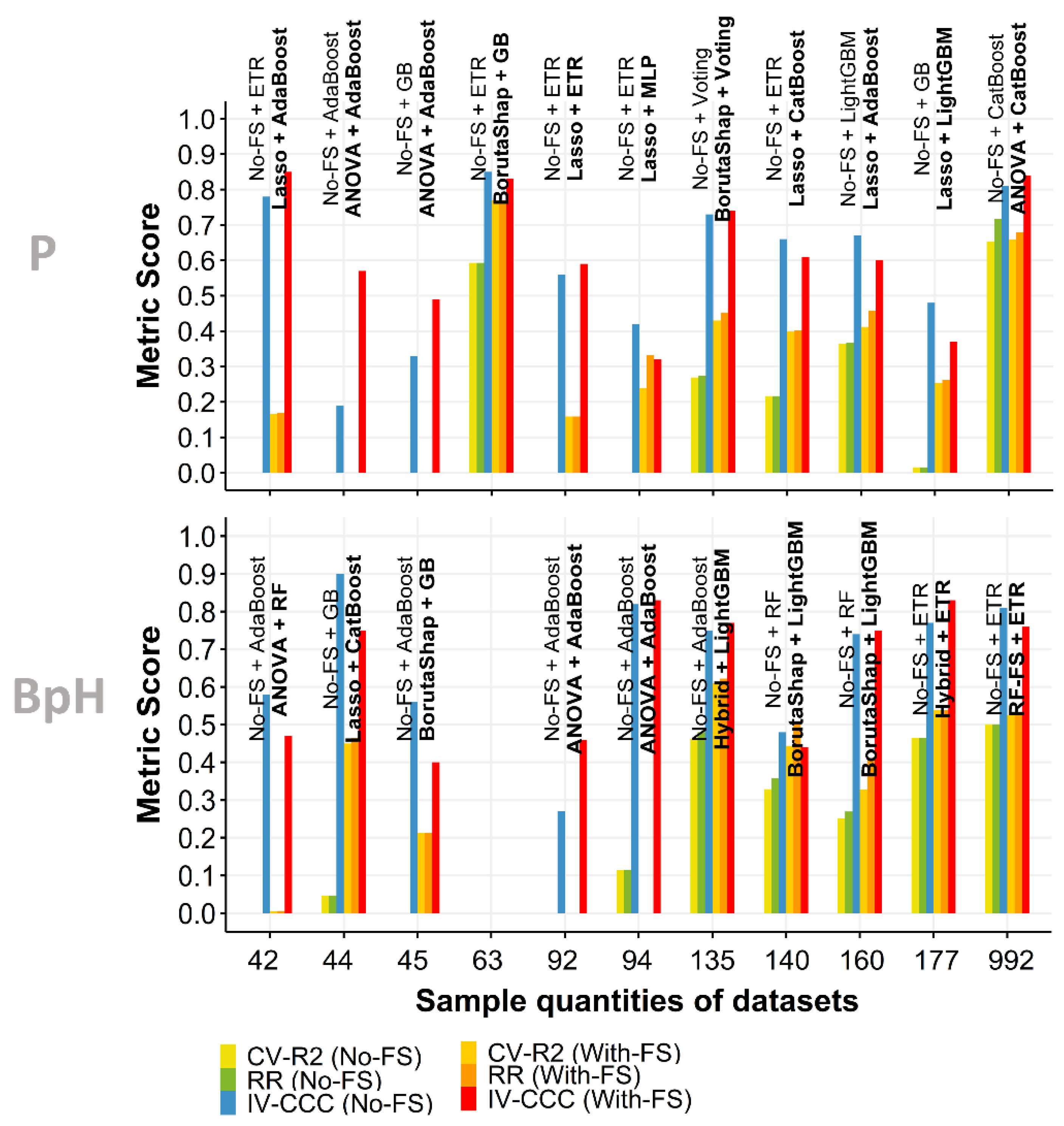
3.4. Independent Validation
3.5. Effect of Sample Quantity
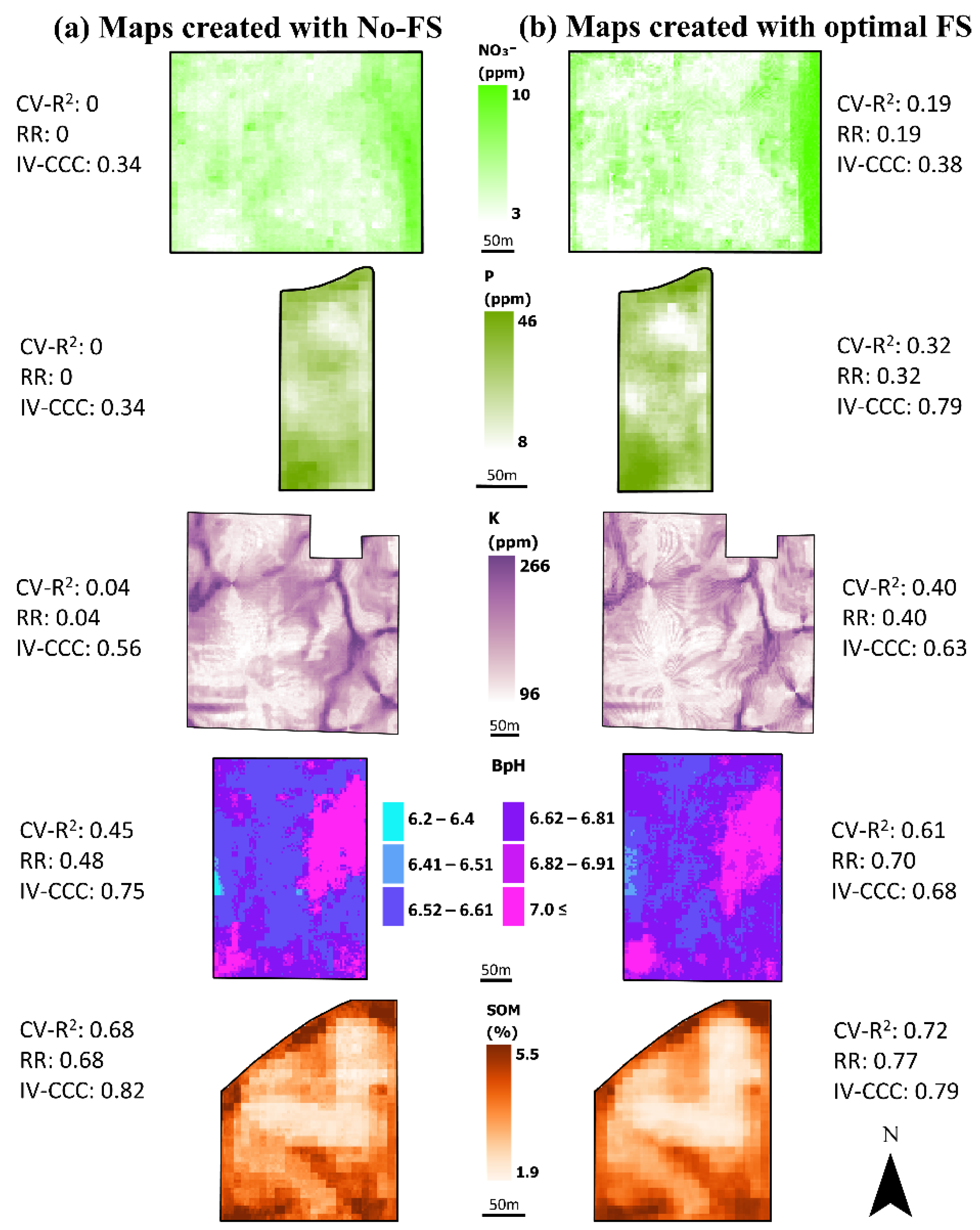
3.6. Comparison of Spatial Patterns in Maps
4. Discussion
4.1. Optimal FS Strategies
4.2. Optimal FS-ML Combinations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minasny, B.; McBratney, A.B. Digital Soil Mapping: A Brief History and Some Lessons. Geoderma 2016, 264, 301–311. [Google Scholar] [CrossRef]
- McBratney, A.B.; Santos, M.L.M.; Minasny, B. On Digital Soil Mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Xiong, X.; Grunwald, S.; Myers, D.B.; Kim, J.; Harris, W.G.; Comerford, N.B. Holistic Environmental Soil-Landscape Modeling of Soil Organic Carbon. Environ. Model. Softw. 2014, 57, 202–215. [Google Scholar] [CrossRef]
- Brungard, C.W.; Boettinger, J.L.; Duniway, M.C.; Wills, S.A.; Edwards, T.C. Machine Learning for Predicting Soil Classes in Three Semi-Arid Landscapes. Geoderma 2015, 239, 68–83. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; ISBN 9781461468493. [Google Scholar]
- Flynn, T.; de Clercq, W.; Rozanov, A.; Clarke, C. High-Resolution Digital Soil Mapping of Multiple Soil Properties: An Alternative to the Traditional Field Survey? S. Afr. J. Plant Soil 2019, 36, 237–247. [Google Scholar] [CrossRef]
- Van Dijk, A.D.J.; Kootstra, G.; Kruijer, W.; de Ridder, D. Machine Learning in Plant Science and Plant Breeding. iScience 2021, 24, 101890. [Google Scholar] [CrossRef]
- Hesami, M.; Jones, A.M.P. Application of Artificial Intelligence Models and Optimization Algorithms in Plant Cell and Tissue Culture. Appl. Microbiol. Biotechnol. 2020, 104, 9449–9485. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [Green Version]
- Bellman, R.; Kalaba, R.E. Dynamic Programming and Modern Control Theory; Citeseer: Princeton, NJ, USA, 1965; Volume 81. [Google Scholar]
- Chandrashekar, G.; Sahin, F. A Survey on Feature Selection Methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature Selection: A Data Perspective. ACM Comput. Surv. 2017, 50, 1–45. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Alonso-Betanzos, A. Ensembles for Feature Selection: A Review and Future Trends. Inf. Fusion 2019, 52, 1–12. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; Minasny, B.; McBratney, A.B. Machine Learning for Digital Soil Mapping: Applications, Challenges and Suggested Solutions. Earth-Sci. Rev. 2020, 210, 103359. [Google Scholar] [CrossRef]
- Yu, L.; Liu, H. Feature Selection for High-Dimensional Data: A Fast Correlation-Based Filter Solution. In Proceedings of the 20th International Conference on Machine Learning (ICML-03), Washington, DC, USA, 21–24 August 2003; pp. 856–863. [Google Scholar]
- De la Iglesia, B. Evolutionary Computation for Feature Selection in Classification Problems. WIREs Data Min. Knowl. Discov. 2013, 3, 381–407. [Google Scholar] [CrossRef]
- Seijo-Pardo, B.; Porto-Díaz, I.; Bolón-Canedo, V.; Alonso-Betanzos, A. Ensemble Feature Selection: Homogeneous and Heterogeneous Approaches. Knowl.-Based Syst. 2017, 118, 124–139. [Google Scholar] [CrossRef]
- Keany, E. BorutaShap: A Wrapper Feature Selection Method Which Combines the Boruta Feature Selection Algorithm with Shapley Values; Zenodo: Geneva, Switzerland, 2020. [Google Scholar]
- Chieregato, M.; Frangiamore, F.; Morassi, M.; Baresi, C.; Nici, S.; Bassetti, C.; Bnà, C.; Galelli, M. A Hybrid Machine Learning/Deep Learning COVID-19 Severity Predictive Model from CT Images and Clinical Data. Sci. Rep. 2022, 12, 4329. [Google Scholar] [CrossRef] [PubMed]
- Keany, E.; Bessardon, G.; Gleeson, E. Using Machine Learning to Produce a Cost-Effective National Building Height Map of Ireland to Categorise Local Climate Zones. Adv. Sci. Res. 2022, 19, 13–27. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. 2017, 2017, 4766–4775. [Google Scholar]
- Shapley, L.S. A Value for N-Person Games. In Contributions to the Theory of Games; Princeton University Press: Princeton, NJ, USA, 1953; Volume 2, pp. 307–317. [Google Scholar]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, J.; Song, X.; Qin, Z.; Wu, L.; Wang, H.; Tang, J. Hyperspectral Band Selection and Modeling of Soil Organic Matter Content in a Forest Using the Ranger Algorithm. PLoS ONE 2021, 16, e0253385. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, L.; Yu, D.; Zhang, H.; Feng, K.; Wang, X.; Song, J. Comparison of Feature Selection Methods for Mapping Soil Organic Matter in Subtropical Restored Forests. Ecol. Indic. 2022, 135, 108545. [Google Scholar] [CrossRef]
- Behrens, T.; Zhu, A.-X.; Schmidt, K.; Scholten, T. Multi-Scale Digital Terrain Analysis and Feature Selection for Digital Soil Mapping. Geoderma 2010, 155, 175–185. [Google Scholar] [CrossRef]
- Campos, A.R.; Giasson, E.; Costa, J.J.F.; Machado, I.R.; da Silva, E.B.; Bonfatti, B.R. Selection of Environmental Covariates for Classifier Training Applied in Digital Soil Mapping. Rev. Bras. Ciênc. Solo 2019, 42, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Chen, S.; Chen, Y.; Linderman, M.; Mouazen, A.M.; Liu, Y.; Guo, L.; Yu, L.; Liu, Y.; Cheng, H.; et al. Comparing Laboratory and Airborne Hyperspectral Data for the Estimation and Mapping of Topsoil Organic Carbon: Feature Selection Coupled with Random Forest. Soil Tillage Res. 2020, 199, 104589. [Google Scholar] [CrossRef]
- Yang, R.-M.; Liu, L.-A.; Zhang, X.; He, R.-X.; Zhu, C.-M.; Zhang, Z.-Q.; Li, J.-G. The Effectiveness of Digital Soil Mapping with Temporal Variables in Modeling Soil Organic Carbon Changes. Geoderma 2022, 405, 115407. [Google Scholar] [CrossRef]
- Luo, C.; Zhang, X.; Wang, Y.; Men, Z.; Liu, H. Regional Soil Organic Matter Mapping Models Based on the Optimal Time Window, Feature Selection Algorithm and Google Earth Engine. Soil Tillage Res. 2022, 219, 105325. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, F.; Zhao, Y.; Song, X.; Zhang, G. An Integrated Method of Selecting Environmental Covariates for Predictive Soil Depth Mapping. J. Integr. Agric. 2019, 18, 301–315. [Google Scholar] [CrossRef]
- Domenech, M.B.; Amiotti, N.M.; Costa, J.L.; Castro-Franco, M. Prediction of Topsoil Properties at Field-Scale by Using C-Band SAR Data. Int. J. Appl. Earth Obs. Geoinf. 2020, 93, 102197. [Google Scholar] [CrossRef]
- Wang, S.-H.; Lu, H.-L.; Zhao, M.-S.; Zhou, L.-M. Assessing soil pH in Anhui Province based on different features mining methods combined with generalized boosted regression models. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2020, 31, 3509–3517. [Google Scholar] [CrossRef]
- Iowa Geospatial Data. Available online: https://geodata.iowa.gov/ (accessed on 28 June 2022).
- Ashley, M.D.; Rea, J. Seasonal Vegetation Differences from ERTS Imagery; American Society of Photogrammetry: Falls Church, VA, USA, 1975; Volume 41. [Google Scholar]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, C. Distinguishing Vegetation from Soil Background Information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Xiaoqin, W.; Miaomiao, W.; Shaoqiang, W.; Yundong, W. Extraction of Vegetation Information from Visible Unmanned Aerial Vehicle Images. Trans. Chin. Soc. Agric. Eng. 2015, 31, 152–159. [Google Scholar]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A Modified Soil Adjusted Vegetation Index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Travis, M.R. VIEWIT: Computation of Seen Areas, Slope, and Aspect for Land-Use Planning; Department of Agriculture, Forest Service, Pacific Southwest Forest and Range Experiment Station: Albany, CA, USA, 1975; Volume 11.
- Tarboton, D.G. A New Method for the Determination of Flow Directions and Upslope Areas in Grid Digital Elevation Models. Water Resour. Res. 1997, 33, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Costa-Cabral, M.C.; Burges, S.J. Digital Elevation Model Networks (DEMON): A Model of Flow over Hillslopes for Computation of Contributing and Dispersal Areas. Water Resour. Res. 1994, 30, 1681–1692. [Google Scholar] [CrossRef]
- Evans, I.S. An Integrated System of Terrain Analysis and Slope Mapping. Z. Für Geomorphol. Suppl. Stuttg. 1980, 36, 274–295. [Google Scholar]
- Heerdegen, R.G.; Beran, M.A. Quantifying Source Areas through Land Surface Curvature and Shape. J. Hydrol. 1982, 57, 359–373. [Google Scholar] [CrossRef]
- Bauer, J.; Rohdenburg, H.; Bork, H. Ein Digitales Reliefmodell als Vorraussetzung für ein Deterministisches Modell der Wasser-und Stoff-Flüsse. Landsch. Landsch. 1985, 10, 1–15. [Google Scholar]
- Zevenbergen, L.W.; Thorne, C.R. Quantitative Analysis of Land Surface Topography. Earth Surf. Process. Landf. 1987, 12, 47–56. [Google Scholar] [CrossRef]
- Haralick, R.M. Ridges and Valleys on Digital Images. Comput. Vis. Graph. Image Process. 1983, 22, 28–38. [Google Scholar] [CrossRef]
- Lin, L.I.-K. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Jonas, R.; Cook, J. Lasso Regression. Br. J. Surg. 2018, 105, 1348. [Google Scholar]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.; Vapnik, V. Support Vector Regression Machines. Adv. Neural Inf. Process. Syst. 1996, 9, 155–161. [Google Scholar]
- Rosenblatt, F. The Perceptron: A Probabilistic Model for Information Storage and Organization in the Brain. Psychol. Rev. 1958, 65, 386. [Google Scholar] [CrossRef] [Green Version]
- Awad, M.; Khanna, R. Support Vector Regression. In Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers; Awad, M., Khanna, R., Eds.; Apress: Berkeley, CA, USA, 2015; pp. 67–80. ISBN 9781430259909. [Google Scholar]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Bagging Predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely Randomized Trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Dorogush, A.V.; Ershov, V.; Gulin, A. CatBoost: Gradient Boosting with Categorical Features Support. arXiv 2018, arXiv:1810.11363. [Google Scholar]
- Freund, Y.; Schapire, R.E. Experiments with a New Boosting Algorithm; Citeseer: Princeton, NJ, USA, 1996; Volume 96, pp. 148–156. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A Highly Efficient Gradient Boosting Decision Tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Oshiro, T.M.; Perez, P.S.; Baranauskas, J.A. How Many Trees in a Random Forest? In Proceedings of the International Workshop on Machine Learning and Data Mining in Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2012; pp. 154–168. [Google Scholar]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-Validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Arlot, S.; Celisse, A. A Survey of Cross-Validation Procedures for Model Selection. Stat. Surv. 2010, 4, 40–79. [Google Scholar] [CrossRef]
- Kelcey, B. Covariate Selection in Propensity Scores Using Outcome Proxies. Multivar. Behav. Res. 2011, 46, 453–476. [Google Scholar] [CrossRef]
- Browne, M.W. Cross-Validation Methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef] [Green Version]
- Berrar, D. Cross-Validation; Tokyo Institute of Technology: Tokyo, Japan, 2019. [Google Scholar]
- Khaledian, Y.; Miller, B.A. Selecting Appropriate Machine Learning Methods for Digital Soil Mapping. Appl. Math. Model. 2020, 81, 401–418. [Google Scholar] [CrossRef]
- Cheng, T.H.; Wei, C.P.; Tseng, S. Feature Selection for Medical Data Mining. In Proceedings of the 19th IEEE International Symposium on Computer-Based Medical Systems (CBMS ’06), Salt Lake City, UT, USA, 22–23 June 2006; pp. 165–170. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An Introduction to Variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Clifton, C. Definition of Data Mining; Encyclopædia Britannica: Chicago, IL, USA, 2010; Volume 9. [Google Scholar]
- Ashtekar, J.M.; Owens, P.R. Remembering Knowledge: An Expert Knowledge Based Approach to Digital Soil Mapping. Soil Horiz. 2013, 54, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Galiano, V.F.; Luque-Espinar, J.A.; Chica-Olmo, M.; Mendes, M.P. Feature Selection Approaches for Predictive Modelling of Groundwater Nitrate Pollution: An Evaluation of Filters, Embedded and Wrapper Methods. Sci. Total Environ. 2018, 624, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.K. Random Decision Forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, pp. 278–282. [Google Scholar]
- Morgan, J.; Daugherty, R.; Hilchie, A.; Carey, B. Sample Size and Modeling Accuracy of Decision Tree Based Data Mining Tools. Acad. Inf. Manag. Sci. J. 2003, 6, 77–91. [Google Scholar]
- Schapire, R.E.; Freund, Y. Boosting: Foundations and Algorithms. Kybernetes 2013, 42, 164–166. [Google Scholar] [CrossRef]
- Meier, M.; de Souza, E.; Francelino, M.R.; Fernandes Filho, E.I.; Schaefer, C.E.G.R. Digital Soil Mapping Using Machine Learning Algorithms in a Tropical Mountainous Area. Rev. Bras. Ciênc. Solo 2018, 42, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Hu, M.Y.; Patuwo, B.E.; Indro, D.C. Artificial Neural Networks in Bankruptcy Prediction: General Framework and Cross-Validation Analysis. Eur. J. Oper. Res. 1999, 116, 16–32. [Google Scholar] [CrossRef] [Green Version]
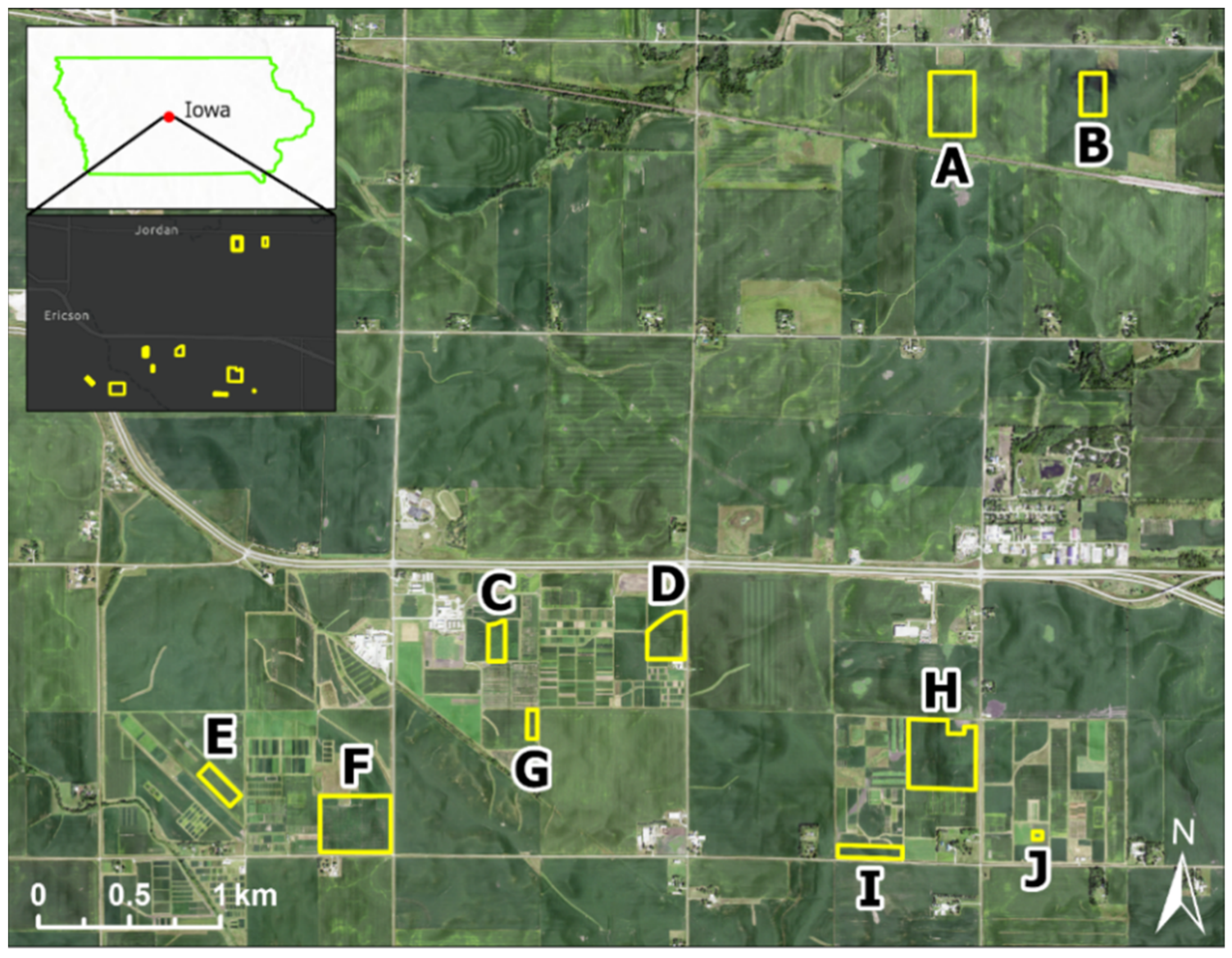


| Soil Property | Field | n | Min | Median | Mean | Max | SD | CoV | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|---|---|
| NO3− (ppm) | A | 135 | 2 | 5 | 5.1 | 13 | 1.94 | 0.38 | 1.05 | 4.51 |
| B | 45 | 4 | 6 | 6.53 | 14 | 2.03 | 0.31 | 1.25 | 5.53 | |
| C | 42 | 3 | 6 | 5.83 | 13 | 1.78 | 0.31 | 1.7 | 7.94 | |
| D | 160 | 2 | 12 | 15.53 | 55 | 11.6 | 0.75 | 1.18 | 3.92 | |
| E | 92 | 2 | 4 | 3.84 | 7 | 1.34 | 0.35 | 0.41 | 2.39 | |
| F | 177 | 3 | 6 | 6.03 | 12 | 1.72 | 0.29 | 0.62 | 3.23 | |
| G | 44 | 2 | 5 | 4.93 | 9 | 1.87 | 0.38 | 0.14 | 2.07 | |
| H | 94 | 3 | 6 | 10.21 | 41 | 8.48 | 0.83 | 1.77 | 5.37 | |
| I | 140 | 3 | 20 | 21.76 | 54 | 8.62 | 0.4 | 0.89 | 3.73 | |
| J | 63 | 8 | 12 | 12.6 | 21 | 2.55 | 0.2 | 0.83 | 3.84 | |
| all fields | 992 | 2 | 7 | 10.23 | 55 | 8.82 | 0.86 | 1.95 | 6.93 | |
| P2O5 (ppm) | A | 135 | 1 | 12 | 13.3 | 38 | 7.51 | 0.56 | 1.1 | 3.87 |
| B | 45 | 11 | 25 | 29.02 | 69 | 15.21 | 0.52 | 1.28 | 3.96 | |
| C | 42 | 8 | 25.5 | 26.21 | 46 | 10.18 | 0.39 | 0.2 | 2.16 | |
| D | 160 | 10 | 41 | 41.95 | 89 | 18.51 | 0.44 | 0.52 | 2.88 | |
| E | 92 | 9 | 23.5 | 25.4 | 76 | 10.61 | 0.42 | 1.55 | 7.47 | |
| F | 177 | 4 | 19 | 21.21 | 54 | 10.34 | 0.49 | 0.82 | 3.12 | |
| G | 44 | 6 | 13.5 | 16.34 | 51 | 9.48 | 0.58 | 1.6 | 5.78 | |
| H | 94 | 1 | 15 | 17.34 | 62 | 11.11 | 0.64 | 1.73 | 7.42 | |
| I | 140 | 3 | 14 | 16.45 | 55 | 9.87 | 0.6 | 1.4 | 5.3 | |
| J | 63 | 1 | 2 | 3.71 | 12 | 2.81 | 0.76 | 1.42 | 4.15 | |
| all fields | 992 | 1 | 18 | 22.07 | 89 | 15.62 | 0.71 | 1.38 | 5.3 | |
| K2O (ppm) | A | 135 | 104 | 174 | 179.47 | 296 | 34.76 | 0.19 | 0.67 | 3.72 |
| B | 45 | 105 | 161 | 166.91 | 298 | 34.41 | 0.21 | 1.26 | 6.13 | |
| C | 42 | 108 | 143 | 147.88 | 219 | 22.64 | 0.15 | 0.76 | 3.73 | |
| D | 160 | 83 | 151 | 164.68 | 438 | 59.26 | 0.36 | 1.68 | 7.32 | |
| E | 92 | 109 | 174 | 180.6 | 302 | 31.4 | 0.17 | 0.67 | 4.46 | |
| F | 177 | 89 | 164 | 162.72 | 241 | 31.16 | 0.19 | 0.14 | 2.56 | |
| G | 44 | 108 | 150.5 | 155.32 | 257 | 26.22 | 0.17 | 1.64 | 7.44 | |
| H | 94 | 96 | 138 | 143.48 | 266 | 34.26 | 0.24 | 1.01 | 3.98 | |
| I | 140 | 99 | 172 | 172.41 | 256 | 36.72 | 0.21 | 0.31 | 2.61 | |
| J | 63 | 133 | 167 | 171.63 | 232 | 23.21 | 0.14 | 0.94 | 3.25 | |
| all fields | 992 | 83 | 163 | 166.32 | 438 | 39.34 | 0.24 | 1.19 | 7.57 | |
| BpH | A | 135 | 6.2 | 6.7 | 6.77 | 7.1 | 0.22 | 0.03 | 0.44 | 2.23 |
| B | 45 | 5.9 | 6.4 | 6.42 | 7.1 | 0.26 | 0.04 | 0.32 | 3.35 | |
| C | 42 | 6.6 | 7.1 | 6.94 | 7.1 | 0.18 | 0.03 | −0.43 | 1.51 | |
| D | 160 | 5.9 | 6.6 | 6.55 | 7.1 | 0.24 | 0.04 | 0.06 | 3.32 | |
| E | 92 | 6.5 | 6.6 | 6.66 | 7.1 | 0.17 | 0.02 | 1.89 | 5.54 | |
| F | 177 | 6 | 6.7 | 6.74 | 7.1 | 0.26 | 0.04 | 0.13 | 2.39 | |
| G | 44 | 6.5 | 6.7 | 6.78 | 7.1 | 0.19 | 0.03 | 0.89 | 2.21 | |
| H | 94 | 0 | 6.4 | 5.08 | 6.8 | 2.66 | 0.52 | −1.38 | 2.95 | |
| I | 140 | 6.4 | 6.75 | 6.8 | 7.1 | 0.18 | 0.03 | 0.51 | 2.35 | |
| J | 63 | 7.1 | 7.1 | 7.1 | 7.1 | 0 | 0 | 0 | 0 | |
| all fields | 992 | 0 | 6.7 | 6.58 | 7.1 | 0.98 | 0.15 | −6.06 | 40.92 | |
| SOM % | A | 135 | 1.5 | 3.6 | 3.76 | 6.7 | 1.16 | 0.31 | 0.45 | 2.71 |
| B | 45 | 2.1 | 3.7 | 3.55 | 4.7 | 0.67 | 0.19 | −0.55 | 2.42 | |
| C | 42 | 2.1 | 2.85 | 2.91 | 4.2 | 0.53 | 0.18 | 0.61 | 2.77 | |
| D | 160 | 1.9 | 3 | 3.06 | 5.5 | 0.7 | 0.23 | 0.57 | 3.1 | |
| E | 92 | 2 | 3.7 | 3.63 | 5.1 | 0.76 | 0.21 | −0.05 | 2.08 | |
| F | 177 | 2.1 | 4.1 | 4.16 | 6.8 | 1.06 | 0.25 | 0.46 | 2.61 | |
| G | 44 | 2.5 | 3.75 | 3.8 | 4.9 | 0.49 | 0.13 | −0.2 | 3.13 | |
| H | 94 | 1.2 | 3.5 | 3.85 | 7.8 | 1.45 | 0.38 | 0.6 | 2.54 | |
| I | 140 | 2 | 3.1 | 3.18 | 6 | 0.7 | 0.22 | 1.13 | 5.06 | |
| J | 63 | 4.4 | 5.4 | 5.37 | 6.3 | 0.47 | 0.09 | −0.38 | 2.56 | |
| all fields | 992 | 1.2 | 3.5 | 3.69 | 7.8 | 1.09 | 0.29 | 0.67 | 3 |
| Environmental Covariates | N | Software | Spatial Resolution (m) | Analysis Scale | Spectral Bands | Date |
|---|---|---|---|---|---|---|
| DTA | ||||||
| Aspect | 65 | GRASS & SAGA | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Cross Sectional Curvature | 51 | GRASS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Longitudinal Curvature | 51 | GRASS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Plan Curvature | 51 | GRASS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Profile Curvature | 51 | GRASS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Relative Elevation | 65 | ArcGIS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Slope | 65 | GRASS & SAGA | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Eastness | 51 | GRASS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Northness | 51 | GRASS | 3, 10 | 9–123 m 130–1010 m | 2009 | |
| Vertical Curvature | 10 | SAGA | 3, 10 | 2009 | ||
| Vertical Distance to Channel | 1 | SAGA | 3 | 2009 | ||
| Saga Wetness Index | 2 | SAGA | 3, 10 | 2009 | ||
| Horizontal Curvature | 10 | SAGA | 3, 10 | 2009 | ||
| Curvature | 10 | SAGA | 3, 10 | 2009 | ||
| Hillshade | 2 | SAGA | 3, 10 | 2009 | ||
| RS | ||||||
| NAIP (spectral bands) | 43 | 1 | R,G,B R,G,B,N | 2005–2019 2010–2019 | ||
| Sentinel-2 (spectral bands) | 312 | 10 | R,G,B,N | 2017–2020 | ||
| NDVI | 7, 25 | NAIP, Sentinel-2 | 1, 10 | 2010–2019 2017–2020 | ||
| SAVI | 7, 25 | NAIP, Sentinel-2 | 1,10 | 2010–2019 2017–2020 | ||
| RVI | 7 | NAIP | 1 | 2010–2019 | ||
| DVI | 7 | NAIP | 1 | 2010–2019 | ||
| VDVI | 5 | NAIP | 1 | 2005–2009 | ||
| MSAVI | 25 | Sentinel-2 | 10 | 2017–2020 | ||
| CI | 25 | Sentinel-2 | 10 | 2017–2020 | ||
| GDVI | 25 | Sentinel-2 | 10 | 2017–2020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferhatoglu, C.; Miller, B.A. Choosing Feature Selection Methods for Spatial Modeling of Soil Fertility Properties at the Field Scale. Agronomy 2022, 12, 1786. https://doi.org/10.3390/agronomy12081786
Ferhatoglu C, Miller BA. Choosing Feature Selection Methods for Spatial Modeling of Soil Fertility Properties at the Field Scale. Agronomy. 2022; 12(8):1786. https://doi.org/10.3390/agronomy12081786
Chicago/Turabian StyleFerhatoglu, Caner, and Bradley A. Miller. 2022. "Choosing Feature Selection Methods for Spatial Modeling of Soil Fertility Properties at the Field Scale" Agronomy 12, no. 8: 1786. https://doi.org/10.3390/agronomy12081786
APA StyleFerhatoglu, C., & Miller, B. A. (2022). Choosing Feature Selection Methods for Spatial Modeling of Soil Fertility Properties at the Field Scale. Agronomy, 12(8), 1786. https://doi.org/10.3390/agronomy12081786






