Transcriptome-Based Weighted Correlation Network Analysis of Maize Leaf Angle Regulation by Exogenous Brassinosteroid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Material BR Treatment
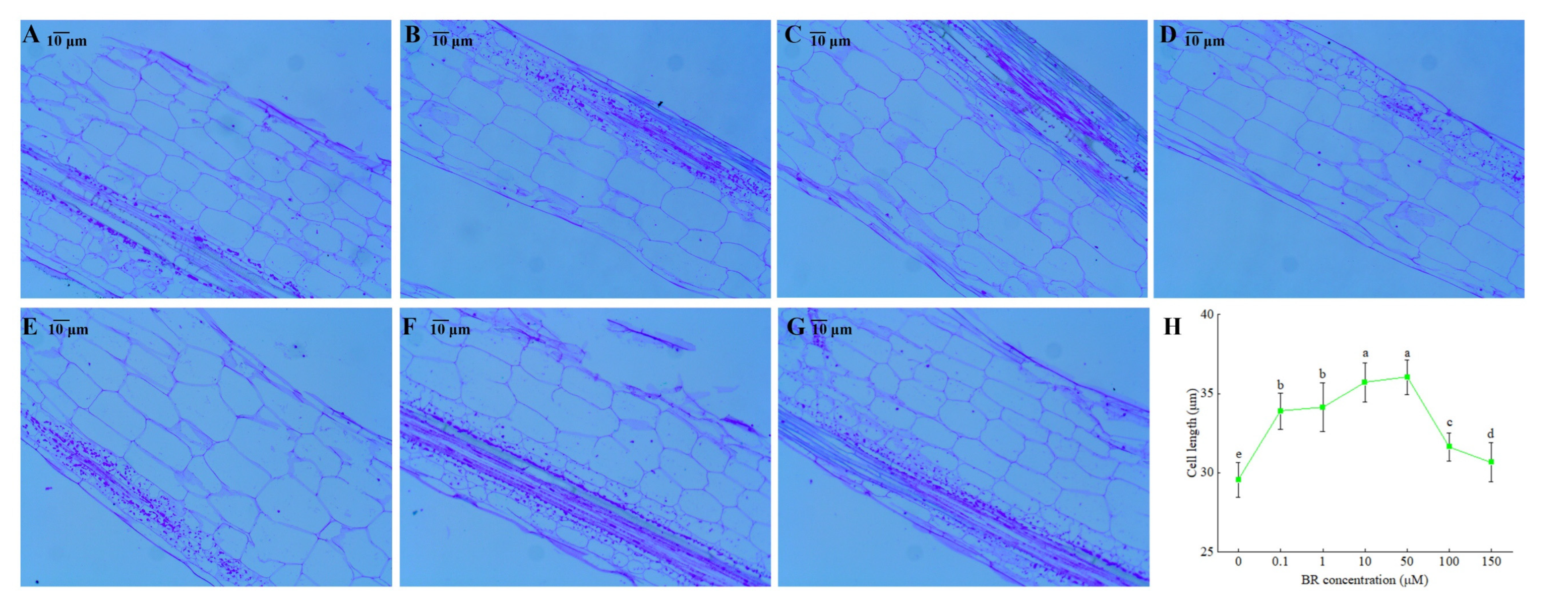
2.2. Ligular Region Longitudinal Structure Analysis
2.3. Total RNA Extractions, cDNA Library Construction, and Sequencing
2.4. Transcriptome Analysis
2.5. Weighted Gene Co-Expression Network Analysis
2.6. qRT-PCR of DEGs
2.7. Statistical Analysis of the Leaf Angle and the Cell Length
3. Results
3.1. Screening of the Optimal Concentration of Exogenous BR
3.2. Cytological Observations of Ligular Region
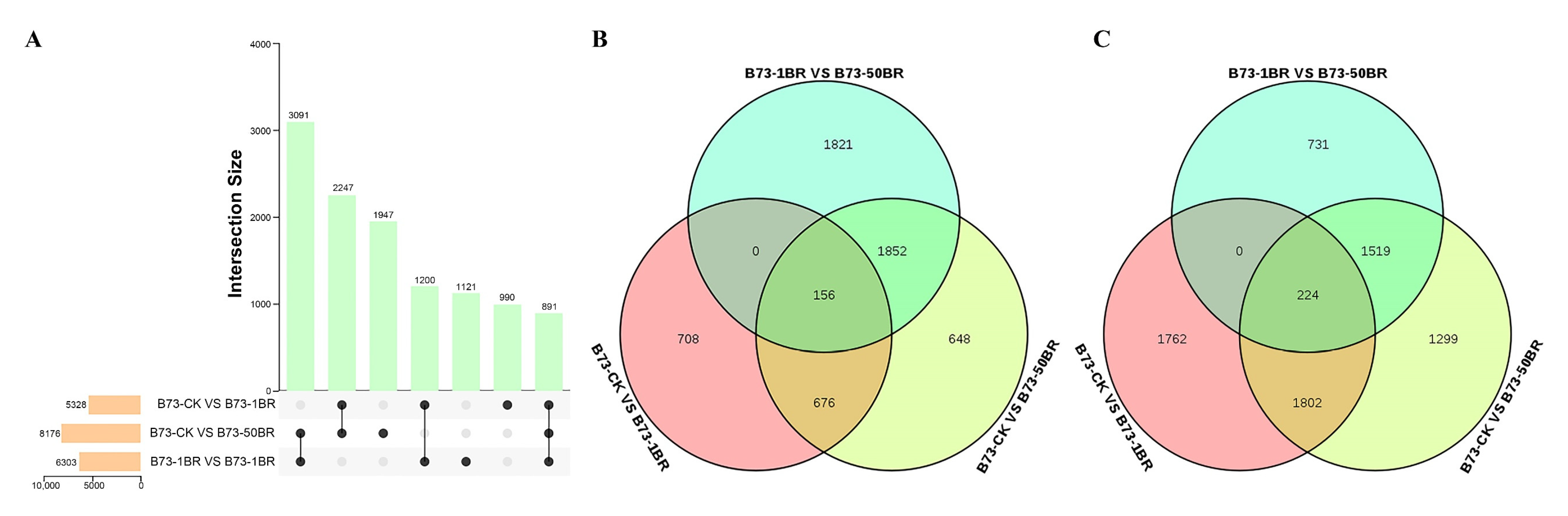
3.3. Analysis of DEGs in B73 under Different Concentrations of BR
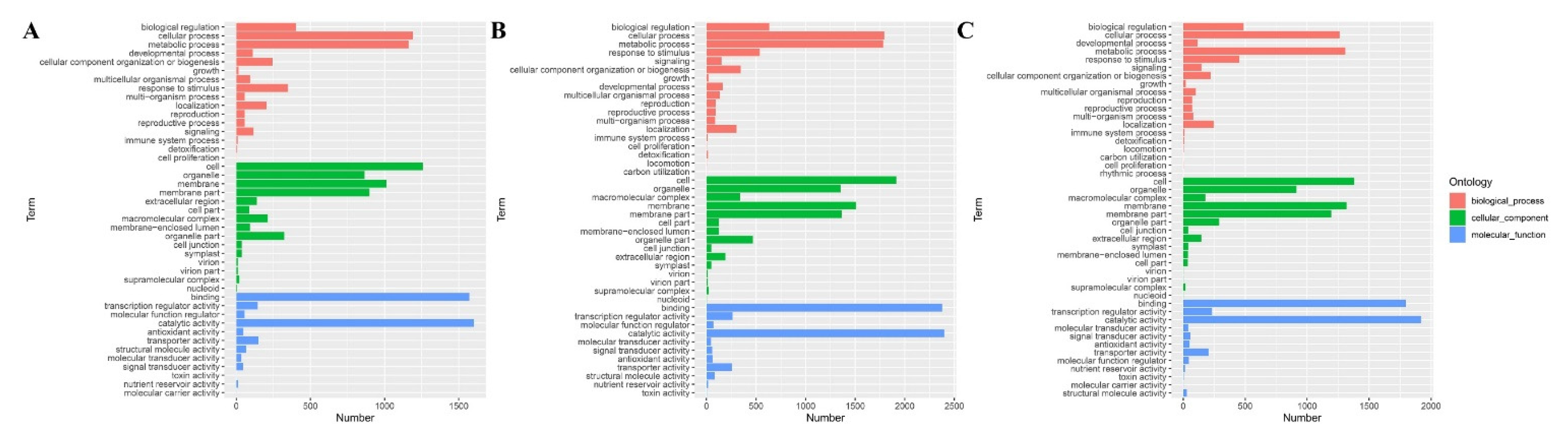
3.4. Gene Ontology Classification Analyses of B73 DEGs under Different BR Concentrations
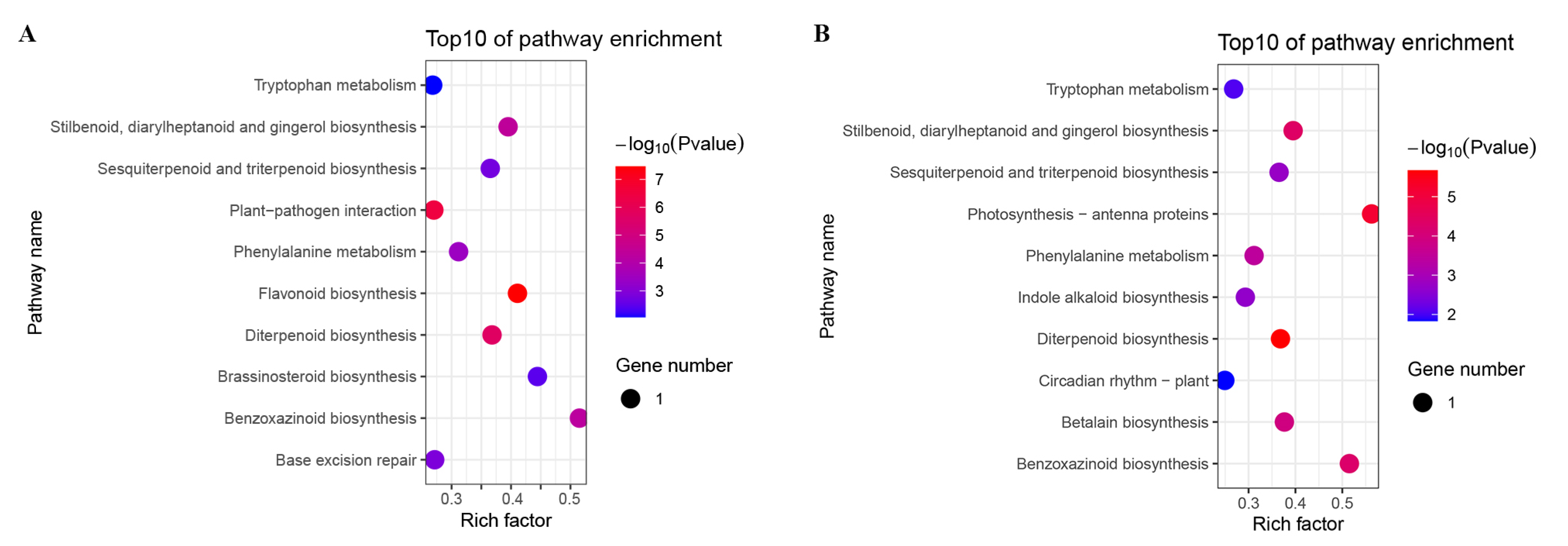
3.5. Pathway Enrichment Analysis of B73 DEGs under Different BR Concentrations
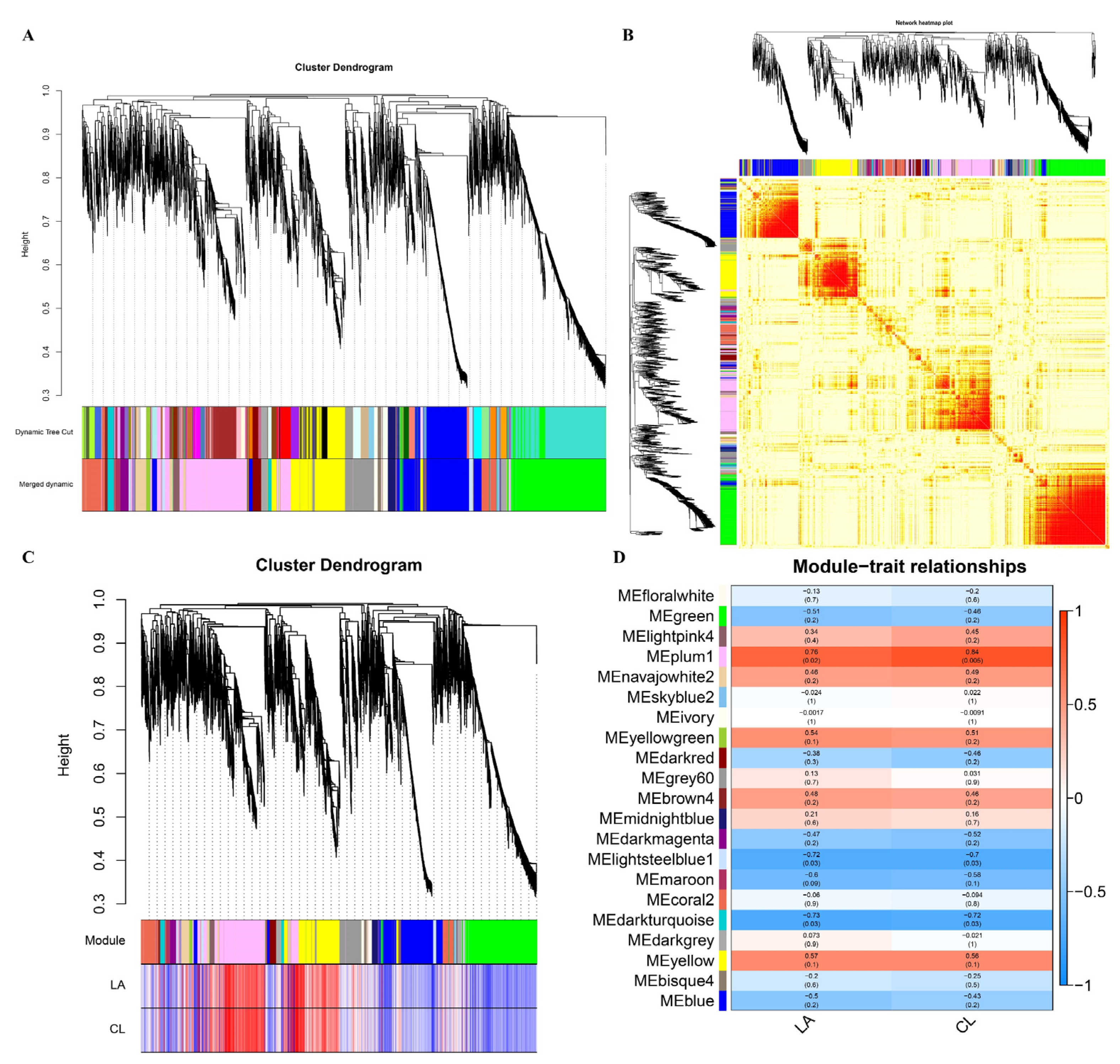
3.6. Gene Co-Expression Network Analysis
3.7. Functional Analysis of Genes in Correlated Modules
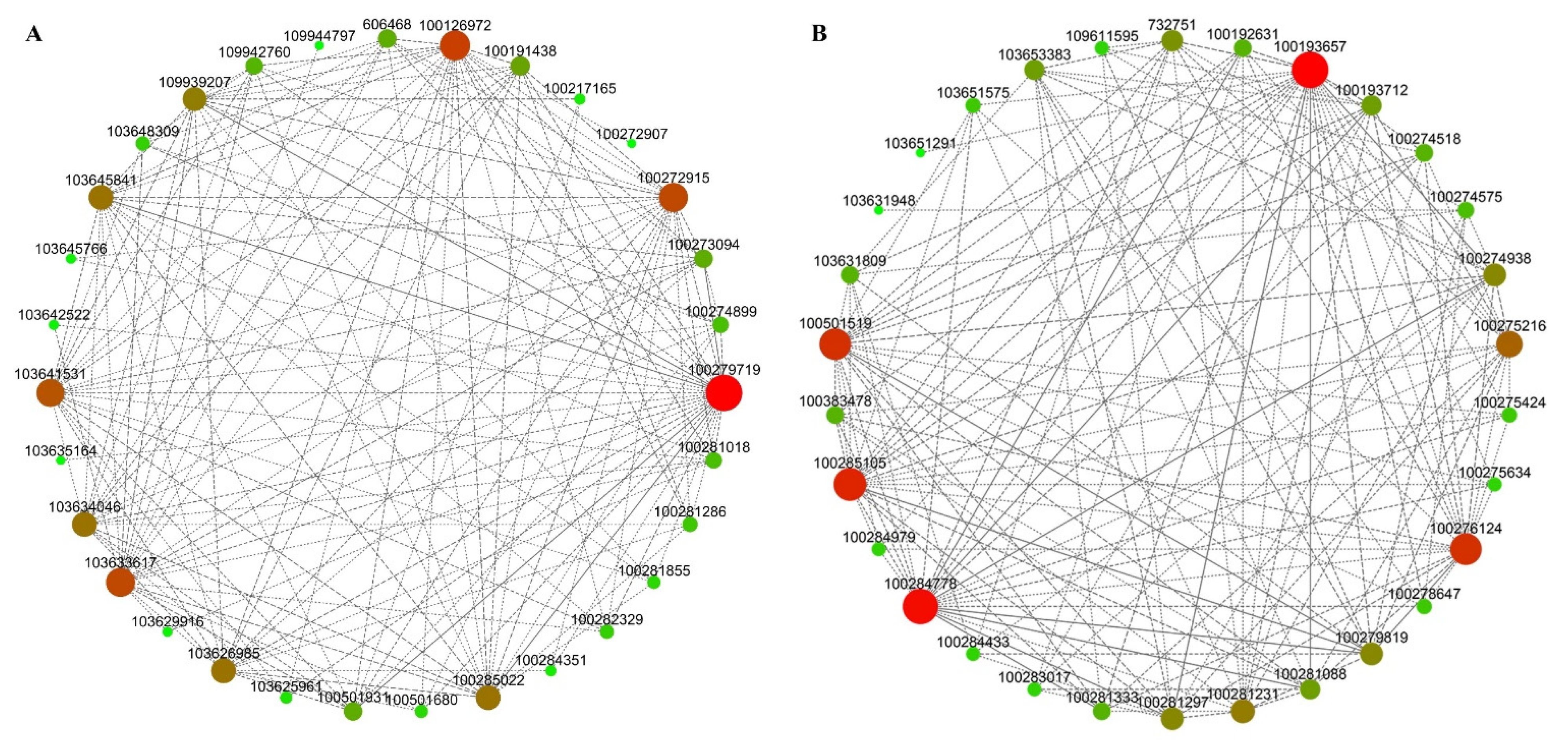
3.8. Analysis of Hub Genes Interaction Network in the Module
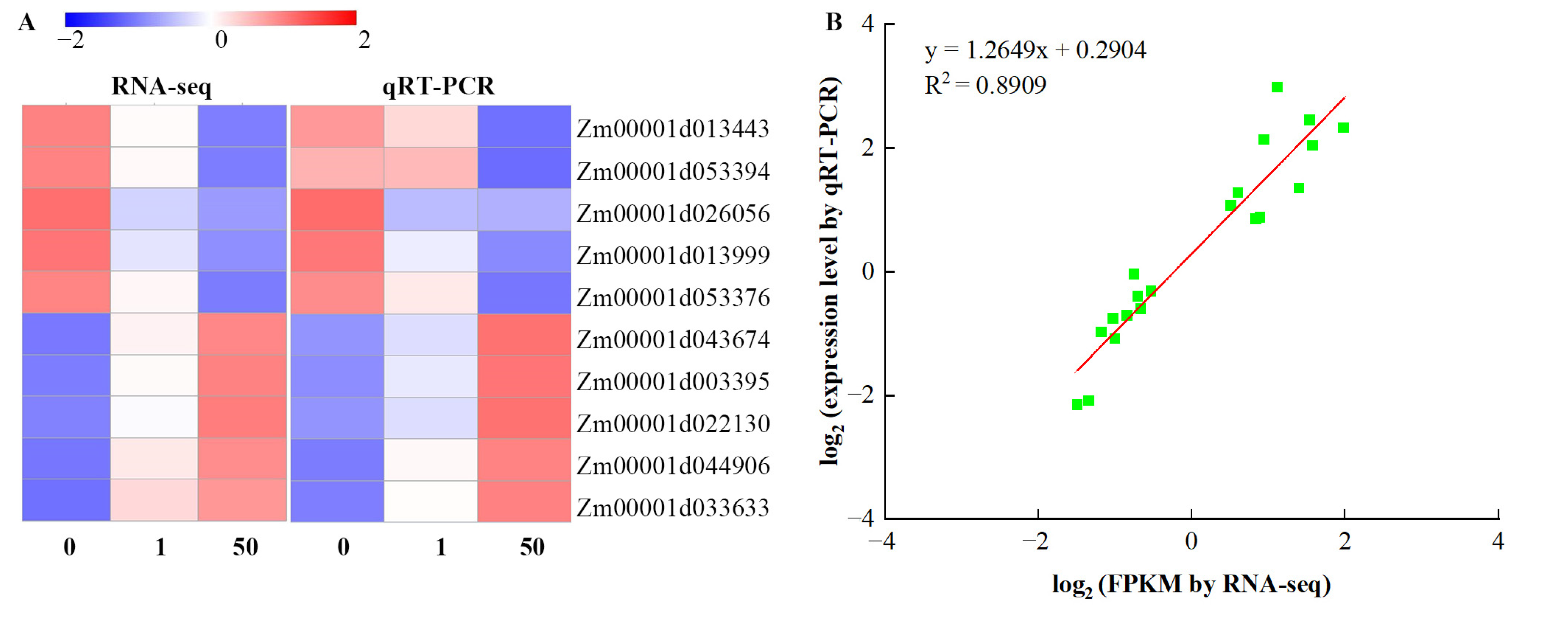
3.9. Validation of DEGs by qRT-PCR Analysis
4. Discussion
4.1. Exogenous BR Promotes the Increase of Maize Leaf Angle
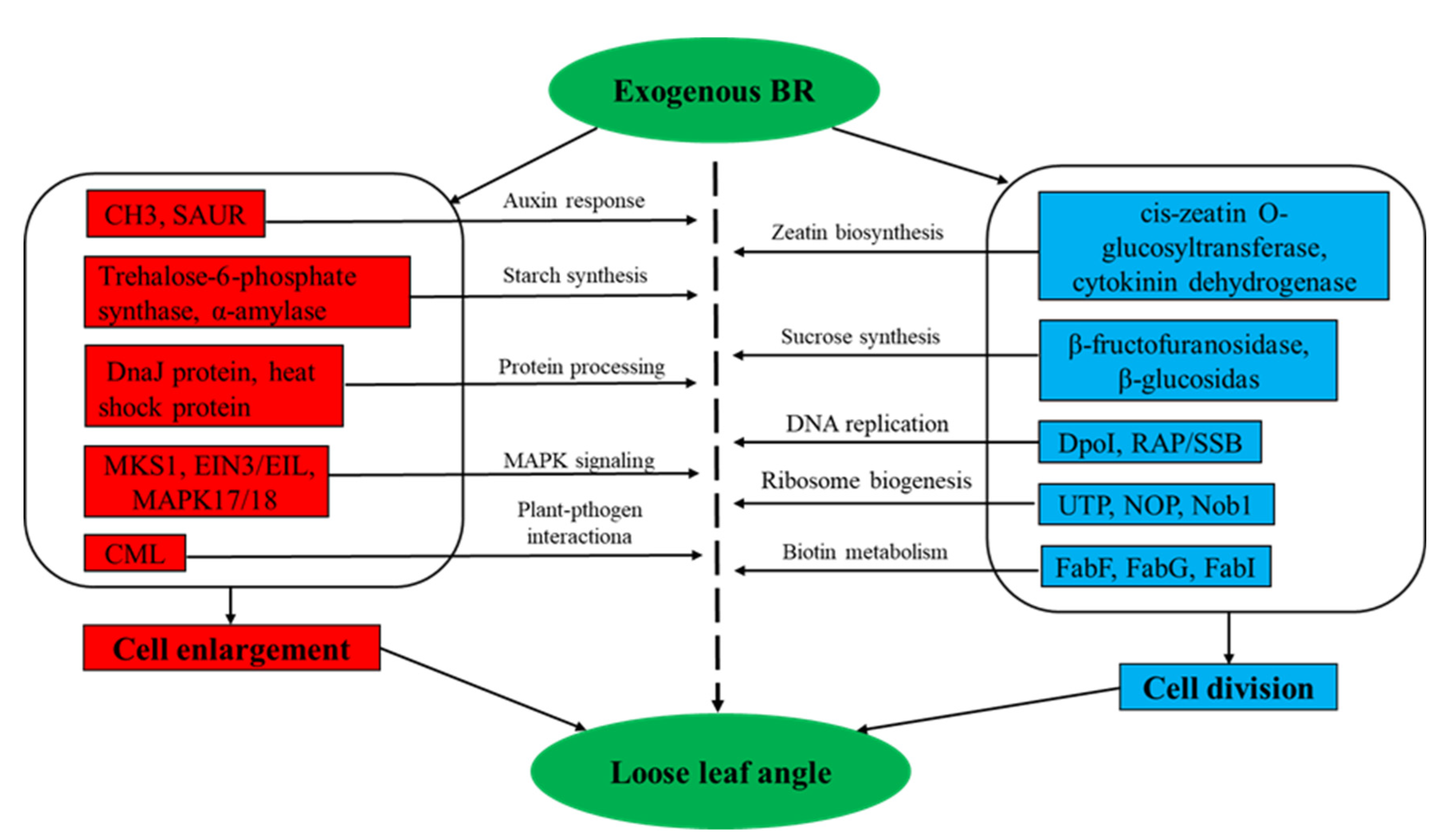
4.2. Analysis of DEGs Regulation by Exogenous BR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Flint-Garcia, S.; Rocheford, T.R.; McMullen, M.D.; Holland, J.B.; Buckler, E.S. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011, 43, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ku, L.X.; Han, Z.P.; Guo, S.L.; Liu, H.J.; Zhang, Z.Z.; Cao, L.R.; Cui, X.J.; Chen, Y.H. The zmcla4 gene in the qla4-1 qtl controls leaf angle in maize (Zea mays L.). J. Exp. Bot. 2014, 65, 5063–5076. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.W.; Costa, C.; Dwyer, L.M. Canopy structure, light interception, and photosynthesis in maize. Agron. J. 2003, 95, 1465–1474. [Google Scholar] [CrossRef]
- Borsari, B.; Vidrine, M.F. Undergraduate agriculture curricula in sustainability: An evaluation across borders. J. Sustain. Agric. 2005, 25, 93–112. [Google Scholar] [CrossRef]
- Ma, D.L.; Xie, R.Z.; Niu, X.K.; Li, S.K.; Long, H.L.; Liu, Y.E. Changes in pthe morphological traits of maize gen-otypes in china between the 1950s and 2000s. Eur. J. Agron. 2014, 58, 1–10. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect leaves caused by brassinosteroid de-ficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Wu, C.; Wang, C.; Roh, J.; Zhang, L.; Chen, J.; Zhang, S.; Zhang, H.; Yang, C.; Hu, J.; et al. Slg controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J. Exp. Bot. 2016, 67, 4241–4253. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zheng, J.; Huang, R.; Huang, Y.; Wang, H.; Jiang, L.; Fang, X. Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep. 2016, 35, 2423–2433. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Hardtke, C.S. Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 2011, 21, R365–R373. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Chu, C. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 2018, 23, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Chen, S. Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul. 1995, 16, 189–196. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Y.; Zhang, C.; Ma, Q.; Joo, S.; Kim, S.; Xu, Z.; Chong, K.; Ausubel, F.M. Oslic, a novel ccch-type zinc finger protein with transcription activation. mediates rice architecture via brassinosteroids signaling. PLoS ONE 2008, 3, e3521. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, Y.; Fujioka, S.; Asami, T.; Li, J.; Li, J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009, 21, 3781–3791. [Google Scholar] [CrossRef] [PubMed]
- Heyman, J.; Cools, T.; Vandenbussche, F.; Heyndrickx, K.S.; Van Leene, J.; Vercauteren, I.; Vanderauwera, S.; Vandepoele, K.; De Jaeger, G.; Van De Straeten, D.; et al. ERF115 controls root quiescent centercell division and stem cell replenishment. Science 2013, 342, 860–863. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Nakamura, A.; Nakajima, N. Arabidopsis aux/iaa genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant Cell Physiol. 2006, 45, 193–205. [Google Scholar]
- Vert, G.; Nemhauser, J.L.; Geldner, N.; Hong, F.; Chory, J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 2005, 21, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, G.; Liu, D.; Niu, M.; Tong, H.; Chu, C. Gsk2 stabilizes ofp3 to suppress brassinosteroid responses in rice. Plant J. 2020, 102, 1187–1201. [Google Scholar] [CrossRef]
- Tao, Y.; Zheng, J.; Xu, Z. Functional analysis of ZmDWF1, a maize homolog of the Arabidopsis brassinosteroids biosyn-thetic DWF1/DIM gene. Plant Sci. 2004, 167, 743–751. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, J.; Wang, M. Expression and functional analysis of ZmDWF4, an ortholog of Arabidopsis DWF4 from maize (Zea mays L.). Plant Cell Rep. 2007, 26, 2091–2099. [Google Scholar] [CrossRef]
- Makarevitch, I.; Thompson, A.; Muehlbauer, G.J. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 2012, 7, e30798. [Google Scholar]
- Tian, J.; Wang, C.; Xia, J.; Wu, L.; Xu, G.; Wu, W.; Li, D.; Qin, W.; Han, X.; Chen, Q.; et al. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 2019, 365, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Tang, S.; Zhang, H.; He, M.; Liu, J.; Zhi, H.; Sui, Y.; Liu, X.; Jia, G.; Zhao, Z.; et al. Droopy leaf1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 21766–21774. [Google Scholar] [CrossRef] [PubMed]
- Kir, G.; Ye, H.; Nelissen, H.; Neelakandan, A.K.; Kusnandar, A.S.; Luo, A.; Inzé, D.; Sylvester, A.W.; Yin, Y.; Becraft, P.W. Rna interference knockdown of brassinosteroid insensitive1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 2015, 169, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, G.; Jin, J.; Liang, J.; Zhang, J.; Peng, H.; Wang, W.; Zhang, Z. Rip2 interacts with rel1 to control leaf architecture by modulating brassinosteroid signaling in rice. Theor. Appl. Genet. 2022, 135, 979–991. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, Y.F.; Sun, H.S.; Zhao, C.L.; Wang, Q.; Wang, Y.S. Optimization of pararafin section for Kentucky bliegress. Acta Agrestia Sin. 2017, 25, 1376–1379. [Google Scholar]
- Chen, F.; Ji, X.; Bai, M. Network Analysis of Different Exogenous Hormones on the Regulation of Deep Sowing Tolerance in Maize Seedlings. Front. Plant Sci. 2021, 12, 739101. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Wgcna: An r package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 55926. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Peng, Y.; Zhao, X.; Wu, B.; Chen, F.; Ren, B. Comparative pro-teomics analysis of the seedling root response of drought-sensitive and drought-tolerant maize varieties to drought stress. Int. J. Mol. Sci. 2019, 20, 2793. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Vanhaeren, H.; Inze, D. Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Divi, U.K.; Krishna, P. Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 2009, 26, 131–136. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, J.; Cao, X. Epigenetic Mutation of RAV6 Affects Leaf Angle and Seed Size in Rice. Plant Physiol. 2015, 169, 2118–2128. [Google Scholar] [CrossRef]
- Hang, L.; Bai, M.; Wu, J. Antagonistic HLH/bHLH Transcription Factors Mediate Brassinosteroid Regulation of Cell Elongation and Plant Development in Rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar]
- Jiang, Y.; Bao, L.; Jeong, S. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 2012, 70, 398–408. [Google Scholar] [CrossRef]
- Tian, Q.; Luan, J.; Guo, C. A bHLH protein, OsBIM1, positively regulates rice leaf angle by promoting brassinosteroid signaling. Biochem. Biophys. Res. Commun. 2021, 578, 129–135. [Google Scholar] [CrossRef]
- Lei, Y.; Xu, Y.; Hettenhausen, C.; Lu, C.; Shen, G.; Zhang, C.; Li, J.; Song, J.; Lin, H.; Wu, J. Comparative analysis of alfalfa (Medicago sativa L.) Leaf transcriptomes reveals genotype-specific salt tolerance mechanisms. BMC Plant Biol. 2018, 18, 35. [Google Scholar] [CrossRef]
- Thiemann, A.; Fu, J.; Schrag, T.A.; Melchinger, A.E.; Frisch, M.; Scholten, S. Correlation between parental transcriptome and field data for the characterization of heterosis in Zea mays L. Theor. Appl. Genet. 2010, 120, 401–413. [Google Scholar] [CrossRef]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef]
- Sun, S.; Chen, D.; Li, X. Brassinosteroid Signaling Regulates Leaf Erectness in Oryza sativa via the Control of a Specific U-Type Cyclin and Cell Proliferation. Dev. Cell 2015, 34, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C. Cellular Responses to Auxin: Division versus Expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Bai, F.; Reinheimer, R.; Durantini, D. TCP transcription factor, Branch Angle Defective 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 12225–12230. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Gao, Q.; Chen, F. Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway. Int. J. Mol. Sci. 2022, 23, 4886. [Google Scholar] [CrossRef]
- Ning, J.; Zhang, B.; Wang, N. Increased Leaf Angle1, a Raf-Like MAPKKK That Interacts with a Nuclear Protein Family, Regulates Mechanical Tissue Formation in the Lamina Joint of Rice. Plant Cell 2011, 23, 4334–4347. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Bartrina, I.; Novák, O. The Cytokinin Status of the Epidermis Regulates Aspects of Vegetative and Reproductive Development in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 613488. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, T.; Jain, M.; Dermastia, M.; Chourey, P.S. Spatial and temporal profiles of cytokinin biosynthesis and accumulation in developing caryopses of maize. Ann. Bot. 2011, 107, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shan, C.; Song, W. Transcriptome analysis of starch and sucrose metabolism change in Gold Queen Hami melons under different storage temperatures. Postharvest. Biol. Technol. 2021, 174, 111445. [Google Scholar] [CrossRef]
- Zhang, M.; Su, J.; Yan, Z. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45 Pt A, 1–10. [Google Scholar] [CrossRef]
- Tian, X.; He, M.; Mei, E. WRKY53 integrates classic brassinosteroid signaling and the mitogen-activated protein kinase pathway to regulate rice architecture and seed size. Plant Cell 2021, 33, 2753–2775. [Google Scholar] [CrossRef] [PubMed]
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Börner, T. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010, 64, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.Q.; Keta, S.; Asai, T. A genetic link between epigenetic repressor AS1–AS2 and DNA replication factors in establishment of adaxial–abaxial leaf polarity of Arabidopsis. Plant Biotechnol. 2018, 35, 39–49. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, X.; Gao, Q.; Zhuang, Z.; Wang, Y.; Zhang, Y.; Peng, Y. Transcriptome-Based Weighted Correlation Network Analysis of Maize Leaf Angle Regulation by Exogenous Brassinosteroid. Agronomy 2022, 12, 1895. https://doi.org/10.3390/agronomy12081895
Ji X, Gao Q, Zhuang Z, Wang Y, Zhang Y, Peng Y. Transcriptome-Based Weighted Correlation Network Analysis of Maize Leaf Angle Regulation by Exogenous Brassinosteroid. Agronomy. 2022; 12(8):1895. https://doi.org/10.3390/agronomy12081895
Chicago/Turabian StyleJi, Xiangzhuo, Qiaohong Gao, Zelong Zhuang, Yinxia Wang, Yunfang Zhang, and Yunling Peng. 2022. "Transcriptome-Based Weighted Correlation Network Analysis of Maize Leaf Angle Regulation by Exogenous Brassinosteroid" Agronomy 12, no. 8: 1895. https://doi.org/10.3390/agronomy12081895
APA StyleJi, X., Gao, Q., Zhuang, Z., Wang, Y., Zhang, Y., & Peng, Y. (2022). Transcriptome-Based Weighted Correlation Network Analysis of Maize Leaf Angle Regulation by Exogenous Brassinosteroid. Agronomy, 12(8), 1895. https://doi.org/10.3390/agronomy12081895




