Enhanced Metabolism Evolved High-Level Resistance to Fenoxaprop-P-Ethyl in Alopecurus japonicus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Herbicide
2.2. Susceptibility Tests of Fenoxaprop-P-Ethyl and other ACCase-Inhibiting Herbicides
2.3. Analysis of ACCase-Resistance Mutations in A. japonicus
2.4. Effect of CYP450 and GST Inhibitors on Fenoxaprop-P-Ethyl Resistance
2.5. Fenoxaprop-P-Ethyl Metabolism Analysis Using HPLC in A. japonicus
2.6. RNA-Seq Analysis
2.7. Differential Gene Expression of the Candidate Glycosyl Transferases (GT) and ABC Transporter Contigs from RNA-Seq
3. Results
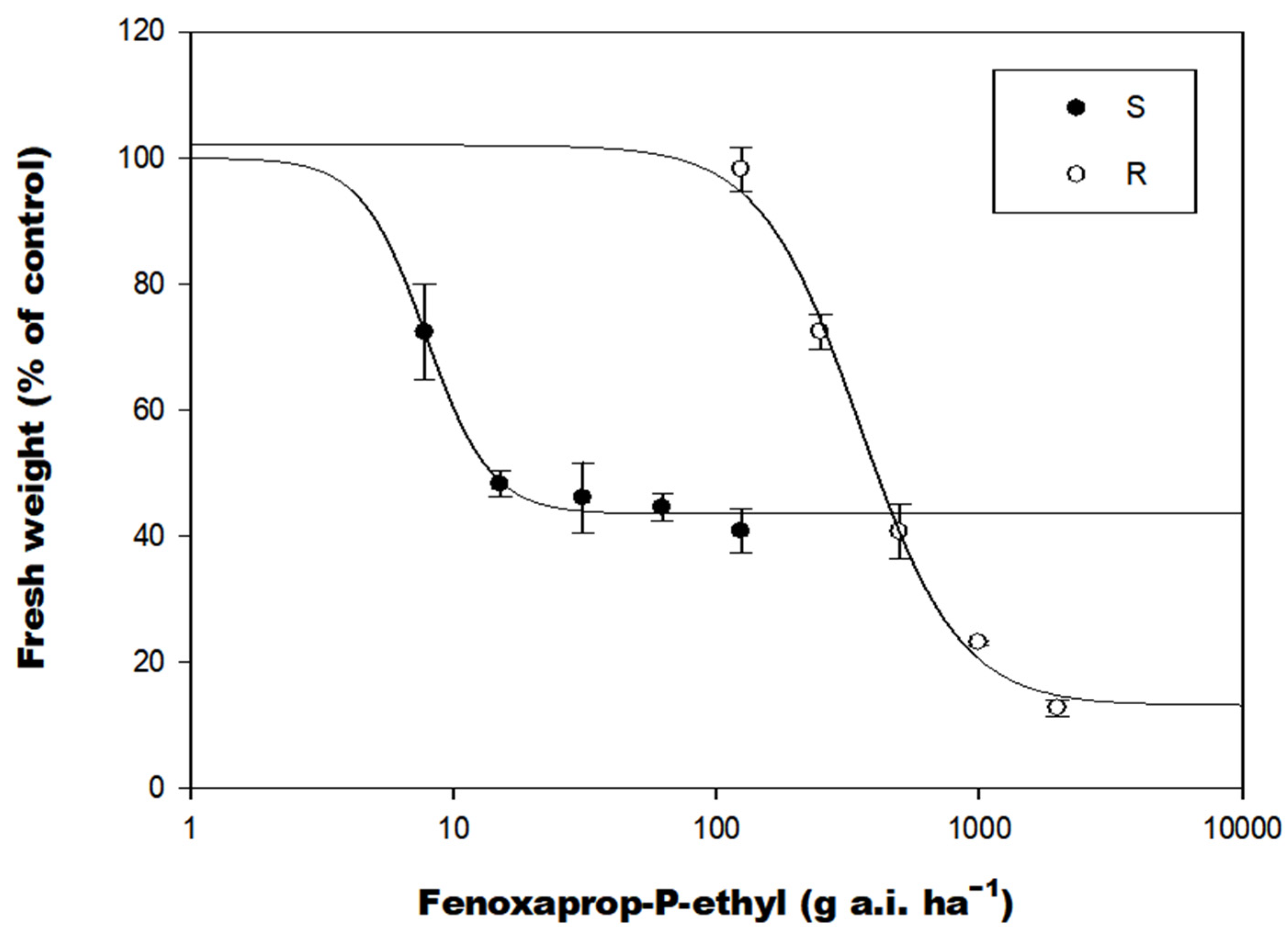
3.1. Fenoxaprop-P-Ethyl Dose–Response and Resistance Testing to Other ACCase-Inhibiting Herbicides in A. japonicus
3.2. ACCase Gene Sequencing in A. japonicus
3.3. Impact of CYP450 and GST Inhibitors on Fenoxaprop-P-Ethyl Resistance in A. japonicus
3.4. Metabolic Rate Difference of Fenoxaprop-P-Ethyl in A. japonicus
3.5. Identifying Differential Expression of GT and ABC Transporter Contigs in A. japonicus
4. Discussion
4.1. NTSR Mechanism Can Confer High-Level ACCase-Inhibiting Herbicide Resistance
4.2. Enhanced Metabolism Leads to a Specific Herbicide Resistance Spectrum in A. japonicus
4.3. Specific Metabolic Enzymes Are Likely to Be Involved in Fenoxaprop-P-Ethyl Resistance in A. japonicus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heap, I. International Survey of Herbicide Resistant Weeds [Online]. Available online: http://www.weedscience.org (accessed on 15 July 2022).
- Kaundun, S.S. Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest. Manag. Sci. 2014, 70, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.P.; Tranel, P.J. Target-Site mutations conferring herbicide resistance. Plants 2019, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.; Duke, S.; Morran, S.; Rigon, C.; Tranel, P.; Kuepper, A.; Dayan, F. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, N.G.; Iwakami, S. CytochromeP450-mediated herbicide metabolism in plants: Current understanding and prospects. Pest. Manag. Sci. 2021, 77, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Gao, H.T.; Xia, W.W.; Zhang, T.; Dong, L.Y. Establishing a herbicide-metabolizing enzyme library in Beckmannia syzigachne to identify genes associated with metabolic resistance. J. Exp. Bot. 2016, 67, 1745–1757. [Google Scholar] [CrossRef]
- Kaundun, S.S.; Bailly, G.C.; Dale, R.P.; Hutchings, S.J.; McIndoe, E. A novel W1999S mutation and non-target site resistance impact on acetyl-CoA carboxylase inhibiting herbicides to varying degrees in a UK Lolium multiflorum population. PLoS ONE 2013, 8, e58012. [Google Scholar] [CrossRef]
- Han, H.; Yu, Q.; Owen, M.J.; Cawthray, G.R.; Powles, S.B. Widespread occurrence of both metabolic and target-site herbicide resistance mechanisms in Lolium rigidum populations. Pest. Manag. Sci. 2016, 72, 255–263. [Google Scholar] [CrossRef]
- Gaines, T.A.; Lorentz, L.; Figge, A.; Herrmann, J.; Maiwald, F.; Ott, M.-C.; Han, H.; Busi, R.; Yu, Q.; Powles, S.B.; et al. RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 2014, 78, 865–876. [Google Scholar] [CrossRef]
- Yu, Q.; Han, H.; Cawthray, G.R.; Wang, S.F.; Powles, S.B. Enhanced rates of herbicide metabolism in low herbicide-dose selected resistant Lolium rigidum. Plant Cell Environ. 2013, 36, 818–827. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-target-site resistance to herbicides: Recent developments. Plants 2019, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Ahmad-Hamdani, M.S.; Yu, Q.; Han, H.; Cawthray, G.R.; Wang, S.F.; Powles, S.B. Herbicide resistance endowed by enhanced rates of herbicide metabolism in Wild Oat (Avena spp.). Weed Sci. 2013, 61, 55–62. [Google Scholar] [CrossRef]
- Zhao, N.; Ge, L.a.; Yan, Y.; Bai, S.; Wang, D.; Liu, W.; Wang, J. Trp-1999-Ser mutation of acetyl-CoA carboxylase and cytochrome P450s-involved metabolism confer resistance to fenoxaprop-P-ethyl in Polypogon fugax. Pest. Manag. Sci. 2019, 75, 3175–3183. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.J.; Gao, Y.; Zhang, Y.; Dong, L.Y.; Li, J. Mechanisms of resistance to pyroxsulam and ACCase inhibitors in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2016, 64, 695–704. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Li, X.; Li, D.; Li, Z.; Cui, H. Pro-197-Ser Mutation in ALS and high-level GST activities: Multiple resistance to ALS and ACCase inhibitors in Beckmannia syzigachne. Front. Plant Sci. 2020, 11, 572610. [Google Scholar] [CrossRef]
- Tang, H.; Li, J.; Dong, L.; Dong, A.; Lu, B.; Zhu, X. Molecular bases for resistance to acetyl-coenzyme A carboxylase inhibitor in Japanese foxtail (Alopecurus japonicus). Pest. Manag. Sci. 2012, 68, 1241–1247. [Google Scholar] [CrossRef]
- Mohamed, I.A.; Li, R.Z.; You, Z.G.; Li, Z.H. Japanese Foxtail (Alopecurus japonicus) resistance to fenoxaprop and pinoxaden in China. Weed Sci. 2012, 60, 167–171. [Google Scholar] [CrossRef]
- Xu, H.; Zhu, X.; Wang, H.; Li, J.; Dong, L. Mechanism of resistance to fenoxaprop in Japanese foxtail (Alopecurus japonicus) from China. Pestic. Biochem. Phys. 2013, 107, 25–31. [Google Scholar] [CrossRef]
- Bi, Y.L.; Liu, W.T.; Li, L.X.; Yuan, G.H.; Jin, T.; Wang, J.X. Molecular basis of resistance to mesosulfuron-methyl in Japanese foxtail, Alopecurus japonicus. J. Pestic. Sci. 2013, 38, 74–77. [Google Scholar] [CrossRef]
- Xu, H.; Li, J.; Zhang, D.; Cheng, Y.; Jiang, Y.; Dong, L. Mutations at codon position 1999 of acetyl-CoA carboxylase confer resistance to ACCase-inhibiting herbicides in Japanese foxtail (Alopecurus japonicus). Pest. Manag. Sci. 2014, 70, 1894–1901. [Google Scholar] [CrossRef]
- Bi, Y.; Liu, W.; Guo, W.; Li, L.; Yuan, G.; Du, L.; Wang, J. Molecular basis of multiple resistance to ACCase- and ALS-inhibiting herbicides in Alopecurus japonicus from China. Pestic. Biochem. Phys. 2016, 126, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, L.; Xu, H.; Wu, X.; Pan, L.; Dong, L. Cross-resistance patterns to acetyl-coA carboxylase inhibitors associated with different mutations in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2017, 65, 444–451. [Google Scholar] [CrossRef]
- Chen, G.Q.; Xu, H.L.; Zhang, T.; Bai, C.Q.; Dong, L.Y. Fenoxaprop-P-ethyl resistance conferred by cytochrome P450s and target site mutation in Alopecurus japonicus. Pest. Manag. Sci. 2018, 74, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Bi, Y.L.; Wu, C.X.; Wang, D.D.; You, L.D.; Liu, W.T.; Wang, J.X. Cross-resistance to acetolactate synthase (ALS) inhibitors associated with different mutations in Japanese foxtail (Alopecurus japonicus). Weed Sci. 2019, 67, 389–396. [Google Scholar] [CrossRef]
- Cummins, I.; Wortley, D.J.; Sabbadin, F.; He, Z.; Coxon, C.R.; Straker, H.E.; Sellars, J.D.; Knight, K.; Edwards, L.; Hughes, D.; et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA 2013, 110, 5812–5817. [Google Scholar] [CrossRef]
- Singh, S.B.; Das, T.K.; Kulshrestha, G. Persistence of herbicide fenoxaprop ethyl and its acid metabolite in soil and wheat crop under Indian tropical conditions. J. Environ. Sci. Health B 2013, 48, 324–330. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Xu, H.; Li, J.; Wu, R.; Su, W.; Wu, X.; Wang, L.; Dong, L. Identification of reference genes for studying herbicide resistance mechanisms in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2017, 65, 557–566. [Google Scholar] [CrossRef]
- Pan, L.; Yu, Q.; Han, H.; Mao, L.; Nyporko, A.; Fan, L.; Bai, L.; Powles, S.B. Aldo-keto reductase metabolizes glyphosate and confers glyphosate resistance in Echinochloa colona. Plant Physiol. 2019, 181, 1519–1534. [Google Scholar] [CrossRef] [PubMed]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A. CYP81A P450s are involved in concomitant cross-resistance to ALS and ACCase herbicides in Echinochloa phyllopogon. New Phytol. 2018, 221, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Collavo, A.; Strek, H.; Beffa, R.; Sattin, M. Management of an ACCase-inhibitor-resistant Lolium rigidum population based on the use of ALS inhibitors: Weed population evolution observed over a 7 year field-scale investigation. Pest. Manag. Sci. 2013, 69, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yu, J.; Pan, L.; Wu, X.; Dong, L. Target-Site resistance to fenoxaprop-P-ethyl in Keng Stiffgrass (Sclerochloa kengiana) from China. Weed Sci. 2017, 65, 452–460. [Google Scholar] [CrossRef]
- Pan, L.; Li, J.; Zhang, W.-N.; Dong, L. Detection of the I1781L mutation in fenoxaprop-p-ethyl-resistant American sloughgrass (Beckmannia syzigachne Steud.), based on the loop-mediated isothermal amplification method. Pest. Manag. Sci. 2015, 71, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Yan, Y.Y.; Ge, L.A.; Zhu, B.L.; Liu, W.T.; Wang, J.X. Target site mutations and cytochrome P450s confer resistance to fenoxaprop-P-ethyl and mesosulfuron-methyl in Alopecurus aequalis. Pest. Manag. Sci. 2019, 75, 204–214. [Google Scholar] [CrossRef]
- Nandula, V.K.; Riechers, D.E.; Ferhatoglu, Y.; Barrett, M.; Duke, S.O.; Dayan, F.E.; Goldberg-Cavalleri, A.; Tétard-Jones, C.; Wortley, D.J.; Onkokesung, N.; et al. Herbicide metabolism: Crop selectivity, bioactivation, weed resistance, and regulation. Weed Sci. 2019, 67, 149–175. [Google Scholar] [CrossRef]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest. Manag. Sci. 2013, 69, 176–187. [Google Scholar] [CrossRef]
- Pan, L.; Yu, Q.; Wang, J.; Han, H.; Mao, L.; Nyporko, A.; Maguza, A.; Fan, L.; Bai, L.; Powles, S. An ABCC-type transporter endowing glyphosate resistance in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2100136118. [Google Scholar] [CrossRef]



| Herbicide | Populations | Treatments | GR50 (g a.i.ha−1) (SE) a | Resistance Index |
|---|---|---|---|---|
| Fenoxaprop-P-ethyl | R | NBD-CI+ Fenoxaprop-P-ethyl | 322.7 (43) | 41.2 |
| Malathion+ Fenoxaprop-P-ethyl | 303.6 (36) | 38.8 | ||
| Fenoxaprop-P-ethyl | 350.6 (33) | 44.8 | ||
| S | NBD-CI+ Fenoxaprop-P-ethyl | 9.7 (1.4) | 1.2 | |
| Malathion+ Fenoxaprop-P-ethyl | 7.6 (0.9) | 1.0 | ||
| Fenoxaprop-P-ethyl | 7.8 (0.5) | - |
| Gene | Family | log2FC a | FC b | t-Test | FC c | t-Test |
|---|---|---|---|---|---|---|
| UDP-GT | 71C | 7.33 | 1.9 | 0.12 | - | - |
| 73C | 5.78 | 2.12 | 0.12 | - | - | |
| 75C | 2.6 | 4.5 | 0.007 | 2.73 | 0.07 | |
| 80B | 1.84 | 1.47 | 0.43 | - | - | |
| 83A | 10.1 | 1.51 | 0.38 | - | - | |
| 86A | 3.83 | >8 | - | >16 | - | |
| 88B | 2.22 | 0.66 | 0.16 | - | - | |
| 89B | 8.97 | 2.04 | 0.18 | - | - | |
| 91A | 7.47 | >8 | - | >8 | - | |
| 92 | 6.98 | 9.42 | 0.02 | 6.35 | 0.24 | |
| ABC | B11 | 1.7 | >16 | - | >16 | - |
| B13 | 4.21 | 5.85 | 0.001 | 2.04 | 0.43 | |
| B28 | 1.26 | 0.73 | 0.18 | - | - | |
| C2 | 1.34 | 0.99 | 0.99 | - | - | |
| C10 | 7.73 | >32 | - | >128 | - | |
| D1 | 1.31 | 0.94 | 0.89 | - | - | |
| E2 | 8.44 | 52.93 | 0.04 | >64 | - | |
| G53 | 2.55 | 1.26 | 0.16 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Liu, H.; Wang, J.; Chen, W.; Bai, L.; Pan, L. Enhanced Metabolism Evolved High-Level Resistance to Fenoxaprop-P-Ethyl in Alopecurus japonicus. Agronomy 2022, 12, 2172. https://doi.org/10.3390/agronomy12092172
Li Z, Liu H, Wang J, Chen W, Bai L, Pan L. Enhanced Metabolism Evolved High-Level Resistance to Fenoxaprop-P-Ethyl in Alopecurus japonicus. Agronomy. 2022; 12(9):2172. https://doi.org/10.3390/agronomy12092172
Chicago/Turabian StyleLi, Zongfang, Haozhe Liu, Junzhi Wang, Wen Chen, Lianyang Bai, and Lang Pan. 2022. "Enhanced Metabolism Evolved High-Level Resistance to Fenoxaprop-P-Ethyl in Alopecurus japonicus" Agronomy 12, no. 9: 2172. https://doi.org/10.3390/agronomy12092172
APA StyleLi, Z., Liu, H., Wang, J., Chen, W., Bai, L., & Pan, L. (2022). Enhanced Metabolism Evolved High-Level Resistance to Fenoxaprop-P-Ethyl in Alopecurus japonicus. Agronomy, 12(9), 2172. https://doi.org/10.3390/agronomy12092172





