Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Isolation and PCR Amplification
2.3. Marker Analysis
2.4. Bioassay against BB Resistance
2.5. Characterization for Morphologic, Quality and Yield Traits
3. Results
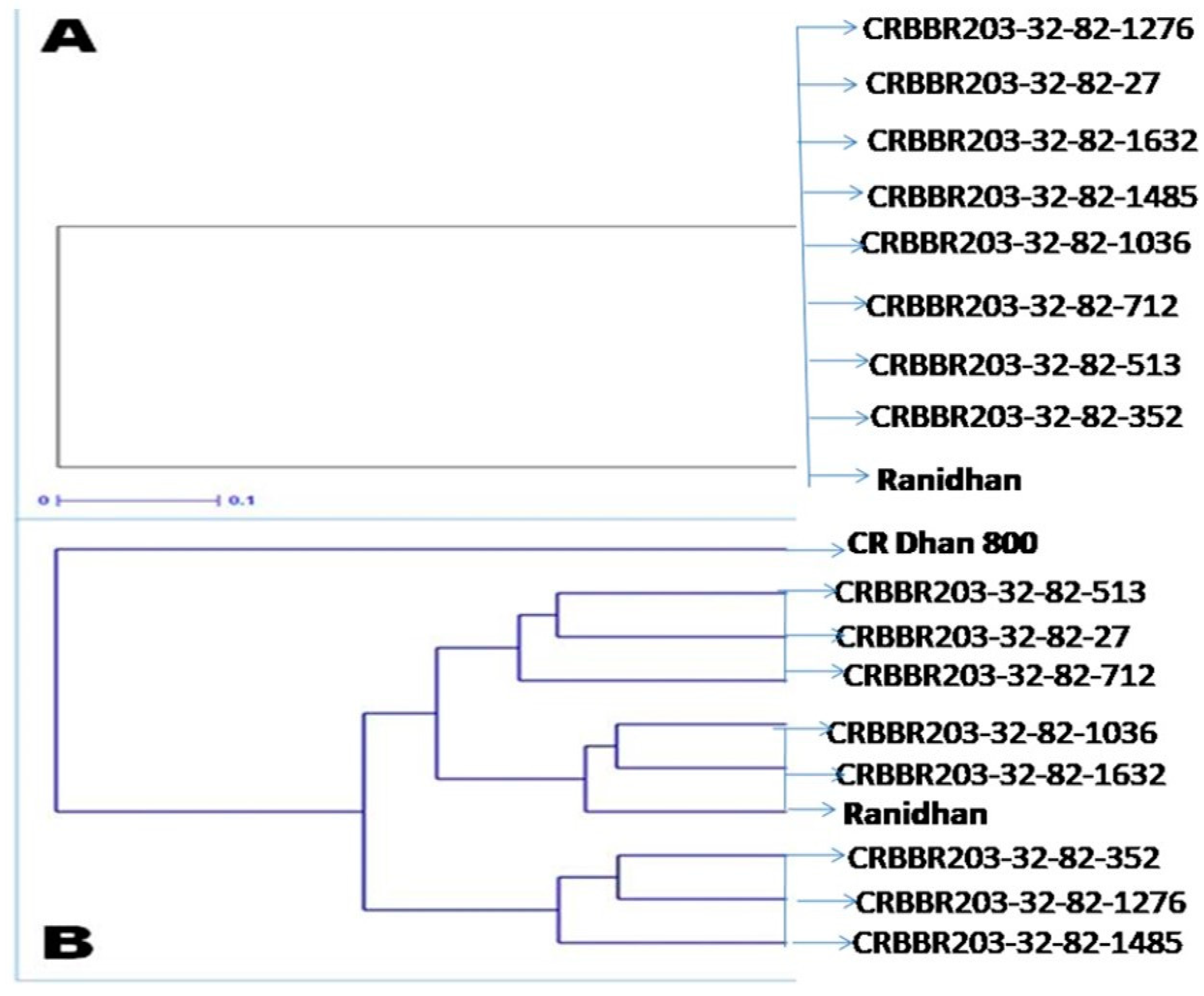
3.1. Selections in the Backcross Progenies during the Forward Breeding
3.2. Bioassays against BB Disease Pathogens
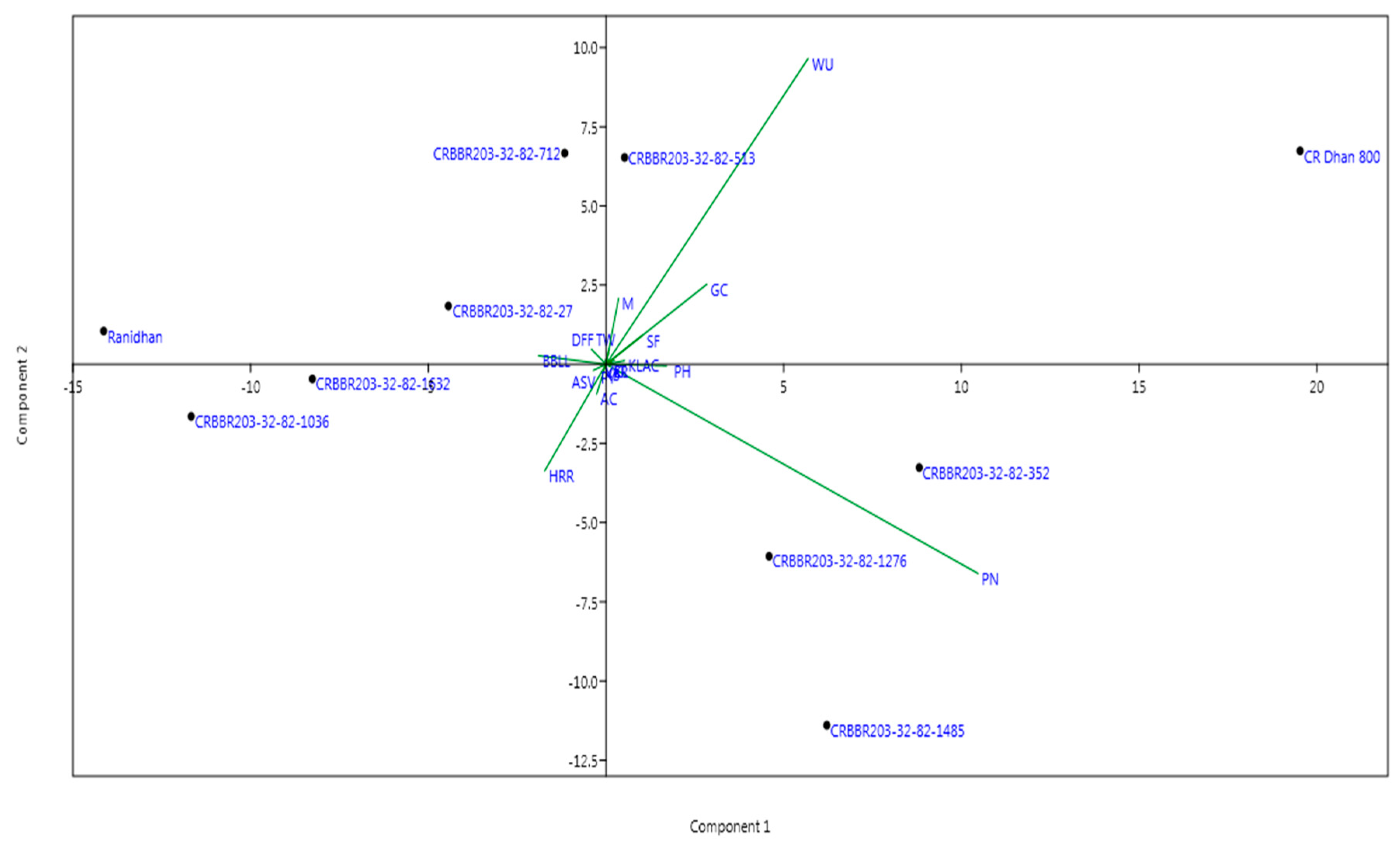
3.3. Grain Yield and Morpho-Quality Traits of the Converted Lines Carrying Four BB Resistance Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declarations
References
- Pandit, E.; Pawar, S.; Barik, S.R.; Mohanty, S.P.; Meher, J.; Pradhan, S.K. Marker-Assisted Backcross Breeding for Improvement of Submergence Tolerance and Grain Yield in the Popular Rice. Variety ‘Maudamani’. Agronomy 2021, 11, 1263. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Sah, R.P.; Behera, L.; Sanghamitra, P.; Bose, L.K.; Das, S.R. Climate-Smart Rice Breeding: Progress and Challenges for the Rain-fed ecologies in India. In Advances in Rice Breeding: StressTolerance, Climate Resilience, Quality and High Yield; Pradhan, S.K., Das, S.R., Patra, B.C., Eds.; ICAR: Odisha, India, 2021; pp. 144–162. Available online: https://icar-nrri.in/wp-content/uploads/2022/05/Book-Advances-in-Rice-Breeding-SKP.pdf (accessed on 12 August 2022).
- Khush, G.S.; Mackill, D.J.; Sidhu, G.S. Breeding Rice for Resistance to Bacterial Leaf Blight; IRRI: Manila, Philippines, 1989; pp. 207–217. [Google Scholar]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Anandan, A.; Reddy, J.N. Characterization of morpho-quality traits and validation of bacterial blight resistance in pyramided rice genotypes under various hotspots of India. Aust. J. Crop Sci. 2015, 9, 127–134. [Google Scholar]
- Mohapatra, S.; Bastia, A.K.; Meher, J.; Sanghamitra, P.; Pradhan, S.K. Development of submergence tolerant, bacterial blight resistant and high yielding near isogenic lines of popular variety, ‘Swarna’ through marker-assisted breeding approach. Front. Plant Sci. 2021, 12, 672618. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Barik, S.R.; Nayak, D.; Pradhan, A.; Pandit, E.; Nayak, P.; Das, S.; Pathak, H.J. Genetics, Molecular Mechanisms and Deployment of Bacterial Blight Resistance Genes in Rice. Crit. Rev. Plant Sci. 2020, 39, 360–385. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Chakraborti, M.; Chakraborty, K.; Behera, L.; Meher, J.; Subudhi, H.N.; Mishra, S.K.; Pandit, E.; Reddy, J.N. Genetic Improvement of Rainfed Shallow low land Rice for Higher Yield and Climate Resilience. In Rice Research for Productivity, Profitability and Climate Resilience; Pathak, H., Nayak, A.K., Jena, M., Singh, O.N., Samal, P., Sharma, S.G., Eds.; ICAR: Odisha, India, 2018; pp. 107–121. Available online: https://icar-nrri.in/wp-content/uploads/2019/02/Rice_Research_book_nrri.pdf (accessed on 12 August 2022).
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef]
- Shanti, M.L.; George, M.L.C.; Vera Cruz, C.M.; Bernardo, M.A.; Nelson, R.J.; Leung, H.; Reddy, J.N.; Sridhar, R. Identification of resistance genes effective against bacterial leaf blight pathogen in eastern India. Plant Dis. 2001, 85, 506–512. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Baksh, S.Y.; Mukherjee, A.K.; Mohanty, S.P. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross breeding. Sci. Rep. 2019, 9, 12810. [Google Scholar] [CrossRef]
- Nayak, D.K.; Pandit, E.; Mohanty, S.; Barik, D.P.; Pradhan, S.K. Marker-assisted selection in back cross progenies for transfer of bacterial leaf blight resistance genes into a popular lowland rice cultivar. Oryza 2015, 52, 163–172. [Google Scholar]
- Sanchez, A.C.; Brar, D.S.; Huang, N.; Khush, G.S. Sequence tagged site markers-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 2002, 40, 792–797. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa-5, xa-13 and Xa-21) using marker-assisted selection into indica rice cultivar PR-106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Joseph, M.; Gopalakrishnan, S.; Sharma, R.K. Combining bacterial blight resistance and basmati quality characteristics by phenotypic and molecular marker assisted selection in rice. Mol. Breed. 2004, 13, 377–387. [Google Scholar] [CrossRef]
- Bharatkumar, S.; Paulraj, R.S.D.; Brindha, P.V.; Kavitha, S.; Gnanamanickam, S.S. Improvent of bacterial blight resistance in rice cultivars ajayothi and IR50 via marker-assisted backcross breeding. J. Crop Improv. 2008, 21, 101–116. [Google Scholar] [CrossRef]
- Hu, K.M.; Qiu, D.Y.; Shen, X.L.; Li, X.H.; Wang, S.P. Isolation and Manipulation of Quantitative trait Loci for Disease Resistance in Rice Using a Candidate Gene Approach. Mol. Plant 2008, 1, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.M.; Redona, E.D.; Mendioro, M.S.; Vera-Cruz, C.M.; Leung, H. Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermo-sensitive genetic male sterile rice (Oryza sativa L.) for the development of two-line hybrids. Euphytica 2008, 164, 627–636. [Google Scholar] [CrossRef]
- Suh, J.P.; Jeung, J.U.; Noh, T.H.; Cho, Y.C.; Park, S.H.; Park, H.S.; Shin, M.S.; Kim, C.K.; Jena, K.K. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 2013, 6, 5. [Google Scholar] [CrossRef]
- Rajpurohit, D.; Kumar, R.; Kumar, M.; Paul, P.; Awasthi, A.A.; Basha, P.O.; Puri, A.; Jhang, T.; Singh, K.; Dhaliwal, H.S. Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 basmati using marker-assisted selection. Euphytica 2011, 178, 111–126. [Google Scholar] [CrossRef]
- Wu, K.S.; Tanksley, S.D. Abundance, polymorphism and genetic mapping of micro satellites in rice. Mol. Gen. Genet. 1993, 241, 225–235. [Google Scholar] [CrossRef]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.G.; McCouch, S.R. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 1997, 95, 553–567. [Google Scholar] [CrossRef]
- Chu, Z.; Fu, B.; Yang, H.; Xu, C.; Li, Z.; Sanchez, A.; Park, Y.J.; Bennetzen, J.L.; Zhang, Q.; Wang, S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2006, 112, 455–461. [Google Scholar] [CrossRef]
- Huang, N.; Angeles, E.R.; Domingo, J.; Magpantay, G.; Singh, S.; Zhang, G.; Kumaravadivel, N.; Bennett, J.; Khush, G.S. Pyramiding of bacterial blight resistance genes in rice: Marker assisted selection using RFLP and PCR. Theor. Appl. Genet. 1997, 95, 313–320. [Google Scholar] [CrossRef]
- Ma, B.J.; Wenming, W.; Bin, Z.; Yongli, Z.; Lihuang, Z.; Wenxue, Z. Studies of PCR marker for the rice bacterial blight resistance gene Xa-4. Hereditas 1999, 21, 9–12. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA mini preparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Mohanty, S.P.; Nayak, D.K.; Pradhan, S.K. Genetic mapping of physiological traits associated with terminal stage drought tolerance in rice. BMC Genet. 2020, 21, 1–12. [Google Scholar] [CrossRef]
- Sahoo, S.; Sanghamitra, P.; Nanda, N.; Pawar, S.; Pandit, E.; Bastia, R.; Muduli, K.C.; Pradhan, S.K. Association of molecular markers with physio-biochemical traits related to seed vigour in rice. Physiol. Mol. Biol. Plants 2020, 26, 1989–2003. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Naveenkumar, R.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Ghritlahre, S.K.; Sanjiba Rao, D.; Reddy, J.N.; et al. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020, 20, 57. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/Darwin (accessed on 10 July 2022).
- Singh, S.; Pradhan, S.K.; Singh, A.K.; Singh, O.N. Marker validation in recombinant inbred lines and random varieties of rice for drought tolerance. Aust. J. Crop Sci. 2012, 6, 606–612. [Google Scholar]
- Pandit, E.; Sahoo, A.; Panda, R.K.; Mohanty, D.P.; Pani, D.R.; Anandan, A.; Pradhan, S.K. Survey of rice cultivars and landraces of upland ecology for phosphorous uptake 1 (pup1) QTL using linked and gene specific molecular markers. Oryza 2016, 53, 1–9. [Google Scholar]
- Sanghamitra, P.; Barik, S.R.; Bastia, R.; Mohanty, S.P.; Pandit, E.; Behera, A.; Mishra, J.; Kumar, G.; Pradhan, S.K. Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping. Plants 2022, 11, 1463. [Google Scholar] [CrossRef]
- Kauffman, H.E.; Reddy, A.P.K.; Hsien, S.P.Y.; Merca, S.D. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Behera, L.; Anandan, A.; Mukherjee, A.K.; Lenka, S.; Barik, D.P. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 2016, 106, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.F.; Li, J.X.; Yu, S.B.; Xing, Y.Z.; Xu, C.G.; Zhang, Q.F. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 1999, 99, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Cagampang, G.B.; Perez, C.M.; Juliano, B.O. A gel consistency test for eating quality in rice. J. Sci. Food Agr. 1973, 24, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Little, R.R.; Hilder, G.B.; Dawson, E.H. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 1958, 35, 111–126. [Google Scholar]
- Juliano, B.O. Rice Quality Screening with the Rapid Visco Analyser; Walker, C.E., Hazelton, J.L., Eds.; Newport Scientific: Sydney, Australia, 1996; pp. 19–24. [Google Scholar]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Singh, S.; Swain, P.; Mohapatra, T. QTL mapping for relative water content trait at reproductive stage drought stress in rice. Indian J. Genet. 2018, 78, 401–408. [Google Scholar]
- Pandit, E.; Panda, R.K.; Pani, D.R.; Chandra, R.; Singh, S.; Pradhan, S.K. Molecular marker and phenotypic analyses for low phosphorus stress tolerance in cultivars and landraces of upland rice under irrigated and drought situations. Indian J. Genet. 2018, 78, 59–68. [Google Scholar] [CrossRef]
- Pawar, S.; Pandit, E.; Mohanty, I.C.; Saha, D.; Pradhan, S.K. Population genetic structure and association mapping for iron toxicity tolerance in rice. PLoS ONE 2021, 16, e0246232. [Google Scholar] [CrossRef]
- Sundaram, R.; Vishnupriya, M.; Biradar, S.; Laha, G.S.; Reddy, G.; Rani, N.; Sarma, N.; Sonti, R. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2007, 160, 411–422. [Google Scholar] [CrossRef]
- Dokku, P.; Das, K.M.; Rao, G.J.N. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica 2013, 192, 87–96. [Google Scholar] [CrossRef]

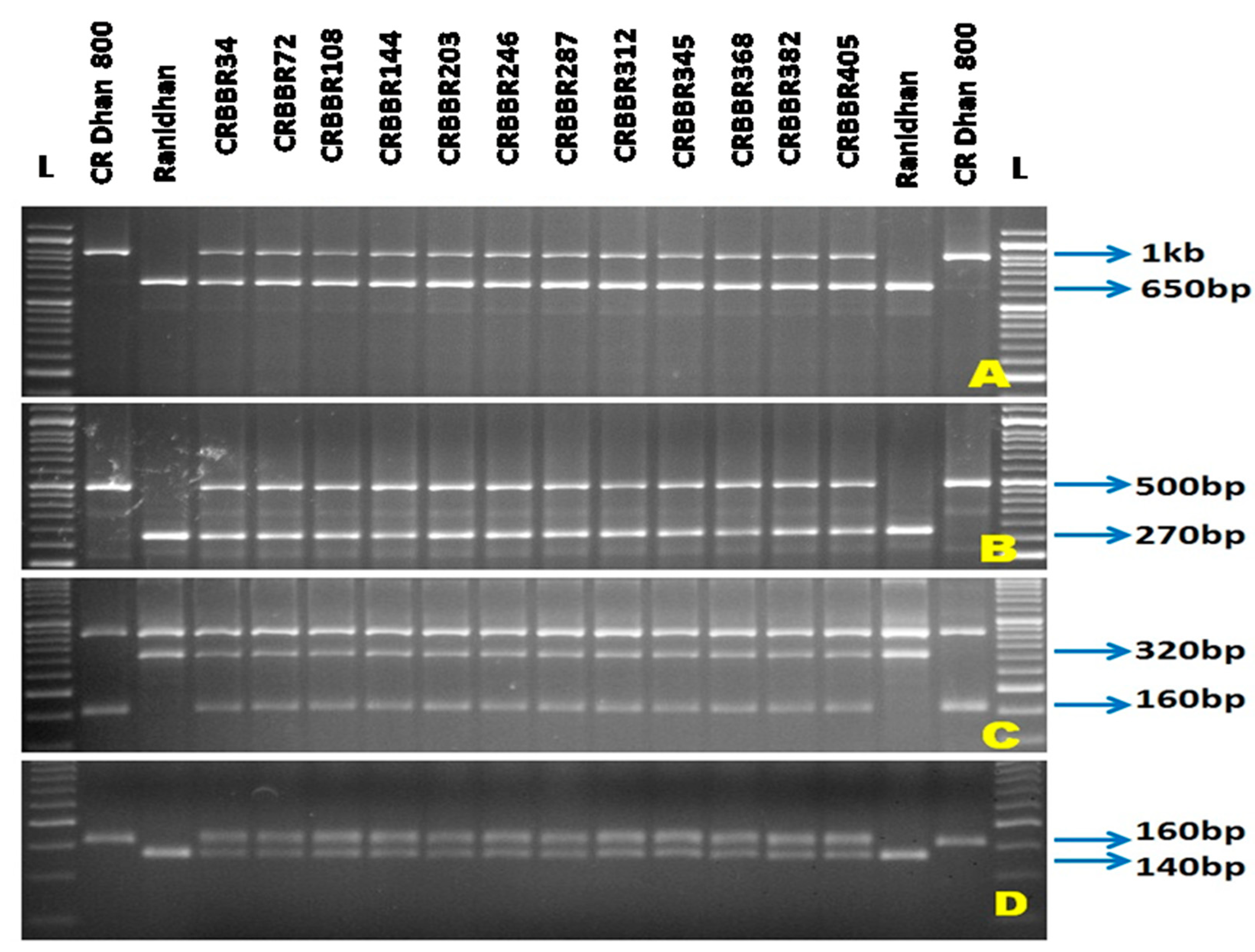
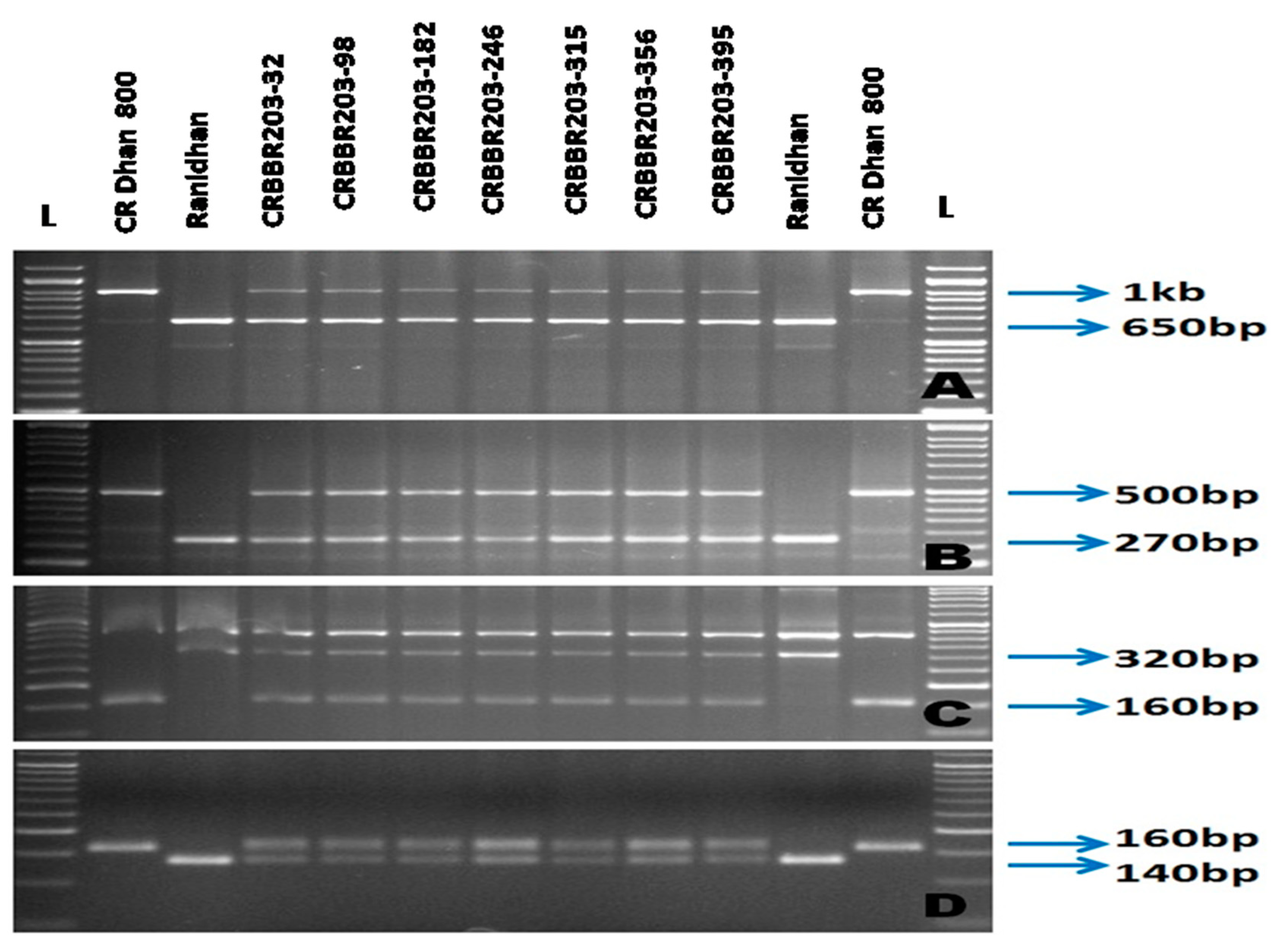

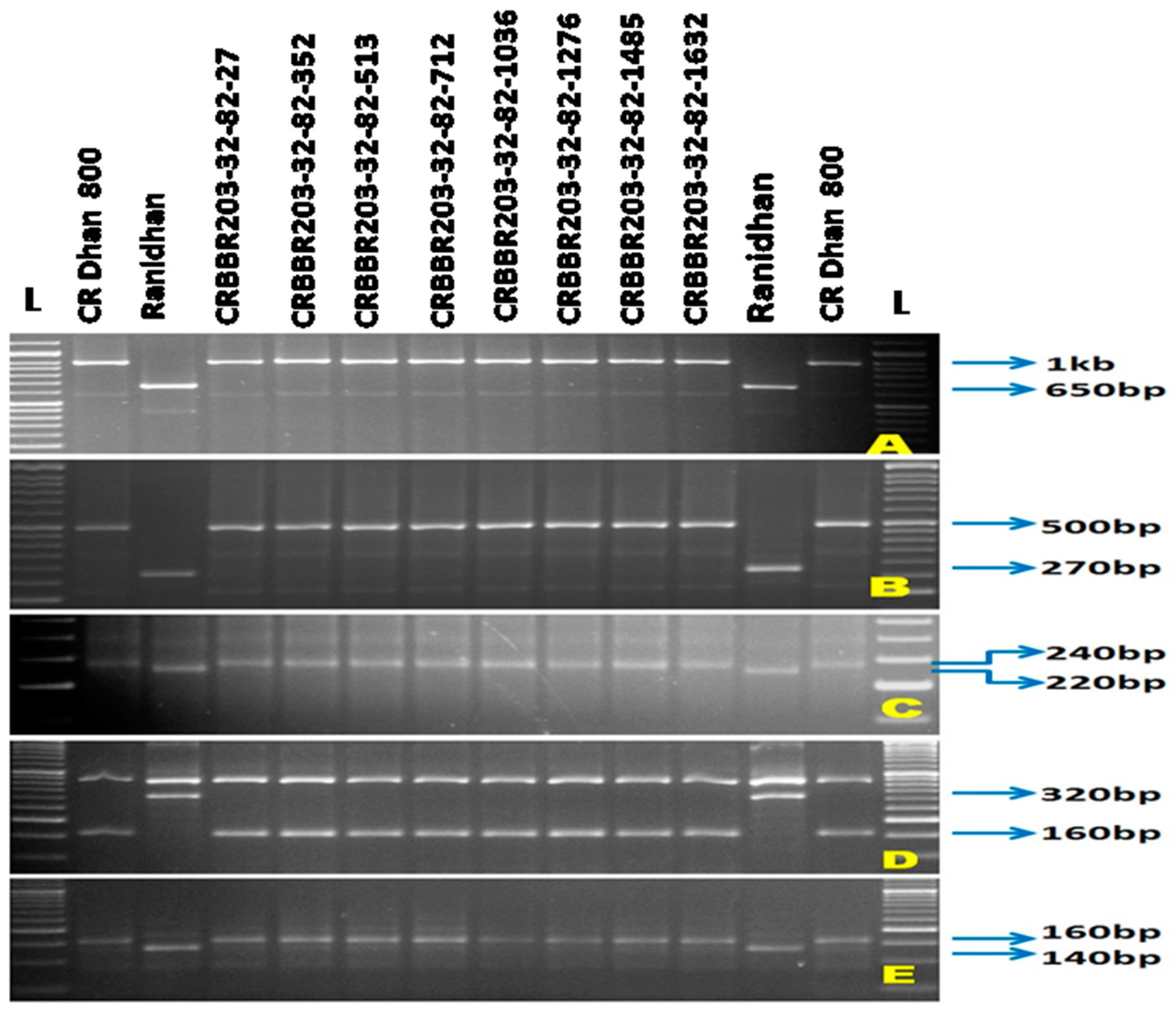
| Resistance Gene | Chromosome Number | Marker | Primer Sequences Used for Gene Detection | Expected Size (bp) | Band Type | References | |
|---|---|---|---|---|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | ||||||
| xa5 | 5 | RM122 | GAGTCGATGTAATGTCATCAGTGC | GAAGGAGGTATCGCTTTGTTGGAC | 260 bp | SSR | [21,22] |
| xa5S (Multiplex) | GTCTGGAATTTGCTCGCGTTCG | TGGTAAAGTAGATACCTTATCAAACTGGA | 160 bp | STS | [10] | ||
| xa5SR/R (Multiplex) | AGCTCGCCATTCAAGTTCTTGAG | TGACTTGGTTCTCCAAGGCTT | |||||
| xa13 | 8 | Xa-13 prom | TCCCAGAAAGCTACTACAGC | GCAGACTCCAGTTTGACTTC | 500 bp | STS | [23] |
| Xa21 | 11 | pTA248 | AGACGCGGAAGGGTGGTTCCCGGA | AGACGCGGTAATCGAAGATGAAA | 1000 bp | STS | [24] |
| Xa4 | 11 | MP-Nbp-131 | ATCGATCGATCTTCACGAGG | TCGTATAAAAGGCATTCGGG- | 160 bp | STS | [25] |
| Sl. No. | Pyramided Lines | Gene Combination | Mean Lesion Length (MLL) in cm (Mean ± Standard Error) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xoo Strains Inoculated | Disease Reaction | |||||||||||
| Xa-17 | Xa-7 | xa-2 | xb-7 | xc-4 | xd-1 | xa-1 | xa-5 | MLL | ||||
| 1 | CRBBR203-32-82-27 | Xa21 + xa13 + xa5 + Xa4 | 2.81 ± 0.83 | 2.53 ± 0.72 | 3.14 ± 0.84 | 2.44 ± 0.68 | 2.73 ± 0.93 | 2.83 ± 0.76 | 2.23 ± 0.95 | 2.34 ± 0.85 | 2.63 ± 0.38 | R |
| 2 | CRBBR203-32-82-352 | Xa21 + xa13 + xa5 + Xa4 | 2.53 ± 0.63 | 2.68 ± 0.64 | 2.42 ± 0.62 | 3.32 ± 0.96 | 2.92 ± 0.93 | 2.62 ± 0.94 | 2.71 ± 77 | 2.41 ± 0.63 | 2.70 ± 0.37 | R |
| 3 | CRBBR203-32-82-513 | Xa21 + xa13 + xa5 + Xa4 | 2.12 ± 0.65 | 2.41 ± 0.62 | 2.53 ± 0.77 | 2.62 ± 0.72 | 2.71 ± 0.85 | 2.59 ± 0.52 | 2.92 ± 0.68 | 2.68 ± 0.85 | 2.57 ± 0.4 | R |
| 4 | CRBBR203-32-82-712 | Xa21 + xa13 + xa5 + Xa4 | 3.39 ± 0.90 | 2.53 ± 0.56 | 2.59 ± 0.72 | 2.72 ± 0.82 | 2.83 ± 0.91 | 2.34 ± 0.53 | 3.24 ± 0.91 | 2.59 ± 0.57 | 2.69 ± 0,38 | R |
| 5 | CRBBR203-32-82-1036 | Xa21 + xa13 + xa5 + Xa4 | 2.72 ± 0.85 | 2.24 ± 0.64 | 2.32 ± 0.56 | 2.43 ± 0.81 | 2.74 ± 0.84 | 2.13 ± 0.78 | 2.81 ± 0.82 | 2.71 ± 0.89 | 2.51 ± 0.4 | R |
| 6 | CRBBR203-32-82-1276 | Xa21 + xa13 + xa5 + Xa4 | 3.38 ± 0.97 | 2.57 ± 0.61 | 2.91 ± 0.73 | 3.14 ± 0.85 | 2.94 ± 0.55 | 2.58 ± 0.59 | 2.53 ± 0.66 | 3.13 ± 0.93 | 2.89 ± 0.36 | R |
| 7 | CRBBR203-32-82-1485 | Xa21 + xa13 + xa5 + Xa4 | 3.13 ± 0.83 | 2.51 ± 0.47 | 3.24 ± 0.92 | 2.83 ± 0.58 | 2.88 ± 0.55 | 2.61 ± 0.46 | 2.52 ± 0.73 | 2.92 ± 0.78 | 2.83 ± 0.35 | R |
| 8 | CRBBR203-32-82-1632 | Xa21 + xa13 + xa5 + Xa4 | 2.62 ± 0.74 | 2.72 ± 0.58 | 2.62 ± 0.73 | 2.71 ± 0.92 | 3.23 ± 0.96 | 3.14 ± 0.93 | 2.54 ± 0.56 | 2.84 ± 0.75 | 2.80 ± 0.35 | R |
| 9 | CR Dhan 800 (donor) | Xa21 + xa13 + xa5 + Xa4 | 2.41 ± 0.74 | 2.13 ± 0.48 | 2.34 ± 0.52 | 2.63 ± 0.67 | 3.14 ± 0.72 | 2.79 ± 0.91 | 2.61 ± 0.76 | 2.52 ± 0.69 | 2.57 ± 0.36 | R |
| 10 | Ranidhan (recipient) | - | 11.78 ± 1.25 | 13.18 ± 1.63 | 13.83 ± 1.52 | 11.71 ± 1.12 | 10.63 ± 1.25 | 14.14 ± 1.23 | 12.32 ± 0.92 | 14.13 ± 1.13 | 12.72 ± 1.26 | S |
| Serial Number | Pyramided Lines | PH (cm) | DFF (Days) | PN | SF (%) | TW (g) | KL (mm) | KB (mm) | Milling (%) | HRR | VER | WU (ml) | KLAC (mm) | ASV | GC | AC (%) | PY (t/ha) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRBBR203-32-82-27 | 108 | 115 | 294 | 87.2 | 19.75 | 5.45 | 2.28 | 68.7 | 63.2 | 4.4 | 162.5 | 8.4 | 6.0 | 59 | 23.25 | 6.325 |
| 2 | CRBBR203-32-82-352 | 107 | 114 | 308 | 87.3 | 19.23 | 5.55 | 2.34 | 68.5 | 64.5 | 5.0 | 165.0 | 8.5 | 5.5 | 63 | 24.75 | 6.075 |
| 3 | CRBBR203-32-82-513 | 109 | 115 | 296 | 88.1 | 19.65 | 5.58 | 2.35 | 69.1 | 61.7 | 4.7 | 170.0 | 8.3 | 6.0 | 57 | 24.55 | 6.385 |
| 4 | CRBBR203-32-82-712 | 106 | 110 | 294 | 86.5 | 20.15 | 5.65 | 2.35 | 68.9 | 64.2 | 4.7 | 168.5 | 8.6 | 5.0 | 63 | 24.75 | 6.250 |
| 5 | CRBBR203-32-82-1036 | 103 | 116 | 290 | 88.3 | 20.25 | 5.75 | 2.26 | 69.2 | 65.1 | 4.8 | 155.5 | 8.4 | 5.0 | 59 | 23.18 | 6.580 |
| 6 | CRBBR203-32-82-1276 | 105 | 112 | 306 | 86.7 | 19.65 | 5.46 | 2.16 | 67.3 | 66.2 | 4.4 | 161.5 | 8.3 | 5.5 | 61 | 25.25 | 6.210 |
| 7 | CRBBR203-32-82-1485 | 110 | 113 | 310 | 86.2 | 19.52 | 5.82 | 2.25 | 66.8 | 65.1 | 4.4 | 158.0 | 9.1 | 5.5 | 57 | 25.05 | 6.385 |
| 8 | CRBBR203-32-82-1632 | 104 | 112 | 292 | 87.5 | 20.35 | 5.48 | 2.15 | 66.7 | 65.5 | 4.8 | 159.5 | 8.3 | 6.0 | 59 | 24.35 | 6.165 |
| 9 | CR Dhan 800 (donor) | 108 | 115 | 312 | 89.2 | 20.65 | 6.36 | 2.14 | 71.1 | 58.1 | 4.7 | 176.5 | 9.9 | 4.5 | 67 | 22.36 | 6.115 |
| 10 | Ranidhan (recipient) | 105 | 116 | 288 | 83.1 | 19.3 | 5.44 | 2.30 | 69.5 | 64.8 | 4.7 | 158.5 | 8.1 | 6.0 | 57 | 25.12 | 5.865 |
| LSD5% | 5.46 | 4.96 | 31.36 | 9.32 | 2.16 | 0.74 | 0.164 | 7.324 | 7.456 | - | 16.38 | 0.754 | - | - | 2.724 | 0.348 | |
| CV% | 3.86 | 1.24 | 10.78 | 5.18 | 4.84 | 7.35 | 8.146 | 6.685 | 9.328 | - | 6.284 | 7.36 | - | - | 6.742 | 10.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pradhan, K.C.; Pandit, E.; Mohanty, S.P.; Moharana, A.; Sanghamitra, P.; Meher, J.; Jena, B.K.; Dash, P.K.; Behera, L.; Mohapatra, P.M.; et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy 2022, 12, 1903. https://doi.org/10.3390/agronomy12081903
Pradhan KC, Pandit E, Mohanty SP, Moharana A, Sanghamitra P, Meher J, Jena BK, Dash PK, Behera L, Mohapatra PM, et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy. 2022; 12(8):1903. https://doi.org/10.3390/agronomy12081903
Chicago/Turabian StylePradhan, Kartik Chandra, Elssa Pandit, Shakti Prakash Mohanty, Arpita Moharana, Priyadarsini Sanghamitra, Jitendriya Meher, Binod Kumar Jena, Prasanta K. Dash, Lambodar Behera, Pavitra Mohan Mohapatra, and et al. 2022. "Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice" Agronomy 12, no. 8: 1903. https://doi.org/10.3390/agronomy12081903






