Nitrogen Distribution and Soil Microbial Community Characteristics in a Legume–Cereal Intercropping System: A Review
Abstract
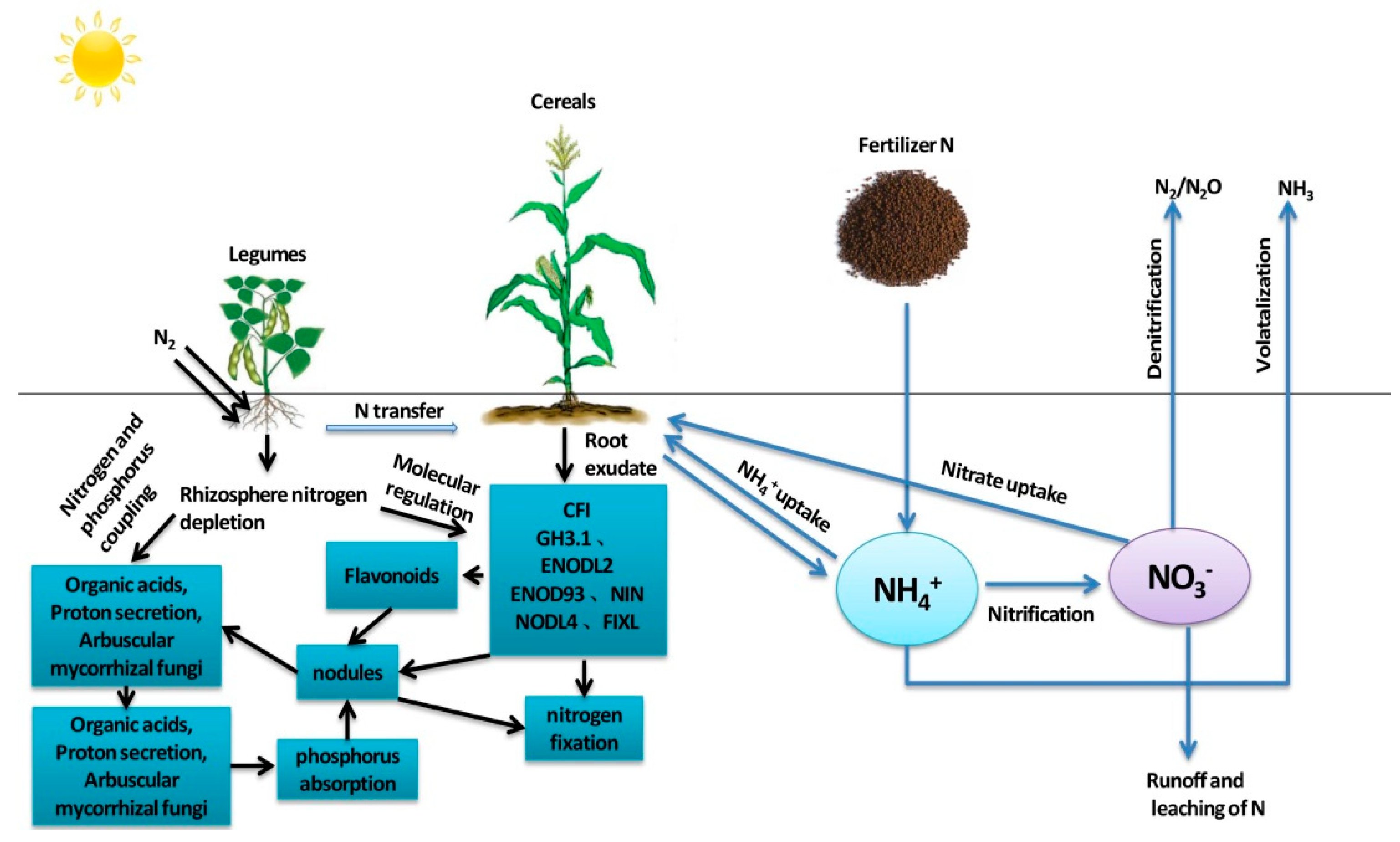
:1. Introduction
2. Transformation and Distribution of Nitrogen in the Legume–Cereal Intercropping System
2.1. Fixation of Nitrogen in Legumes
2.2. Nitrogen Uptake by Cereals in Intercropping Systems
3. Soil Microbial Community Characteristics in the Legume–Cereal Intercropping System
3.1. Soil Microbial Composition and Diversity in the Legume–Cereal Intercropping System
| Types of Intercropping Crops | Soil Type | Microorganisms with Increased Proportion | Microorganisms with Reduced Proportion | Microorganisms with No Significant Change | Reference |
|---|---|---|---|---|---|
| Soybean–maize | Brown soil | Actinomycetes, Firmicutes, Corynebacter, Cysts, Halophile bacillus, Phagocytes | Proteobacteria, Bacteroidetes, Burkholderia, Desulphurobacter, Neisseria | [58] | |
| Soybean–sugarcane | Red soil | Streptomyces, Bacillus, Pantomyces, Enterobacter, Arthrobacter, Symbiotic bacteria rhizobia, Chinese rhizobia | [59] | ||
| Peanut–sugarcane | Red soil | Streptomyces, Bacillus, Pantomyces, Enterobacter, Arthrobacter, Symbiotic bacteria rhizobia, Chinese rhizobia | [59,60] | ||
| Pea–wheat | Loamy soil | Acidobacteria, Proteobacteria, Bacteroidetes, Chlorobacteria | [45] | ||
| Soybean–sugarcane | Red soil | Proteobacteria, Acidobacteria, Chlorobacteria, Actinomycetes, Bacteroidetes | [61] | ||
| Peanut–maize | Hydragric anthrosol | Actinobacteria, Acidobacteria | Gammaproteobacteria, Bacteroidetes, Firmicutes | Alphaproteobacteria | [62] |
| Fata beans–maize | Clay loam | Firmicutes, Bacteroidetes, Bacillus, Clostridium, Sporobacteria, Desulphuria, Alicyclobacter | Methylobacter,Sphingomonas | Acidobacteria, Floating mold, Blastomonas phylum | [63] |
| Mung bean–proso millet | Loess-like | Proteobacteria, Chlorobacteria, Blastomonas, Acidobacteria, Helicobacter nitrifying, Firmicutes | Actinomycetes | Phylum fungi, Ascomycetes, Morpita, basidiomycetes | [64] |
| Wild soybean–sorghum | Salined fluvo-aquic soil | Proteobacteria, Bacteroidetes | Firmicutes, Gemmatimonadetes | [65] |
| Types of Intercropping Crops | Soil Type | Microorganisms with Increased Proportion | Microorganisms with Reduced Proportion | Microorganisms with No Significant Change | Reference |
|---|---|---|---|---|---|
| Soybean–maize | Brown soil | Actinomycetes, Firmicutes, Desulphurobacter, Erythrococcus, Kinetospora, Bacillus, Bacillus, phagocyte | Proteobacteria, Achromatobacteria, Burkholderia, Pseudomonas, Aikenella | Bacteroidetes | [58] |
| Soybean–sugarcane | Red soil | Streptomyces, Bacillus, Pantomyces, Enterobacter, Arthrobacter, Symbiotic bacteria rhizobia, Chinese rhizobia | [59] | ||
| Peanut–sugarcane | Red soil | Streptomyces, Bacillus, Pantomyces, Enterobacter, Arthrobacter, Symbiotic bacteria rhizobia, Chinese rhizobia, Acidobacteria, Chloroflexi | Proteobacteria, Actinobacteria | [59,60] | |
| Pea–wheat | Loamy Soil | Acidobacteria, Proteobacteria, Bacteroidetes, Chlorobacteria | [45] | ||
| Soybean–sugarcane | Red soil | Proteobacteria, Acidobacteria, Chlorobacteria, Actinomycetes, Bacteroidetes | [61] | ||
| Peanut–maize | Hydragric anthrosol | Deltaproteobacteria, Acidobacteria, Chloroflexi, Gemmatimonadetes | Actinobacteria, | Alphaproteobacteria | [62] |
| Fata beans–maize | Clay loam | Firmicutes, Bacteroidetes, Bacillus, Clostridium, Sporobacteria, Desulphuria, Alicyclobacter | Methylobacter,Sphingomonas | Acidobacteria, Floating mold, Blastomonas phylum | [63] |
| Mung bean–proso millet | Loess-like | Proteobacteria, Chlorobacteria, Blastomonas, Acidobacteria, Helicobacter nitrifying, Firmicutes | Actinomycetes | Phylum fungi, Ascomycetes, Morpita, basidiomycetes | [64] |
| Wild soybean–sorghum | Salined fluvo-aquic soil | Proteobacteria, Bacteroidetes | Firmicutes, Gemmatimonadetes | [65] |
3.2. Soil Microbial Activity in the Legume–Cereal Intercropping System
3.3. Mechanism of Plant–Soil–Microorganism Interactions in the Legume–Cereal Intercropping System
3.4. Application and Development of Modern Biological Detection Technology in Soil Microorganisms
4. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ladha, J.K.; Peoples, M.B.; Reddy, P.M.; Biswas, J.C.; Bennett, A.; Jat, M.L.; Krupnik, T.J. Biological nitrogen fixation and prospects for ecological intensification in cereal-based cropping systems. Field Crops Res. 2022, 283, 108541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.S.; Wang, J.Q.; Zhang, W.F.; Cui, Z.L.; Ma, W.Q.; Chen, X.P.; Jiang, R.F. Nutrient use efficiencies of major cereal crops in China and measures for improvement. Acta Pedol. Sin. 2008, 45, 915–924. [Google Scholar]
- Zhang, F.S.; Li, L. Using competitive and facilitative interactions in intercropping systems enhances crop productivity and nutrient-use efficiency. Plant Soil 2003, 248, 1–2. [Google Scholar] [CrossRef]
- Li, S.J.; Jensen, E.S.; Liu, N.; Zhang, Y.J.; Martensson, L.M.D. Species Interactions and Nitrogen Use during Early Intercropping of Intermediate Wheatgrass with a White Clover Service Crop. Agronomy 2021, 11, 388. [Google Scholar] [CrossRef]
- Hauggaard-Nielsen, H.; Gooding, M.; Ambus, P.; Corre-Hellou, G.; Crozat, Y.; Dahlmann, C.; Dibet, A.; von Fragstein, P.; Pristeri, A.; Monti, M.; et al. Pea-barley intercropping for efficient symbiotic N-2-fixation, soil N acquisition and use of other nutrients in European organic cropping systems. Field Crops Res. 2009, 113, 64–71. [Google Scholar] [CrossRef]
- Thilakarathna, M.S.; McElroy, M.S.; Chapagain, T.; Papadopoulos, Y.A.; Raizada, M.N. Belowground nitrogen transfer from legumes to non-legumes under managed herbaceous cropping systems. A review. Agron. Sustain. Dev. 2016, 36, 58. [Google Scholar] [CrossRef]
- Reilly, E.C.; Gutknecht, J.L.; Tautges, N.E.; Sheaffer, C.C.; Jungers, J.M. Nitrogen transfer and yield effects of legumes intercropped with the perennial grain crop intermediate wheatgrass. Field Crops Res. 2022, 286, 108627. [Google Scholar] [CrossRef]
- Ashworth, A.J.; West, C.P.; Allen, F.L.; Keyser, P.D.; Weiss, S.A.; Tyler, D.D.; Taylor, A.M.; Warwick, K.L.; Beamer, K.P. Biologically Fixed Nitrogen in Legume Intercropped Systems: Comparison of Nitrogen-Difference and Nitrogen-15 Enrichment Techniques. Agron. J. 2015, 107, 2419–2430. [Google Scholar] [CrossRef]
- Zhou, M.H.; Butterbach-Bahl, K. Assessment of nitrate leaching loss on a yield-scaled basis from maize and wheat cropping systems. Plant Soil 2014, 374, 977–991. [Google Scholar] [CrossRef]
- Mus, F.; Crook, M.B.; Garcia, K.; Costas, A.G.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.H.; Oldroyd, G.E.D.; Poole, P.S.; et al. Symbiotic Nitrogen Fixation and the Challenges to Its Extension to Nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef]
- Glaze-Corcoran, S.; Hashemi, M.; Sadeghpour, A.; Jahanzad, E.; Afshar, R.K.; Liu, X.B.; Herbert, S.J. Understanding intercropping to improve agricultural resiliency and environmental sustainability. Adv. Agron. 2020, 162, 199–256. [Google Scholar]
- Li, C.J.; Hoffland, E.; Kuyper, T.W.; Yu, Y.; Zhang, C.C.; Li, H.G.; Zhang, F.S.; van der Werf, W. Syndromes of production in intercropping impact yield gains. Nat. Plants 2020, 6, 653–660. [Google Scholar] [CrossRef]
- Li, X.F.; Wang, Z.G.; Bao, X.G.; Sun, J.H.; Yang, S.C.; Wang, P.; Wang, C.B.; Wu, J.P.; Liu, X.R.; Tian, X.L.; et al. Long-term increased grain yield and soil fertility from intercropping. Nat. Sustain. 2021, 4, 943–950. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef]
- Udvardi, M.; Poole, P.S. Transport and Metabolism in Legume-Rhizobia Symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [PubMed]
- Schipanski, M.E.; Drinkwater, L.E.; Russelle, M.P. Understanding the variability in soybean nitrogen fixation across agroecosystems. Plant Soil 2010, 329, 379–397. [Google Scholar] [CrossRef]
- Li, L.; Yang, S.C.; Li, X.L.; Zhang, F.S.; Christie, P. Interspecific complementary and competitive interactions between intercropped maize and faba bean. Plant Soil 1999, 212, 105–114. [Google Scholar] [CrossRef]
- Wang, X.; Gao, Y.; Zhang, H.; Shao, Z.; Sun, B.; Gao, Q. Enhancement of rhizosphere citric acid and decrease of NO3−/NH4+ ratio by root interactions facilitate N fixation and transfer. Plant Soil 2019, 447, 169–182. [Google Scholar] [CrossRef]
- Fan, F.L.; Zhang, F.S.; Song, Y.N.; Sun, J.H.; Bao, X.G.; Guo, T.W.; Li, L. Nitrogen fixation of faba bean (Vicia faba L.) interacting with a non-legume in two contrasting intercropping systems. Plant Soil 2006, 283, 275–286. [Google Scholar] [CrossRef]
- Nyfeler, D.; Huguenin-Elie, O.; Suter, M.; Frossard, E.; Lüscher, A. Grass-legume mixtures can yield more nitrogen than legume pure stands due to mutual stimulation of nitrogen uptake from symbiotic and non-symbiotic sources. Agric. Ecosyst. Environ. 2011, 140, 155–163. [Google Scholar] [CrossRef]
- Schipanski, M.E.; Drinkwater, L.E. Nitrogen fixation in annual and perennial legume-grass mixtures across a fertility gradient. Plant Soil 2012, 357, 147–159. [Google Scholar] [CrossRef]
- Tian, J.H.; Tang, M.T.; Xu, X.; Luo, S.S.; Condron, L.M.; Lambers, H.; Cai, K.Z.; Wang, J.W. Soybean (Glycine max (L.) Merrill) intercropping with reduced nitrogen input influences rhizosphere phosphorus dynamics and phosphorus acquisition of sugarcane (Saccharum officinarum). Biol. Fertil. Soils 2020, 56, 1063–1075. [Google Scholar] [CrossRef]
- Fan, Z.; Zhao, Y.; Chai, Q.; Zhao, C.; Yu, A.; Coulter, J.A.; Gan, Y.; Cao, W. Synchrony of nitrogen supply and crop demand are driven via high maize density in maize/pea strip intercropping. Sci. Rep. 2019, 9, 10954. [Google Scholar] [CrossRef] [PubMed]
- Hei, Z.W.; Xiang, H.M.; Zhang, J.E.; Liang, K.M.; Zhong, J.W.; Li, M.J.; Lu, Y.Q. Rice intercropping with water mimosa (Neptunia oleracea Lour.) can facilitate soil N utilization and alleviate apparent N loss. Agric. Ecosyst. Environ. 2021, 313, 107378. [Google Scholar] [CrossRef]
- Wang, X.Y.; Gao, Y.Z. Advances in the mechanism of cereal/legume intercropping promotion of symbiotic nitrogen fixation. Chin. Sci. Bull. 2019, 65, 142–149. [Google Scholar] [CrossRef]
- Francisquini, A.; Calonego, J.C.; Rosolem, C.A.; dos Santos, C.H.; Tiritan, C.S. Increase of nitrogen-use efficiency by phosphorus fertilization in grass-legume pastures. Nutr. Cycl. Agroecosystems 2020, 118, 165–175. [Google Scholar] [CrossRef]
- Li, L.; Li, S.M.; Sun, J.H.; Zhou, L.L.; Bao, X.G.; Zhang, H.G.; Zhang, F.S. Diversity enhances agricultural productivity via rhizosphere phosphorus facilitation on phosphorus-deficient soils. Proc. Natl. Acad. Sci. USA 2007, 104, 11192–11196. [Google Scholar] [CrossRef] [PubMed]
- Salgado, G.C.; Ambrosano, E.J.; Rossi, F.; Otsuk, I.P.; Ambrosano, G.M.B.; Santana, C.A.; Muraoka, T.; Trivelin, P.C.O. Biological N Fixation and N Transfer in an Intercropping System between Legumes and Organic Cherry Tomatoes in Succession to Green Corn. Agriculture 2021, 11, 690. [Google Scholar] [CrossRef]
- Boddey, R.M.; Sa, J.C.D.; Alves, B.J.R.; Urquiaga, S. The contribution of biological nitrogen fixation for sustainable agricultural systems in the tropics. Soil Biol. Biochem. 1997, 29, 787–799. [Google Scholar] [CrossRef]
- Tripathi, S.C.; Venkatesh, K.; Meena, R.P.; Chander, S.; Singh, G.P. Sustainable intensification of maize and wheat cropping system through pulse intercropping. Sci. Rep. 2021, 11, 18805. [Google Scholar] [CrossRef]
- Willey, R. Intercropping Its Importance And Research Needs Part 1. Competition And Yield Advantages. Field Crop Abstr. 1979, 32, 1–10. [Google Scholar]
- Wahbi, S.; Maghraoui, T.; Hafidi, M.; Sanguin, H.; Oufdou, K.; Prin, Y.; Duponnois, R.; Galiana, A. Enhanced transfer of biologically fixed N from faba bean to intercropped wheat through mycorrhizal symbiosis. Appl. Soil Ecol. 2016, 107, 91–98. [Google Scholar] [CrossRef]
- Xiao, Y.B.; Li, L.; Zhang, F.S. Effect of root contact on interspecific competition and N transfer between wheat and fababean using direct and indirect N-15 techniques. Plant Soil 2004, 262, 45–54. [Google Scholar] [CrossRef]
- Yong, T.W.; Liu, X.M.; Yang, F.; Song, C.; Wang, X.C.; Liu, W.G.; Su, B.Y.; Zhou, L.; Yang, W.Y. Characteristics of Nitrogen Uptake, Use and Transfer in a Wheat-Maize-Soybean Relay Intercropping System. Plant Prod. Sci. 2015, 18, 388–397. [Google Scholar] [CrossRef]
- Ledgard, S.F.; Freney, J.R.; Simpson, J.R. Assessing nitrogen transfer from legumes to associated grasses. Soil Biol. Biochem. 1985, 17, 575–577. [Google Scholar] [CrossRef]
- Shao, Z.; Wang, X.; Gao, Q.; Zhang, H.; Gao, Y. Root Contact between Maize and Alfalfa Facilitates Nitrogen Transfer and Uptake Using Techniques of Foliar 15N-Labeling. Agronomy 2020, 10, 360. [Google Scholar] [CrossRef]
- Yu, L.; Tang, Y.; Wang, Z.; Gou, Y.; Wang, J. Nitrogen-cycling genes and rhizosphere microbial community with reduced nitrogen application in maize/soybean strip intercropping. Nutr. Cycl. Agroecosyst. 2018, 113, 35–49. [Google Scholar] [CrossRef]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013, 7, 2248–2258. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gomez-Cadenas, A.; Perez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Hu, L.F.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.F.; Shen, Q.R.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Zhang, R.Z.; Mu, Y.; Li, X.R.; Li, S.M.; Sang, P.; Wang, X.R.; Wu, H.L.; Xu, N. Response of the arbuscular mycorrhizal fungi diversity and community in maize and soybean rhizosphere soil and roots to intercropping systems with different nitrogen application rates. Sci. Total Environ. 2020, 740, 139810. [Google Scholar] [CrossRef] [PubMed]
- Bainard, L.D.; Koch, A.M.; Gordon, A.M.; Klironomos, J.N. Growth response of crops to soil microbial communities from conventional monocropping and tree-based intercropping systems. Plant Soil 2013, 363, 345–356. [Google Scholar] [CrossRef]
- Pivato, B.; Semblat, A.; Guegan, T.; Jacquiod, S.; Martin, J.; Deau, F.; Moutier, N.; Lecomte, C.; Burstin, J.; Lemanceau, P. Rhizosphere Bacterial Networks, but Not Diversity, Are Impacted by Pea-Wheat Intercropping. Front. Microbiol. 2021, 12, 674556. [Google Scholar] [CrossRef] [PubMed]
- Morrien, E.; Hannula, S.E.; Snoek, L.B.; Helmsing, N.R.; Zweers, H.; de Hollander, M.; Soto, R.L.; Bouffaud, M.L.; Buee, M.; Dimmers, W.; et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 2017, 8, 14349. [Google Scholar] [CrossRef]
- Li, S.; Wu, F.Z. Diversity and Co-occurrence Patterns of Soil Bacterial and Fungal Communities in Seven Intercropping Systems. Front. Microbiol. 2018, 9, 1521. [Google Scholar] [CrossRef]
- Hai, B.; Diallo, N.H.; Sall, S.; Haesler, F.; Schauss, K.; Bonzi, M.; Assigbetse, K.; Chotte, J.L.; Munch, J.C.; Schloter, M. Quantification of Key Genes Steering the Microbial Nitrogen Cycle in the Rhizosphere of Sorghum Cultivars in Tropical Agroecosystems. Appl. Environ. Microbiol. 2009, 75, 4993–5000. [Google Scholar] [CrossRef]
- Rodriguez, C.; Carlsson, G.; Englund, J.E.; Flohr, A.; Pelzer, E.; Jeuffroy, M.H.; Makowski, D.; Jensen, E.S. Grain legume-cereal intercropping enhances the use of soil-derived and biologically fixed nitrogen in temperate agroecosystems. A meta-analysis. Eur. J. Agron. 2020, 118, 126077. [Google Scholar] [CrossRef]
- Zeng, H.L.; Yu, L.L.; Liu, P.; Wang, Z.G.; Chen, Y.; Wang, J.W. Nitrogen fertilization has a stronger influence than cropping pattern on AMF community in maize/soybean strip intercropping systems. Appl. Soil Ecol. 2021, 167, 104034. [Google Scholar] [CrossRef]
- Zhao, R.T.; Li, X.; Bei, S.K.; Li, D.D.; Li, H.G.; Christie, P.; Bender, S.F.; Zhang, J.L. Enrichment of nosZ-type denitrifiers by arbuscular mycorrhizal fungi mitigates N2O emissions from soybean stubbles. Environ. Microbiol. 2021, 23, 6587–6602. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.Q.; Fallah, N.; Weng, P.Y.; Zhou, Y.M.; Tang, X.M.; Tayyab, M.; Liu, Y.M.; Liu, Q.; Xiao, Y.J.; Hu, C.H.; et al. Sugarcane-Peanut Intercropping System Enhances Bacteria Abundance, Diversity, and Sugarcane Parameters in Rhizospheric and Bulk Soils. Front. Microbiol. 2022, 12, 815129. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.Y.; Huang, L.K.; Liu, Q.Z.; Xu, S.N.; Wen, Z.Y.; Qin, S.; Li, T.Q.; Feng, Y. Positive effects of applying endophytic bacteria in eggplant-Sedum intercropping system on Cd phytoremediation and vegetable production in cadmium polluted greenhouse. J. Environ. Sci. 2022, 115, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Mu, Y.H.; Cheng, Y.B.; Liu, X.G.; Nian, H. Effects of intercropping sugarcane and soybean on growth, rhizosphere soil microbes, nitrogen and phosphorus availability. Acta Physiol. Plant. 2013, 35, 1113–1119. [Google Scholar] [CrossRef]
- Zhang, R.Z. Study on Productivity, Nutrient Uptake and Mechanism of Soil Microbial Activity in Maize/Soybean Intercropping by Nitrogen Fertilizer. Ph.D. Thesis, Northeast Agricultural University, Harbin, China, 2020. [Google Scholar]
- Chung, H.G.; Zak, D.R.; Reich, P.B.; Ellsworth, D.S. Plant species richness, elevated CO2, and atmospheric nitrogen deposition alter soil microbial community composition and function. Glob. Change Biol. 2007, 13, 980–989. [Google Scholar] [CrossRef]
- Duchene, O.; Vian, J.-F.; Celette, F. Intercropping with legume for agroecological cropping systems: Complementarity and facilitation processes and the importance of soil microorganisms. A review. Agric. Ecosyst. Environ. 2017, 240, 148–161. [Google Scholar] [CrossRef]
- Lin, W.W.; Li, N.; Chen, L.S.; Wu, Z.Y.; Lin, W.X.; Shen, L.H. Effects of maize and sobean interspecific interactions on rhizospheric bacteria community structure and diversity. Chin. J. Eco Agric. 2022, 30, 26–37. [Google Scholar]
- Solanki, M.K.; Wang, F.Y.; Wang, Z.; Li, C.N.; Lan, T.J.; Singh, R.K.; Singh, P.; Yang, L.T.; Li, Y.R. Rhizospheric and endospheric diazotrophs mediated soil fertility intensification in sugarcane-legume intercropping systems. J. Soils Sediments 2019, 19, 1911–1927. [Google Scholar] [CrossRef]
- Tang, X.M.; Jiang, J.; Huang, Z.P.; Wu, H.N.; Wang, J.; He, L.Q.; Xiong, F.Q.; Zhong, R.C.; Liu, J.; Han, Z.Q.; et al. Sugarcane/peanut intercropping system improves the soil quality and increases the abundance of beneficial microbes. J. Basic Microbiol. 2021, 61, 165–176. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, H.; Liu, Q.; Zhao, X.; Chen, B. Effect of Two Different Sugarcane Cultivars on Rhizosphere Bacterial Communities of Sugarcane and Soybean Upon Intercropping. Front. Microbiol. 2021, 11, 3404. [Google Scholar] [CrossRef]
- Chen, P.; He, W.; Shen, Y.; Zhu, L.Y.; Yao, X.Z.; Sun, R.B.; Dai, C.C.; Sun, B.; Chen, Y. Interspecific Neighbor Stimulates Peanut Growth Through Modulating Root Endophytic Microbial Community Construction. Front. Plant Sci. 2022, 13, 830666. [Google Scholar] [CrossRef]
- Liao, H.; Li, Y.Y.; Yao, H.Y. Biochar Amendment Stimulates Utilization of Plant-Derived Carbon by Soil Bacteria in an Intercropping System. Front. Microbiol. 2019, 10, 1361. [Google Scholar] [CrossRef] [PubMed]
- Xga, B.; Cl, C.; Jing, L.; Yan, L.; Qya, B.; Wza, B.; Pu, Y.; Bfa, B. Responses of rhizosphere soil properties, enzyme activities and microbial diversity to intercropping patterns on the Loess Plateau of China—ScienceDirect. Soil Tillage Res. 2018, 195, 104355. [Google Scholar]
- Zhu, Y.H.; Song, X.L.; Wang, X.F.; Chen, W.F.; Niu, X.C. The yield increase and land improvement effects of different sorghum/wild soybean intercropping patterns on reclaimed coastal salt pans. J. Soils Sediments 2022, 22, 731–744. [Google Scholar] [CrossRef]
- Ma, X.J.; Li, Y.F. Soil Microbial Biomass and Enzyme Activities during Revegetation Process in the Southeastern Fringe of the Tengger Desert. J. Desert Res. 2019, 39, 159–166. [Google Scholar]
- Maynur, Y.K.M.; Zhang, B.C.; Mamtimin, S.L.Y.M. Seaonal Variations of Microbial Biomass and Soil Enzyme Activity in Biological Soil Crusts in the Gurbantunggut Desert. J. Desert Res. 2013, 33, 1091–1097. [Google Scholar]
- Wu, X.S.; Zhou, X.L.; Cao, F.M.; Zhu, B.C.; Zhao, T.K.; Shen, D.L. Effects of different fertilization on the corn yield and soil enzyme activity in corn growth period. Soil Fert. Sci. China 2015, 1, 44–49. [Google Scholar]
- Silva, L.S.; Laroca, J.V.D.; Coelho, A.P.; Gonsalves, E.C.; Gomes, R.P.; Pacheco, L.P.; Carvalho, P.C.D.; Pires, G.C.; Oliveira, R.L.; de Souza, J.M.A.; et al. Does grass-legume intercropping change soil quality and grain yield in integrated crop-livestock systems? Appl. Soil Ecol. 2022, 170, 104257. [Google Scholar] [CrossRef]
- Kumar, A.; Blagodaskaya, E.; Dippold, M.A.; Temperton, V.M. Positive intercropping effects on biomass production are species-specific and involve rhizosphere enzyme activities: Evidence from a field study. Soil Ecol. Lett. 2021, 1–10. [Google Scholar] [CrossRef]
- Dai, C.C.; Chen, Y.; Wang, X.X.; Li, P.D. Effects of intercropping of peanut with the medicinal plant Atractylodes lancea on soil microecology and peanut yield in subtropical China. Agrofor. Syst. 2013, 87, 417–426. [Google Scholar] [CrossRef]
- Yao, H.Y.; Jiao, X.D.; Wu, F.Z. Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 2006, 284, 195–203. [Google Scholar] [CrossRef]
- Jing, J.; Cong, W.-F.; Bezemer, T.M. Legacies at work: Plant-soil-microbiome interactions underpinning agricultural sustainability. Trends Plant Sci. 2022, 27, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Oram, N.J.; Ravenek, J.M.; Barry, K.E.; Weigelt, A.; Chen, H.M.; Gessler, A.; Gockele, A.; de Kroon, H.; van der Paauw, J.W.; Scherer-Lorenzen, M.; et al. Below-ground complementarity effects in a grassland biodiversity experiment are related to deep-rooting species. J. Ecol. 2018, 106, 265–277. [Google Scholar] [CrossRef]
- Yang, H.; Xu, H.S.; Zhang, W.P.; Li, Z.X.; Fan, H.X.; Lambers, H.; Li, L. Overyielding is accounted for partly by plasticity and dissimilarity of crop root traits in maize/legume intercropping systems. Funct. Ecol. 2022. [Google Scholar] [CrossRef]
- Vora, S.M.; Joshi, P.; Belwalkar, M.; Archana, G. Root exudates influence chemotaxis and colonization of diverse plant growth promoting rhizobacteria in the pigeon pea-maize intercropping system. Rhizosphere 2021, 18, 100331. [Google Scholar] [CrossRef]
- Liu, Y.C.; Qin, X.M.; Xiao, J.X.; Tang, L.; Wei, C.Z.; Wei, J.J.; Zheng, Y. Intercropping influences component and content change of flavonoids in root exudates and nodulation of Faba bean. J. Plant Interact. 2017, 12, 187–192. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.Y.; Wu, H.M.; Zhang, F.F.; Li, C.J.; Li, X.X.; Lambers, H.; Li, L. Root exudates drive interspecific facilitation by enhancing nodulation and N-2 fixation. Proc. Natl. Acad. Sci. USA 2016, 113, 6496–6501. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Lateif, K.; Bogusz, D.; Hocher, V. The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal. Behav. 2012, 7, 636–641. [Google Scholar] [CrossRef]
- Concha, C.; Doerner, P. The impact of the rhizobia-legume symbiosis on host root system architecture. J. Exp. Bot. 2020, 71, 3902–3921. [Google Scholar] [CrossRef]
- Kumar, A.; Patel, J.S.; Meena, V.S.; Ramteke, P.W. Plant growth-promoting rhizobacteria: Strategies to improve abiotic stresses under sustainable agriculture. J. Plant Nutr. 2019, 42, 1402–1415. [Google Scholar] [CrossRef]
- Chamkhi, I.; Cheto, S.; Geistlinger, J.; Zeroual, Y.; Kouisni, L.; Bargaz, A.; Ghoulam, C. Legume-based intercropping systems promote beneficial rhizobacterial community and crop yield under stressing conditions. Ind. Crops Prod. 2022, 183, 114958. [Google Scholar] [CrossRef]
- Shukla, P.S.; Agarwal, P.K.; Jha, B. Improved Salinity Tolerance of Arachis hypogaea (L.) by the Interaction of Halotolerant Plant-Growth-Promoting Rhizobacteria. J. Plant Growth Regul. 2012, 31, 195–206. [Google Scholar] [CrossRef]
- Nautiyal, C.S.; Srivastava, S.; Chauhan, P.S.; Seem, K.; Mishra, A.; Sopory, S.K. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Bioch. 2013, 66, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.M.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Zhang, R.F.; Vivanco, J.M.; Shen, Q.R. The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 2017, 37, 8–14. [Google Scholar] [CrossRef]
- Volkov, I.; Banavar, J.R.; Maritan, A. Comment on “Computational improvements reveal great bacterial diversity and high metal toxicity in soil”. Science 2006, 313, 918. [Google Scholar] [CrossRef]
- Whiteley, A.S.; Manefield, M.; Lueders, T. Unlocking the ’microbial black box’ using RNA-based stable isotope probing technologies. Curr. Opin. Biotechnol. 2006, 17, 67–71. [Google Scholar] [CrossRef]
- Li, Z.; Yao, Q.M.; Guo, X.; Crits-Christoph, A.; Mayes, M.A.; Hervey, W.J.; Lebeis, S.L.; Banfield, J.F.; Hurst, G.B.; Hettich, R.L.; et al. Genome-Resolved Proteomic Stable Isotope Probing of Soil Microbial Communities Using (CO2)-C-13 and C-13-Methanol. Front. Microbiol. 2019, 10, 2706. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, W.Q.; He, H.L.; Wang, Z.T.; Cao, Y.H. Effects of Sugarcane and Soybean Intercropping on the Nitrogen-Fixing Bacterial Community in the Rhizosphere. Front. Microbiol. 2021, 12, 2846. [Google Scholar] [CrossRef]
- Sertse, D.; You, F.M.; Ravichandran, S.; Cloutier, S. The Complex Genetic Architecture of Early Root and Shoot Traits in Flax Revealed by Genome-Wide Association Analyses. Front. Plant Sci. 2019, 10, 1483. [Google Scholar] [CrossRef]
- Chiewattanakul, M.; McAleer, A.D.A.; Reay, M.K.; Griffiths, R.I.; Buss, H.L.; Evershed, R.P. Compound-specific amino acid 15N-stable isotope probing for the quantification of biological nitrogen fixation in soils. Soil Biol. Biochem. 2022, 169, 108654. [Google Scholar] [CrossRef]
- Ouyang, W.Y.; Su, J.Q.; Richnow, H.H.; Adrian, L. Identification of dominant sulfamethoxazole-degraders in pig farm-impacted soil by DNA and protein stable isotope probing. Environ. Int. 2019, 126, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Koechli, C.; Campbell, A.N.; Pepe-Ranney, C.; Buckley, D.H. Assessing fungal contributions to cellulose degradation in soil by using high-throughput stable isotope probing. Soil Biol. Biochem. 2019, 130, 150–158. [Google Scholar] [CrossRef]
- Meier-Augenstein, W. From stable isotope ecology to forensic isotope ecology—Isotopes’ tales. Forensic Sci. Int. 2019, 300, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.W.; Zhao, J.; Zheng, Y.; Zhang, H.M.; Zhang, J.B.; Chen, R.R.; Lin, X.G.; Jia, Z.J. Active Soil Nitrifying Communities Revealed by In Situ Transcriptomics and Microcosm-Based Stable-Isotope Probing. Appl. Environ. Microbiol. 2020, 86, e01807-20. [Google Scholar] [CrossRef]
- Luo, D.; Meng, X.T.; Zheng, N.G.; Li, Y.Y.; Yao, H.Y.; Chapman, S.J. The anaerobic oxidation of methane in paddy soil by ferric iron and nitrate, and the microbial communities involved. Sci. Total Environ. 2021, 788, 147773. [Google Scholar] [CrossRef]
- Wang, J.; Chapman, S.J.; Ye, Q.F.; Yao, H.Y. Limited effect of planting transgenic rice on the soil microbiome studied by continuous (CO2)-C-13 labeling combined with high-throughput sequencing. Appl. Microbiol. Biotechnol. 2019, 103, 4217–4227. [Google Scholar] [CrossRef]
- Wang, F.; Shi, N.; Jiang, R.F.; Zhang, F.S.; Feng, G. In situ stable isotope probing of phosphate-solubilizing bacteria in the hyphosphere. J. Exp. Bot. 2016, 67, 1689–1701. [Google Scholar] [CrossRef]
- Gou, Y.G.; Yu, L.L.; Xu, X.; Wang, J.W. Identification of 15N-DNA enrichment sites in DNA-SIP to reveal functional genes by qPCR from sugarcane-soybean intercropping soil. J. Agro Environ. Sci. 2019, 38, 140–147. [Google Scholar]
- Wang, J.; Zhang, X.; Yao, H.Y. Optimizing ultracentrifugation conditions for DNA-based stable isotope probing (DNA-SIP). J. Microbiol. Methods 2020, 173, 105938. [Google Scholar] [CrossRef]
- Ding, T.T.; Yan, Z.C.; Zhang, W.Z.; Duan, T.Y. Green Manure Crops Affected Soil Chemical Properties and Fungal Diversity and Community of Apple Orchard in the Loess Plateau of China. J. Soil Sci. Plant Nutr. 2021, 21, 1089–1102. [Google Scholar] [CrossRef]
- Li, H.; Luo, L.Y.; Tang, B.; Guo, H.L.; Cao, Z.Y.; Zeng, Q.; Chen, S.L.; Chen, Z.H. Dynamic changes of rhizosphere soil bacterial community and nutrients in cadmium polluted soils with soybean-corn intercropping. BMC Microbiol. 2022, 22, 57. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.K.; Wang, Z.; Wang, F.Y.; Li, C.N.; Gupta, C.L.; Singh, R.K.; Malviya, M.K.; Singh, P.; Yang, L.T.; Li, Y.R. Assessment of DiazotrophicProteobacteriain Sugarcane Rhizosphere When Intercropped With Legumes (Peanut and Soybean) in the Field. Front. Microbiol. 2020, 11, 1814. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Naorem, A.K.; Lal, R.; Dalal, R.C.; Sinha, N.K.; Patra, A.K.; Chaudhari, S.K. Disease-Suppressive Soils-Beyond Food Production: A Critical Review. J. Soil Sci. Plant Nutr. 2021, 21, 1437–1465. [Google Scholar] [CrossRef] [PubMed]
- Alawiye, T.; Babalola, O. Metagenomic Insight into the Community Structure and Functional Genes in the Sunflower Rhizosphere Microbiome. Agriculture 2021, 11, 167. [Google Scholar] [CrossRef]
- Iquebal, M.A.; Jagannadham, J.; Jaiswal, S.; Prabha, R.; Rai, A.; Kumar, D. Potential Use of Microbial Community Genomes in Various Dimensions of Agriculture Productivity and Its Management: A Review. Front. Microbiol. 2022, 13, 708335. [Google Scholar] [CrossRef]
- Kaushal, M.; Tumuhairwe, J.B.; Kaingo, J.; Richard, M.; Nakamanya, F.; Taulya, G.; Coyne, D. Compositional Shifts in Microbial Diversity under Traditional Banana Cropping Systems of Sub-Saharan Africa. Biology 2022, 11, 756. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Zeng, Y.; Tang, L.; Xiao, J.X.; Zeng, J.; Zhang, K.X. Rhizosphere Biological Processes of Legume//Cereal Intercropping Systems: A Review. J. Agric. Resour. Environ. 2016, 33, 407–415. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, H.; Gao, F.; Su, H.; Zheng, P.; Li, Y.; Yao, H. Nitrogen Distribution and Soil Microbial Community Characteristics in a Legume–Cereal Intercropping System: A Review. Agronomy 2022, 12, 1900. https://doi.org/10.3390/agronomy12081900
Lai H, Gao F, Su H, Zheng P, Li Y, Yao H. Nitrogen Distribution and Soil Microbial Community Characteristics in a Legume–Cereal Intercropping System: A Review. Agronomy. 2022; 12(8):1900. https://doi.org/10.3390/agronomy12081900
Chicago/Turabian StyleLai, Huiling, Fuyun Gao, Hao Su, Peng Zheng, Yaying Li, and Huaiying Yao. 2022. "Nitrogen Distribution and Soil Microbial Community Characteristics in a Legume–Cereal Intercropping System: A Review" Agronomy 12, no. 8: 1900. https://doi.org/10.3390/agronomy12081900
APA StyleLai, H., Gao, F., Su, H., Zheng, P., Li, Y., & Yao, H. (2022). Nitrogen Distribution and Soil Microbial Community Characteristics in a Legume–Cereal Intercropping System: A Review. Agronomy, 12(8), 1900. https://doi.org/10.3390/agronomy12081900






