Forest Gaps Modulate the Composition and Co-Occurrence Network of Soil Bacterial Community in Larix principis-rupprechtii Mayr Plantation
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Experimental Design
2.3. Data Collection
2.3.1. Soil Bacterial Community
2.3.2. Gap-Related Microenvironmental Monitoring
2.3.3. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
2.4. High-Throughput Analysis
2.5. Statistical Analysis
3. Results
3.1. Light Properties of Forest Gaps
3.2. Soil Physicochemical Properties of Forest Gaps
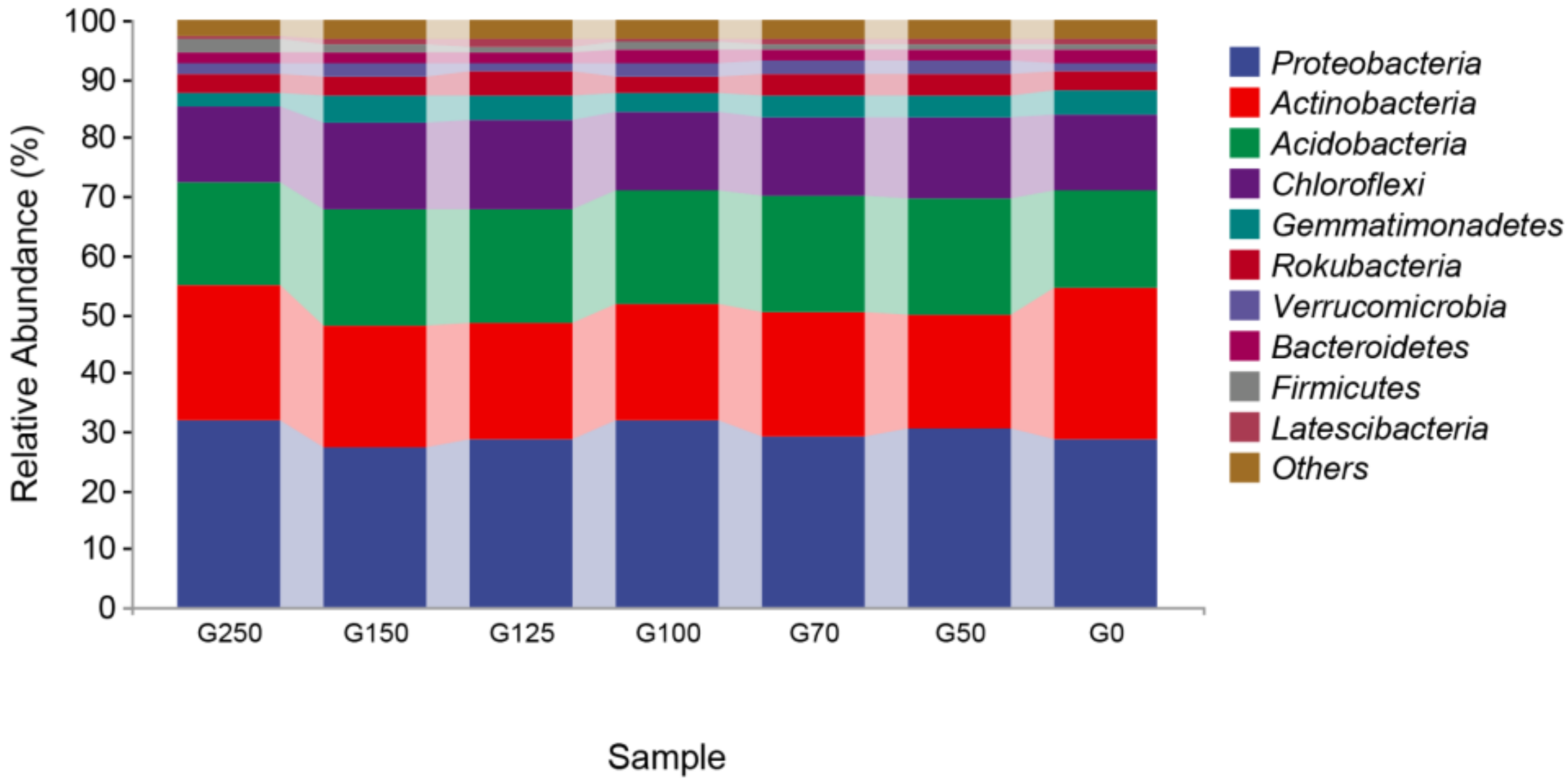
3.3. The Composition of Soil Bacterial Communities in Forest Gaps
3.4. Soil Bacterial Community Diversity in Forest Gaps
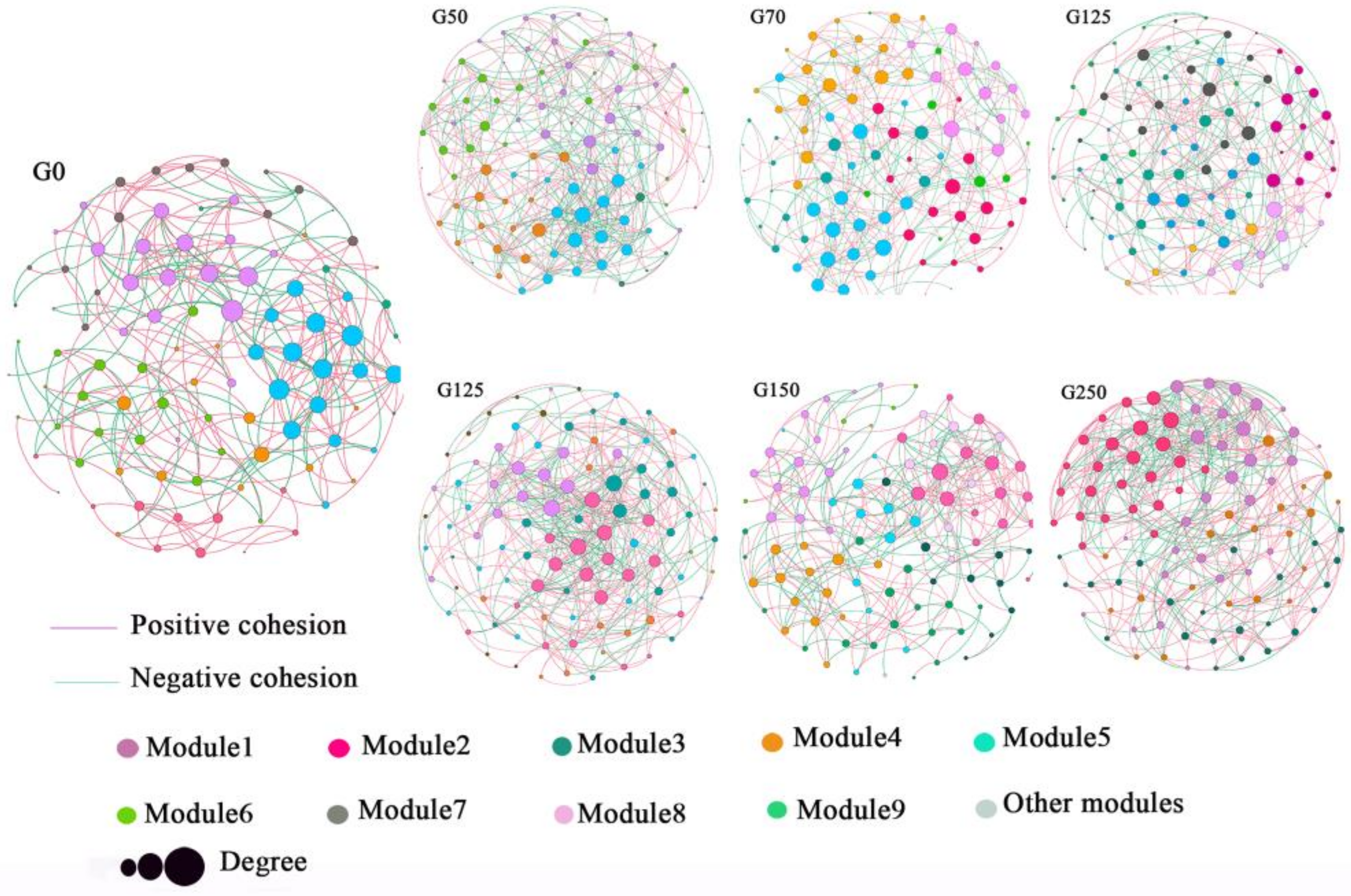
3.5. Co-Occurrence Network Structure of Soil Bacterial Community in Forest Gaps
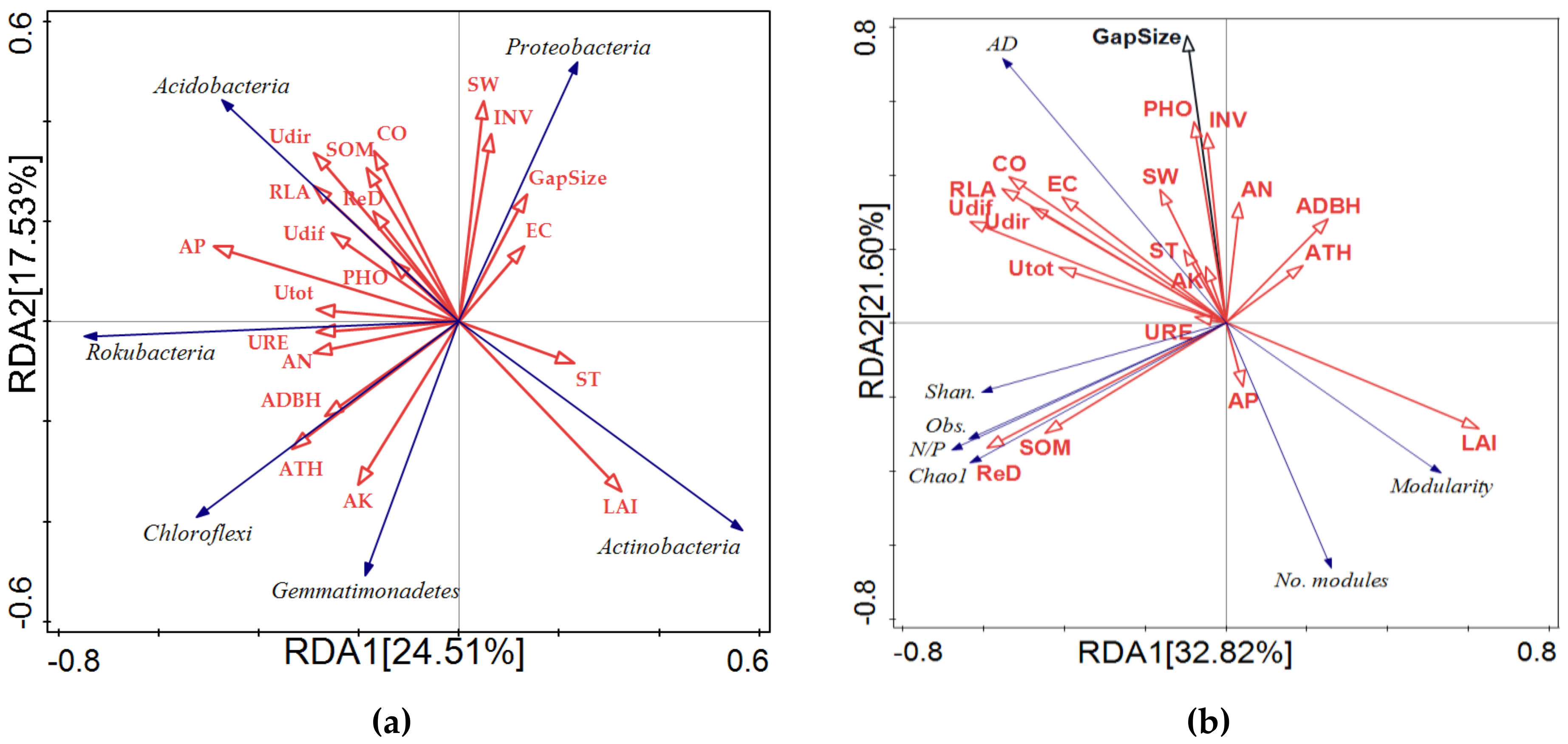
3.6. Correlation between Gap-Related Microenvironmental Factors and Soil Bacterial Community Composition and Network Structure
3.7. Linkages of Soil Bacterial Composition and Co-Occurrence Network Complexity to Forest Gap Sizes
4. Discussion
4.1. Gap-Related Microenvironments
4.2. Effects of Gap-Related Microenvironments on Soil Bacterial Composition and Co-Occurrence Network Structure
4.3. Effects of Regeneration in L. principis-rupprechtii Plantations on the Soil Bacterial Communities under Forest Gaps
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muscolo, A.; Bagnato, S.; Sidari, M.; Mercurio, R. A review of the roles of forest canopy gaps. J. For. Res. 2014, 25, 725–736. [Google Scholar] [CrossRef]
- Lu, D.; Zhu, J.; Wang, X.; Hao, G.; Wang, G.G. A systematic evaluation of gap size and within-gap position effects on seedling regeneration in a temperate secondary forest, Northeast China. For. Ecol. Manag. 2021, 490, 119140. [Google Scholar] [CrossRef]
- Muscolo, A.; Sidari, M.; Mercurio, R. Influence of gap size on organic matter decomposition, microbial biomass and nutrient cycle in Calabrian pine (Pinus laricio, Poiret) stands. For. Ecol. Manag. 2007, 242, 412–418. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; He, Z.; Xing, C.; Zhu, J.; Gu, X.; Lan, Y.; Wu, Z.; Liao, P.; Zhu, D. Forest gaps mediate the structure and function of the soil microbial community in a Castanopsis kawakamii forest. Ecol. Indic. 2021, 122, 107288. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; Su, Y.; Li, X.; Yin, H.; Li, X.; Guo, M.; He, Y. Forest gaps influence fungal community assembly in a weeping cypress forest. Appl. Microbiol. Biotechnol. 2019, 103, 3215–3224. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Tang, M.; Shi, B.; Jin, G. Effect of canopy gap size on soil respiration in a mixed broadleaved-Korean pine forest: Evidence from biotic and abiotic factors. Eur. J. Soil Biol. 2020, 99, 103194. [Google Scholar] [CrossRef]
- Dupuy, J.M.; Chazdon, R. Interacting effects of canopy gap, understory vegetation and leaf litter on tree seedling recruitment and composition in tropical secondary forests. For. Ecol. Manag. 2008, 255, 3716–3725. [Google Scholar] [CrossRef]
- Schliemann, S.A.; Bockheim, J.G. Influence of gap size on carbon and nitrogen biogeochemical cycling in Northern hardwood forests of the Upper Peninsula, Michigan. Plant Soil 2014, 377, 323–335. [Google Scholar] [CrossRef]
- Yang, Y.; Geng, Y.; Zhou, H.; Zhao, G.; Wang, L. Effects of gaps in the forest canopy on soil microbial communities and enzyme activity in a Chinese pine forest. Pedobiologia 2017, 61, 51–60. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, G.; Zhang, X.; He, N.; Wang, Q.; Wang, S.; Wang, R.; Zhao, N.; Jia, Y.; Wang, C. Soil enzyme activity and stoichiometry in forest ecosystems along the North-South Transect in eastern China (NSTEC). Soil Biol. Biochem. 2017, 104, 152–163. [Google Scholar] [CrossRef]
- Yu, X.; Yang, L.; Fei, S.; Ma, Z.; Hao, R.; Zhao, Z. Effect of Soil Layer and Plant–Soil Interaction on Soil Microbial Diversity and Function after Canopy Gap Disturbance. Forests 2018, 9, 680. [Google Scholar] [CrossRef]
- Julie, S.D. Supplement: Tropical Succession || Gap Partitioning among Tropical Rainforest Trees. Biotropica 1980, 12, 47–55. [Google Scholar] [CrossRef]
- Brokaw, N.; Busing, R.T. Niche versus chance and tree diversity in forest gaps. Trends Ecol. Evol. 2000, 15, 183–188. [Google Scholar] [CrossRef] [PubMed]
- McClure, J.W.; Lee, T.D.; Leak, W.B. Gap capture in northern hardwoods: Patterns of establishment and height growth in four species. For. Ecol. Manag. 2000, 127, 181–189. [Google Scholar] [CrossRef]
- Webster, C.R.; Lorimer, C.G. Minimum opening sizes for canopy recruitment of mid-tolerant tree species: A retrospective approach. Ecol. Appl. 2005, 15, 1245–1262. [Google Scholar] [CrossRef]
- Stavridou, E.; Webster, R.J.; Robson, P.R.H. Novel Miscanthus genotypes selected for different drought tolerance phenotypes show enhanced tolerance across combinations of salinity and drought treatments. Ann. Bot. 2019, 124, 653–674. [Google Scholar] [CrossRef]
- Li, S.; Wu, F. Diversity and Co-occurrence Patterns of Soil Bacterial and Fungal Communities in Seven Intercropping Systems. Front. Microbiol. 2018, 9, 1521. [Google Scholar] [CrossRef]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis: Part 2 Chemical and Microbiological Properties; Page, A.L., Ed.; Soil Science Society of America Inc.: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Shi, Y.; Li, Y.; Xiang, X.; Sun, R.; Yang, T.; He, D.; Zhang, K.; Ni, Y.; Zhu, Y.-G.; Adams, J.; et al. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 2018, 6, 27. [Google Scholar] [CrossRef]
- Gianello, C.; Bremner, J.M. Comparison of chemical methods of assessing potentially available organic nitrogen in soil. Commun. Soil Sci. Plant Anal. 1986, 17, 215–236. [Google Scholar] [CrossRef]
- Guan, Y.S. Soil Enzyme and Its Study Method; China Agriculture Press: Beijing, China, 1986; pp. 274–320. (In Chinese) [Google Scholar]
- Zhou, L.K. Soil Enzymology; Science Press: Beijing, China, 1987. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Ma, J.; Lu, Y.; Chen, F.; Li, X.; Xiao, D.; Wang, H. Molecular Ecological Network Complexity Drives Stand Resilience of Soil Bacteria to Mining Disturbances among Typical Damaged Ecosystems in China. Microorganisms 2020, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.J.; David, A.S.; Menges, E.S.; Searcy, C.A.; Afkhami, M.E. Environmental stress destabilizes microbial networks. ISME J. 2021, 15, 1722–1734. [Google Scholar] [CrossRef]
- Herren, C.M.; McMahon, K.D. Cohesion: A method for quantifying the connectivity of microbial communities. ISME J. 2017, 11, 2426–2438. [Google Scholar] [CrossRef]
- Zhang, S.; Kong, J.; Chen, L.; Guo, K.; Zhou, X. Increased Tea Saponin Content Influences the Diversity and Function of Plantation Soil Microbiomes. Microbiol. Spectr. 2022, 10, e02324-21. [Google Scholar] [CrossRef]
- Upton, R.N.; Sielaff, A.C.; Hofmockel, K.S.; Xu, X.; Polley, H.W.; Wilsey, B.J. Soil depth and grassland origin cooperatively shape microbial community co-occurrence and function. Ecosphere 2020, 11, e02973. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Fahey, C.; Koyama, A.; Antunes, P.M.; Dunfield, K.; Flory, S.L. Plant communities mediate the interactive effects of invasion and drought on soil microbial communities. ISME J. 2020, 14, 1396–1409. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.P.; Wang, S.L. Coupling relationship of structure and tree diversity between upper and lower canopy layer based on structural equation model. J. Beijing For. Univ. 2015, 37, 9–16. [Google Scholar] [CrossRef]
- Muscolo, A.; Sidari, M.; Bagnato, S.; Mallamaci, C.; Mercurio, R. Gap size effects on above- and below-ground processes in a silver fir stand. Eur. J. For. Res. 2009, 129, 355–365. [Google Scholar] [CrossRef]
- Lu, X.; Xu, N.; Chen, Y.; Li, Y.; Gan, X. Effects of Light Intensity and Ground Cover on Seedling Regeneration of Tetracentron sinense Oliv. J. Plant Growth Regul. 2020, 40, 736–748. [Google Scholar] [CrossRef]
- Abebe, A. Effect of soil factors on net N-mineralization and decomposition rate of organic nutrient sources. J. Soil Sci. Environ. Manag. 2018, 9, 59–67. [Google Scholar] [CrossRef]
- Hartman, W.H.; Richardson, C.J.; Vilgalys, R.; Bruland, G.L. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc. Natl. Acad. Sci. USA 2008, 105, 17842–17847. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Yergeau, E.; Wong, L.C.; Pijl, A.S.; Van Veen, J.A.; Kowalchuk, G.A. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 2011, 79, 12–24. [Google Scholar] [CrossRef]
- Ugarelli, K.; Laas, P.; Stingl, U. The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites. Microorganisms 2018, 7, 4. [Google Scholar] [CrossRef]
- Zhang, X.; Johnston, E.R.; Barberán, A.; Ren, Y.; Wang, Z.; Han, X. Effect of intermediate disturbance on soil microbial functional diversity depends on the amount of effective resources. Environ. Microbiol. 2018, 20, 3862–3875. [Google Scholar] [CrossRef]
- Piwosz, K.; Vrdoljak, A.; Frenken, T.; González-Olalla, J.M.; Šantić, D.; McKay, R.M.; Spilling, K.; Guttman, L.; Znachor, P.; Mujakić, I.; et al. Light and Primary Production Shape Bacterial Activity and Community Composition of Aerobic Anoxygenic Phototrophic Bacteria in a Microcosm Experiment. mSphere 2020, 5, e00354-20. [Google Scholar] [CrossRef]
- Ali, R.S.; Poll, C.; Kandeler, E. Dynamics of soil respiration and microbial communities: Interactive controls of temperature and substrate quality. Soil Biol. Biochem. 2018, 127, 60–70. [Google Scholar] [CrossRef]
- Latif, Z.A.; Blackburn, G.A. The effects of gap size on some microclimate variables during late summer and autumn in a temperate broadleaved deciduous forest. Int. J. Biometeorol. 2009, 54, 119–129. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.L.; Gibbons, S.; Owens, S.M.; Hampton-Marcell, J.; Johnston, E.; Jastrow, J.; Gilbert, J.A.; Meyer, F.; Antonopoulos, D.A. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 2016, 18, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xue, L.; Su, Z. Impacts of Forest Gaps on Soil Properties after a Severe Ice Storm in a Cunninghamia lanceolata Stand. Pedosphere 2016, 26, 408–416. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Trojan, D.; Roux, S.; Herbold, C.; Rattei, T.; Woebken, D. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 2018, 20, 1041–1063. [Google Scholar] [CrossRef]
- Yuste, J.C.; Barba, J.; Fernandez-Gonzalez, A.J.; Fernandez-Lopez, M.; Mattana, S.; Martinez-Vilalta, J.; Nolis, P.; Lloret, F. Changes in soil bacterial community triggered by drought-induced gap succession preceded changes in soil C stocks and quality. Ecol. Evol. 2012, 2, 3016–3031. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant Acidification in Major Chinese Croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef]
- Ullah, S.; He, P.; Ai, C.; Zhao, S.; Ding, W.; Song, D.; Zhang, J.; Huang, S.; Abbas, T.; Zhou, W. How Do Soil Bacterial Diversity and Community Composition Respond under Recommended and Conventional Nitrogen Fertilization Regimes? Microorganisms 2020, 8, 1193. [Google Scholar] [CrossRef]
- Zhao, H.; Li, X.; Zhang, Z.; Yang, J.; Zhao, Y.; Yang, Z.; Hu, Q. Effects of natural vegetative restoration on soil fungal and bacterial communities in bare patches of the southern Taihang Mountains. Ecol. Evol. 2019, 9, 10432–10441. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Chang. Biol. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Ling, N.; Chen, D.; Guo, H.; Wei, J.; Bai, Y.; Shen, Q.; Hu, S. Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 2017, 292, 25–33. [Google Scholar] [CrossRef]
- Turner, B.L.; Engelbrecht, B.M.J. Soil organic phosphorus in lowland tropical rain forests. Biogeochemistry 2010, 103, 297–315. [Google Scholar] [CrossRef]
- Jankowiak, J.G.; Gobler, C.J. The Composition and Function of Microbiomes Within Microcystis Colonies Are Significantly Different Than Native Bacterial Assemblages in Two North American Lakes. Front. Microbiol. 2020, 11, 1016. [Google Scholar] [CrossRef]
- Dahroug, Z.; Santana, N.F.; Pagioro, T.A. Eichhornia azurea decomposition and the bacterial dynamic: An experimental research. Braz. J. Microbiol. 2016, 47, 279–286. [Google Scholar] [CrossRef]
- Ji, L.; Shen, F.; Liu, Y.; Yang, Y.; Wang, J.; Purahong, W.; Yang, L. Contrasting altitudinal patterns and co-occurrence networks of soil bacterial and fungal communities along soil depths in the cold-temperate montane forests of China. Catena 2021, 209, 105844. [Google Scholar] [CrossRef]
 : Gap border trees
: Gap border trees  : Regeneration individual in L. principis-rupprechtii plantations
: Regeneration individual in L. principis-rupprechtii plantations  : Acquisition points for hemispherical photographs. Abbreviations: A, expanded gap: crown projection of border trees. B, canopy gap: vertical projection of the canopy opening. C = center point of the gap; N = northern transition; E = eastern transition; S = southern transition; W = western transition; NE = northeast; SE = southeast; NW = northwest; SW = southwest.
: Acquisition points for hemispherical photographs. Abbreviations: A, expanded gap: crown projection of border trees. B, canopy gap: vertical projection of the canopy opening. C = center point of the gap; N = northern transition; E = eastern transition; S = southern transition; W = western transition; NE = northeast; SE = southeast; NW = northwest; SW = southwest.
 : Gap border trees
: Gap border trees  : Regeneration individual in L. principis-rupprechtii plantations
: Regeneration individual in L. principis-rupprechtii plantations  : Acquisition points for hemispherical photographs. Abbreviations: A, expanded gap: crown projection of border trees. B, canopy gap: vertical projection of the canopy opening. C = center point of the gap; N = northern transition; E = eastern transition; S = southern transition; W = western transition; NE = northeast; SE = southeast; NW = northwest; SW = southwest.
: Acquisition points for hemispherical photographs. Abbreviations: A, expanded gap: crown projection of border trees. B, canopy gap: vertical projection of the canopy opening. C = center point of the gap; N = northern transition; E = eastern transition; S = southern transition; W = western transition; NE = northeast; SE = southeast; NW = northwest; SW = southwest.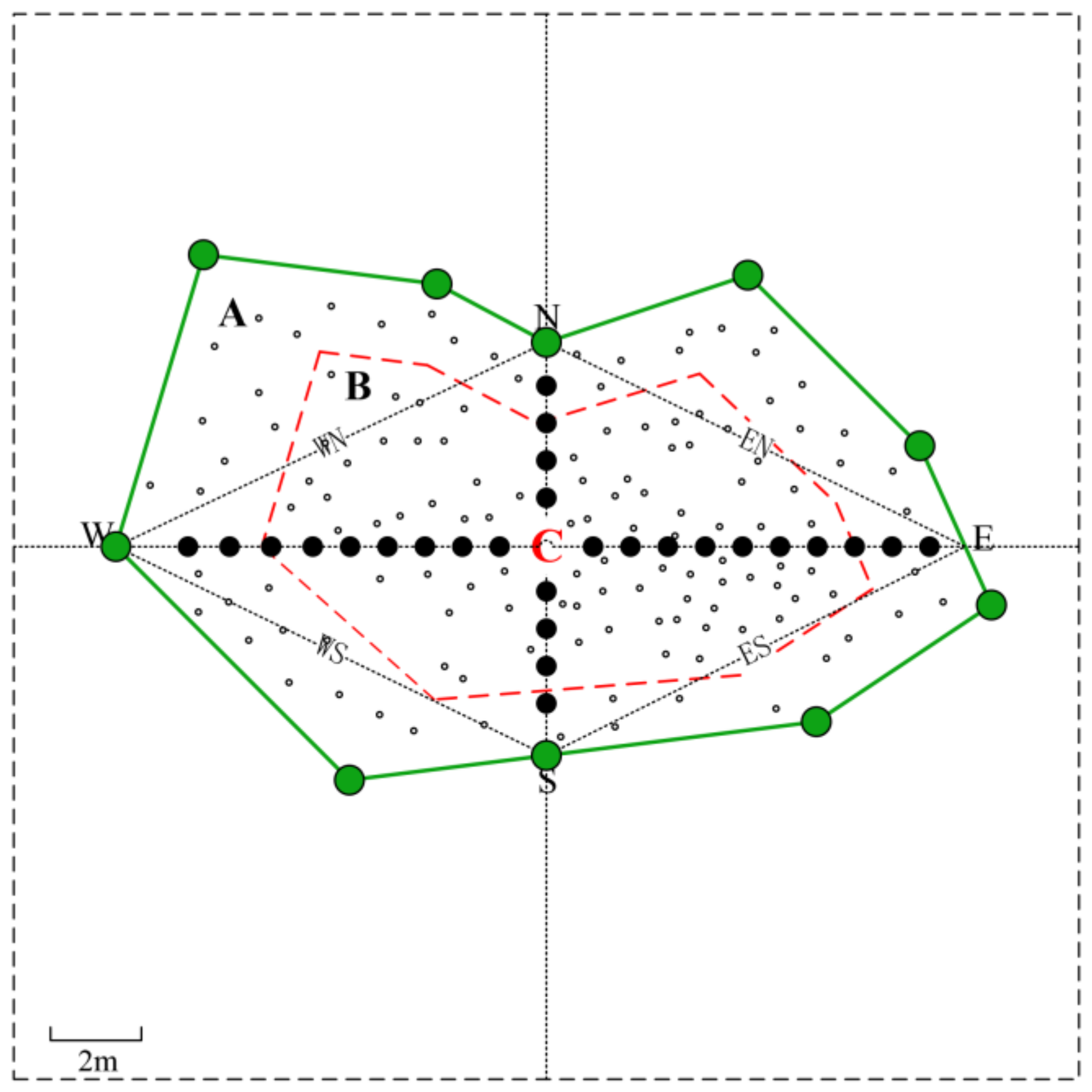


| Type of Gap | Expand Gap Size (m2) | Altitude (m) | Slope (°) | Number of Gap Border Trees | Mean DBH of Border Trees | Mean Age of Regenerated Individuals(a) | Number of Regenerated Individuals (Stems per Gap/Closed Forest) | Regenerated Individual Density (Stems per ha) |
|---|---|---|---|---|---|---|---|---|
| G50 (50–70 m2) | 63.2 ± 7.7 a | 2053 ± 15.0 | 20–22 | 6 ± 1 a | 26.5 ± 7.93 ab | 4.6 ± 2.5 ab | 115 c | 18,367 d |
| G70 (70–100 m2) | 73.4 ± 3.2 ab | 2042 ± 11.0 | 20–23 | 9 ± 2 ab | 21.2 ± 10.24 a | 4.4 ± 2.5 a | 91 bc | 12,354 c |
| G100 (100–125 m2) | 115.7 ± 8.1 ab | 2066 ± 3.9 | 19–21 | 6 ± 1 a | 33.5 ± 1.83 b | 4.9 ± 3.4 ab | 164 d | 14,302 c |
| G125 (125–150 m2) | 137.7 ± 6.3 c | 2056 ± 18.5 | 19–23 | 11 ± 2 c | 19.4 ± 6.79 a | 8.0 ± 0.5 c | 74 ab | 5405 b |
| G150 (150–250 m2) | 173.5 ± 21.6 d | 2061 ± 10.6 | 19–22 | 7 ± 2 a | 34.2 ± 2.3 b | 4.8 ± 1.9 ab | 77 b | 4009 ab |
| G250 (>250 m2) | 330.6 ± 29.6 e | 2066 ± 24.9 | 22–26 | 10 ± 3 ab | 32.9 ± 3.9 b | 6.5 ± 0.3 c | 49 a | 1463 a |
| G0 (10 m × 10 m) | - | 2032 ± 5.9 | 21–24 | - | - | 7.1 ± 1.0 c | 141 cd | 3533 ab |
| Group | CO (%) | Utot (W/m2) | Udir (W/m2) | Udif (W/m2) | LAI | RLA (%) |
|---|---|---|---|---|---|---|
| G0 | 22.45 ± 2.69 a | 3.19 ± 0.66 a | 2.9 ± 0.57 a | 6.1 ± 1.35 a | 2.30 ± 0.09 a | 19.68 ± 3.86 a |
| G50 | 43.16 ± 2.32 b | 5.79 ± 0.57 b | 8.22 ± 0.49 b | 14.01 ± 1.17 bc | 1.01 ± 0.07 b | 47.98 ± 3.34 b |
| G70 | 48.54 ± 2.33 bc | 7.41 ± 0.54 bc | 8.92 ± 0.48 bc | 16.34 ± 1.16 bcd | 0.79 ± 0.07 bc | 57.5 ± 3.3 bc |
| G100 | 52.01 ± 2.69 c | 7.59 ± 0.57 bc | 9.64 ± 0.49 bc | 17.24 ± 1.17 cd | 0.71 ± 0.06 cd | 60.62 ± 3.1 c |
| G125 | 51.55 ± 2.33 c | 8.01 ± 0.53 c | 10.13 ± 0.49 cd | 18.15 ± 1.16 d | 0.66 ± 0.09 cd | 63.49 ± 3.2 c |
| G150 | 48.19 ± 2.69 bc | 6.68 ± 0.65 bc | 10.31 ± 0.57 cd | 13.27 ± 1.35 b | 0.81 ± 0.08 bc | 55.21 ± 3.86 bc |
| G250 | 62.16 ± 2.69 d | 7.66 ± 0.68 bc | 11.56 ± 0.57 d | 19.21 ± 1.35 d | 0.47 ± 0.08 d | 74.36 ± 3.8 d |
| p value | 0.005 ** | 0.02 * | 0.024 * | 0.037 * | 0.078 | 0.026 * |
| Group | SW (%) | ST (°C) | EC (mS/m) | SOM (g/kg) | AN (mg/kg) | AP (mg/kg) | AK (mg/kg) | PHO (mg) | URE (mg) | INV (mg) |
|---|---|---|---|---|---|---|---|---|---|---|
| G0 | 13.95 ± 1.39 ab | 17.06 ± 0.98 a | 69.15 ± 4.76 ab | 18.66 ± 1.23 ab | 60.9 ± 11.63 a | 5.13 ± 0.38 a | 138.24 ± 36.65 ab | 29.59 ± 9.71 a | 151.41 ± 35.03 ab | 2.93 ± 0.74 a |
| G50 | 13.78 ± 1.2 ab | 17.73 ± 0.85 a | 82.12 ± 4.12 bc | 19.89 ± 1.19 ab | 59.85 ± 10.07 a | 4.93 ± 0.33 a | 106.51 ± 31.74 a | 27.12 ± 8.41 a | 110.77 ± 32.94 ab | 2.36 ± 0.64 a |
| G70 | 14.86 ± 1.3 ab | 17.41 ± 0.81 a | 76.29 ± 4.03 abc | 22.12 ± 1.03 bc | 60.55 ± 10.01 a | 5.09 ± 0.31 a | 138.36 ± 31.62 ab | 26.44 ± 8.44 a | 124.12 ± 29.65 ab | 2.96 ± 0.64 a |
| G100 | 16.12 ± 1.22 bc | 17.24 ± 0.8 a | 75.31 ± 4.09 abc | 24.46 ± 1.16 c | 69.3 ± 9.06 a | 5.11 ± 0.33 a | 147.7 ± 31.74 ab | 34.11 ± 8.6 a | 151.26 ± 30.87 ab | 2.26 ± 0.63 a |
| G125 | 12.47 ± 1.17 ab | 16.48 ± 0.86 a | 76.34 ± 4.23 abc | 20.66 ± 1.07 b | 76.65 ± 10.03 ab | 5.63 ± 0.34 a | 244.16 ± 30.28 b | 43.88 ± 9.03 a | 194.34 ± 38.22 b | 2.75 ± 0.52 a |
| G150 | 11.66 ± 1.39 a | 17.18 ± 0.98 a | 66.5 ± 4.76 a | 16.47 ± 1.37 a | 85.87 ± 11.63 ab | 5.43 ± 0.39 a | 84.79 ± 26.65 a | 23.16 ± 9.7 a | 87.27 ± 28.42 a | 2.35 ± 0.73 a |
| G250 | 18.55 ± 1.4 c | 18.9 ± 0.99 a | 84.47 ± 4.86 c | 18.12 ± 1.21 ab | 104.3 ± 11.51 b | 4.75 ± 0.41 a | 148.16 ± 36.65 ab | 71.28 ± 9.73 b | 133.72 ± 22.41 ab | 5.05 ± 0.74 b |
| p value | 0.002 ** | 0.695 | 0.101 | 0.016 * | 0.064 | 0.661 | 0.037 * | 0.004 ** | 0.283 | 0.031 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Y.; Liang, W.; Wei, X.; Han, Y. Forest Gaps Modulate the Composition and Co-Occurrence Network of Soil Bacterial Community in Larix principis-rupprechtii Mayr Plantation. Agronomy 2023, 13, 38. https://doi.org/10.3390/agronomy13010038
Niu Y, Liang W, Wei X, Han Y. Forest Gaps Modulate the Composition and Co-Occurrence Network of Soil Bacterial Community in Larix principis-rupprechtii Mayr Plantation. Agronomy. 2023; 13(1):38. https://doi.org/10.3390/agronomy13010038
Chicago/Turabian StyleNiu, Yajie, Wenjun Liang, Xi Wei, and Youzhi Han. 2023. "Forest Gaps Modulate the Composition and Co-Occurrence Network of Soil Bacterial Community in Larix principis-rupprechtii Mayr Plantation" Agronomy 13, no. 1: 38. https://doi.org/10.3390/agronomy13010038
APA StyleNiu, Y., Liang, W., Wei, X., & Han, Y. (2023). Forest Gaps Modulate the Composition and Co-Occurrence Network of Soil Bacterial Community in Larix principis-rupprechtii Mayr Plantation. Agronomy, 13(1), 38. https://doi.org/10.3390/agronomy13010038





