Effects of the Continuous Cropping of Amomum villosum on Rhizosphere Soil Physicochemical Properties, Enzyme Activities, and Microbial Communities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiments and Soil Sampling
2.2. Analysis of Soil Physicochemical Properties
2.3. Analysis of Soil Enzyme Activities
2.4. DNA Extraction and MiSeq Sequencing
2.5. Bioinformatic Analyses
2.6. Statistical Analyses
3. Results
3.1. Soil Physicochemical Properties
3.2. Enzyme Activities
3.3. Description of the Bacterial and Fungal Communities
3.4. Microbial Diversity
3.4.1. Alpha Diversity Indices of the Bacterial and Fungal Communities
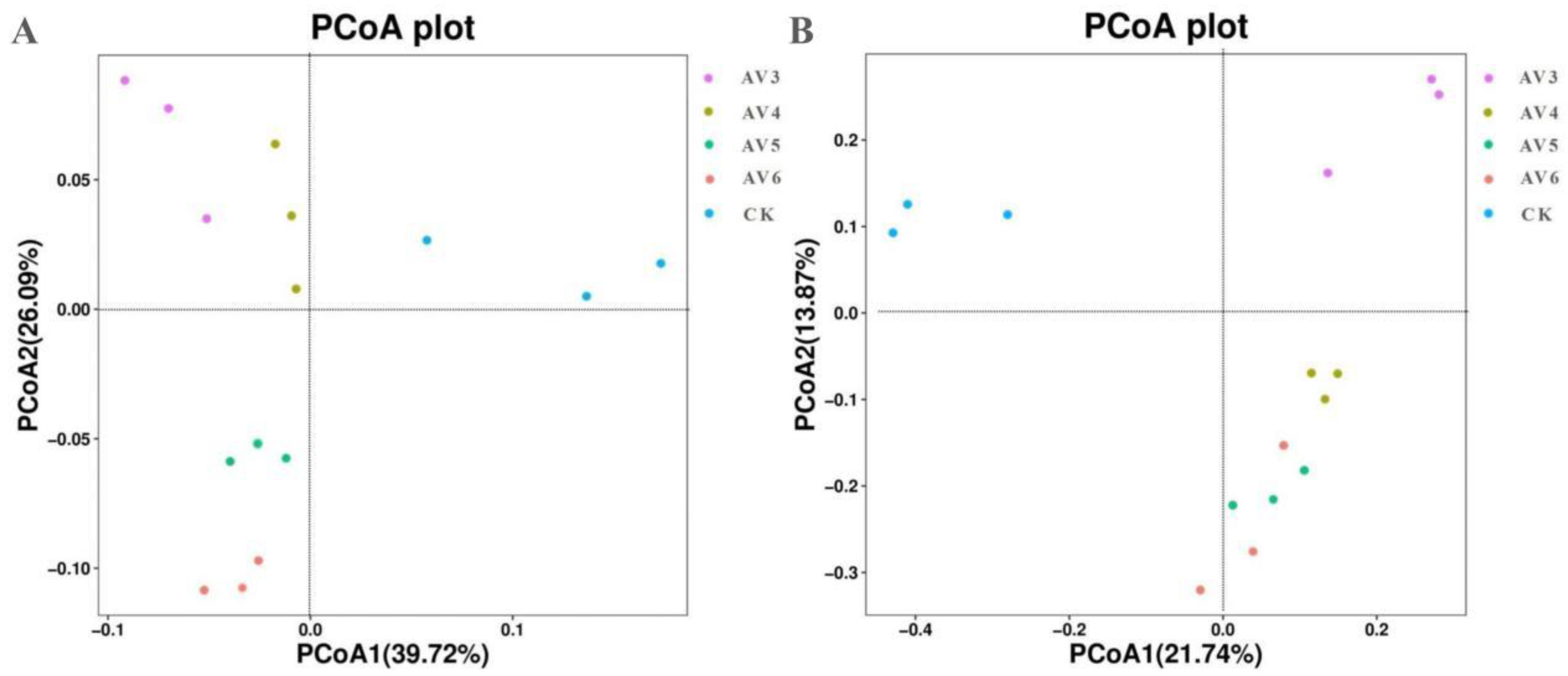
3.4.2. Beta Diversity Indices of the Bacterial and Fungal Communities
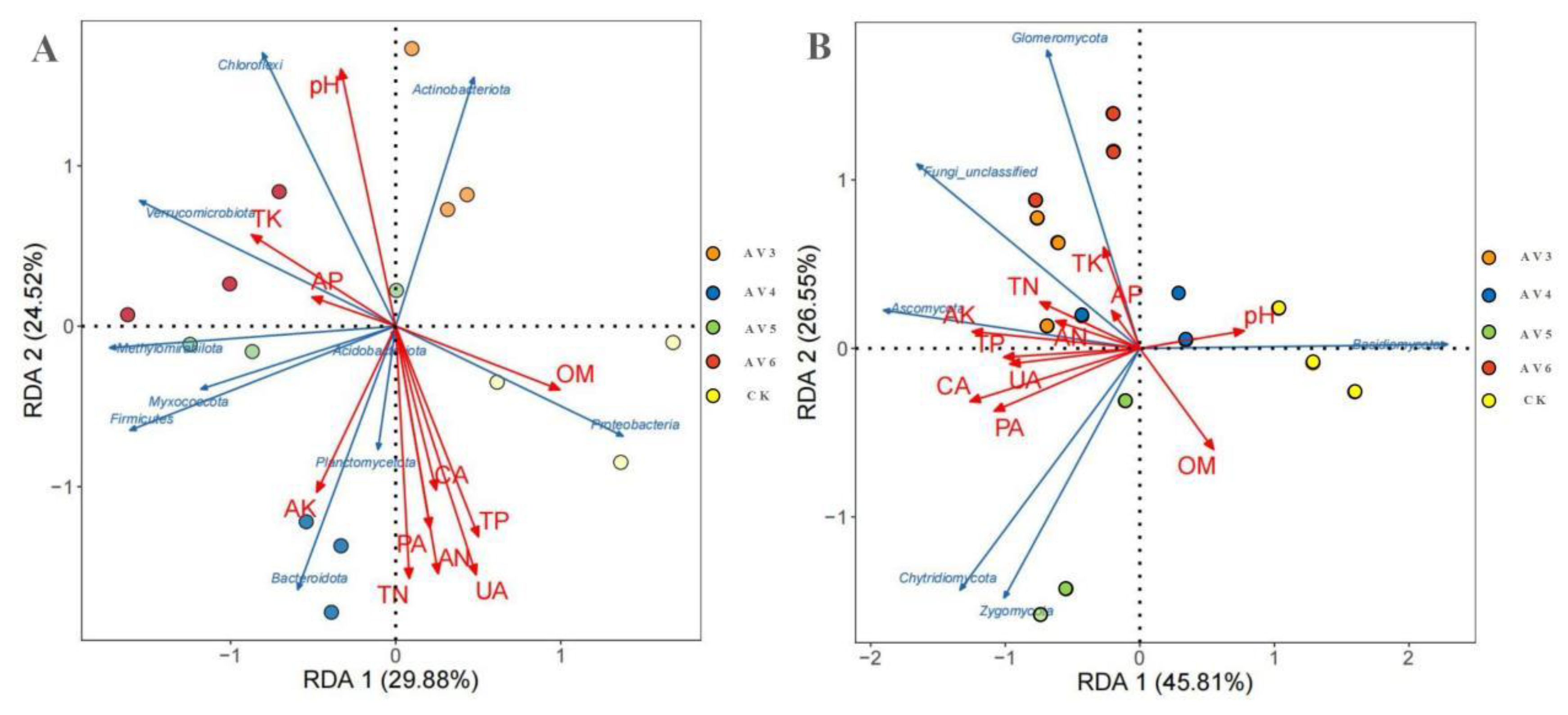
3.5. Effects of Environmental Factors on Bacterial and Fungal Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, Y.; Chen, X.; Nie, L.; Sun, W.; Hu, H.; Lin, Y.; Li, H.; Zheng, X.; Song, J.; Yao, H. Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int. J. Mol. Sci. 2019, 20, 4040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doh, E.J.; Kim, J.-H.; Lee, G. Identification and monitoring of Amomifructus and its adulterants based on DNA barcoding analysis and designed DNA markers. Molecules 2019, 24, 4193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-L.; Lee, S.-K.; Min, D.-E.; Choi, B.-K.; Lee, D.-R. Anti-obesity effects of a mixture of Atractylodes macrocephala and Amomum villosum extracts on 3T3-L1 adipocytes and high-fat diet-induced obesity in mice. Molecules 2022, 27, 906. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, X.; Wang, X.; Shi, D.; Zhang, S.; Yin, Y.; Zhang, H.; Liu, B.; Song, N.; Zhang, Y. Vanillic acid as a promising xanthine oxidase inhibitor: Extraction from Amomum villosum Lour and biocompatibility improvement via extract nanoemulsion. Foods 2022, 11, 968. [Google Scholar] [CrossRef]
- Cho, J.-H.; Lee, J.-S.; Kim, H.-G.; Lee, H.W.; Fang, Z.; Kwon, H.-H.; Kim, D.W.; Lee, C.-M.; Jeong, J.-W. Ethyl acetate fraction of Amomum villosum var. xanthioides attenuates hepatic endoplasmic reticulum stress-induced non-alcoholic steatohepatitis via improvement of antioxidant capacities. Antioxidants 2021, 10, 998. [Google Scholar] [CrossRef] [PubMed]
- Ao, H.; Wang, J.; Chen, L.; Li, S.; Dai, C. Comparison of volatile oil between the fruits of Amomum villosum Lour. and Amomum villosum Lour. var. xanthioides T. L. Wu et Senjen based on GC-MS and chemometric techniques. Molecules 2019, 24, 1663. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Duan, Z.; Yang, J.; Ma, X.; Zhan, R.; Xu, H.; Chen, W. SNP typing for germplasm identification of Amomum villosum Lour. based on DNA barcoding markers. PLoS ONE 2014, 9, e114940. [Google Scholar] [CrossRef] [Green Version]
- Alami, M.M.; Xue, J.; Ma, Y.; Zhu, D.; Gong, Z.; Shu, S.; Wang, X. Structure, Diversity, and composition of bacterial communities in rhizospheric soil of Coptis chinensis Franch under continuously cropped fields. Diversity 2020, 12, 57. [Google Scholar] [CrossRef] [Green Version]
- Alami, M.M.; Xue, J.; Ma, Y.; Zhu, D.; Abbas, A.; Gong, Z.; Wang, X. Structure, function, diversity, and composition of fungal communities in rhizospheric soil of Coptis chinensis Franch under a successive cropping system. Plants 2020, 9, 244. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; He, Y.; Zhou, W.; Ai, L.; Liu, H.; Chen, L.; Xie, Y. Effects of continuous cropping of Codonopsis tangshen on rhizospheric soil bacterial community as determined by pyrosequencing. Diversity 2021, 13, 317. [Google Scholar] [CrossRef]
- Tang, L.; He, G.; Su, J.; Xu, H. The strategy to promote the development of industry of genuine medicinal material of Amomum villosum. Chin. Agric. Sci. Bull. 2012, 28, 94–99. [Google Scholar]
- Cheng, J.; Jin, H.; Zhang, J.; Xu, Z.; Yang, X.; Liu, H.; Xu, X.; Min, D.; Lu, D.; Qin, B. Effects of allelochemicals, soil enzyme activities, and environmental factors on rhizosphere soil microbial community of Stellera chamaejasme L. along a growth-coverage gradient. Microorganisms 2022, 10, 158. [Google Scholar] [CrossRef]
- Shi, R.; Gu, H.; He, S.; Xiong, B.; Huang, Y.; Horowitz, A.R.; He, X. Comparative metagenomic and metabolomic profiling of rhizospheres of Panax notoginseng grown under forest and field conditions. Agronomy 2021, 11, 2488. [Google Scholar] [CrossRef]
- Bogati, K.; Walczak, M. The impact of drought stress on soil microbial community, enzyme activities and plants. Agronomy 2022, 12, 189. [Google Scholar] [CrossRef]
- Muhammad, I.; Lv, J.Z.; Yang, L.; Ahmas, S.; Farooq, S.; Zeeshan, M.; Zhou, X.B. Low irrigation water minimizes the nitrate nitrogen losses without compromising the soil fertility, enzymatic activities and maize growth. BMC Plant Biol. 2022, 22, 159. [Google Scholar] [CrossRef]
- Bueis, T.; Turrión, M.B.; Bravo, F.; Pando, V.; Muscolo, A. Factors determining enzyme activities in soils under Pinus halepensis and Pinus sylvestris plantations in Spain: A basis for establishing sustainable forest management strategies. Ann. For. Sci. 2018, 75, 34. [Google Scholar] [CrossRef] [Green Version]
- Alami, M.M.; Pang, Q.; Gong, Z.; Yang, T.; Tu, D.; Zhen, O.; Yu, W.; Alami, M.J.; Wang, X. Continuous cropping changes the composition and diversity of bacterial communities: A meta-analysis in nine different fields with different plant cultivation. Agriculture 2021, 11, 1224. [Google Scholar] [CrossRef]
- Ishida, J.K.; Bini, A.P.; Creste, S.; Sluys, M.V. Towards defining the core Saccharum microbiome: Input from five genotypes. BMC Microbiol. 2022, 22, 193. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Firl, A.; Hamran, H.; Imaniar, N.I.; Crow, T.M.; Forbes, S.J. Impacts of shade trees on the adjacent cacao rhizosphere in a young diversified agroforestry system. Agronomy 2022, 12, 195. [Google Scholar] [CrossRef]
- Chen, J.; Huang, X.; Tong, B.; Wang, D.; Liu, J.M.; Liao, X.F.; Sun, Q.W. Effects of rhizosphere fungi on the chemical composition of fruits of the medicinal plant Cinnamomum migao endemic to southwestern China. BMC Microbiol. 2021, 21, 206. [Google Scholar] [CrossRef]
- Zuo, J.; Zu, M.; Liu, L.; Song, X.M.; Yuan, Y.D. Composition and diversity of bacterial communities in the rhizosphere of the Chinese medicinal herb Dendrobium. BMC Plant Biol. 2021, 21, 127. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Zhong, C.F.; Gao, X.; Tan, C.Y.; Bai, H.; Ning, K. Glycyrrhiza uralensis Fisch. Root-associated microbiota: The multifaceted hubs associated with environmental factors, growth status and accumulation of secondary metabolites. Environ. Microbiome 2022, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Yang, N.; Zhao, Y.; Liu, W.H.; Li, T.F. Long-term watermelon continuous cropping leads to drastic shifts in soil bacterial and fungal community composition across gravel mulch fields. BMC Microbiol. 2022, 22, 189. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Xu, J.; Zhang, L.; Yang, J.; Liao, B.S.; Li, X.W.; Chen, S.L. High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin. Med. 2017, 12, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Bian, X.; Zhao, Y.; Yang, H.; Xu, Y.H.; Han, Y.Z.; Zhang, L.X. Rhizosphere analysis of field-grown Panax ginseng with different degrees of red skin provides the basis for preventing red skin syndrome. BMC Microbiol. 2022, 22, 12. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, Z.; He, Y.; Li, G.F.; Lv, X.H.; Zhuang, L. High-throughput sequencing analysis of the rhizosphere arbuscular mycorrhizal fungi (AMF) community composition associated with Ferula sinkiangensis. BMC Microbiol. 2020, 20, 335. [Google Scholar] [CrossRef]
- Bao, S.D. Analysis Method of Soil and Agricultural Chemistry, 3rd ed.; China Agricultural Press: Beijing, China, 2000; pp. 25–108. [Google Scholar]
- Guan, S.Y. Soil Enzyme and Its Study Method; China Agriculture Press: Beijing, China, 1986; pp. 270–285. [Google Scholar]
- Logue, J.B.; Stedmon, C.A.; Kellerman, A.M.; Nielsen, N.J.; Andersson, A.F.; Laudon, H.; Lindstrom, E.S.; Kritzberg, E.S. Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 2016, 10, 533–545. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Matthew, R.D.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Guo, L.; Chen, X.; Li, Z.; Wang, M.; Che, Y.; Zhang, L.; Jiang, Z.; Jie, S. Effects of continuous cropping on bacterial community and diversity in rhizosphere soil of industrial hemp: A five-year experiment. Diversity 2022, 14, 250. [Google Scholar] [CrossRef]
- Xiong, W.; Li, Z.; Liu, H.; Xue, C.; Zhang, R.; Wu, H.; Li, R.; Shen, Q. The Effect of long-term continuous cropping of black pepper on soil bacterial communities as determined by 454 Pyrosequencing. PLoS ONE 2015, 10, e0136946. [Google Scholar] [CrossRef] [Green Version]
- Barak, P.; Jobe, B.O.; Krueger, A.R.; Peterson, L.A.; Laird, D.A. Effects of long-term soil acidification due to nitrogen fertilizer inputs in Wisconsin. Plant Soil 1997, 197, 61–69. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G.; Ravit, B.; Elgersma, K. Feedback in the plant-soil system. Annu. Rev. Environ. Resour. 2005, 30, 75–115. [Google Scholar] [CrossRef] [Green Version]
- Baweja, P.; Kumar, S.; Kumar, G. Fertilizers and Pesticides: Their Impact on Soil Health and Environment; Springer Nature: Berne, Switzerland, 2020; p. 270. [Google Scholar]
- Pang, Z.; Dong, F.; Liu, Q.; Lin, W.; Hu, C.; Yuan, Z. Soil Metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 2021, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Tang, C.X.; Severi, J.; Butterly, C.R.; Baldock, J.A. Rhizosphere priming effect on soil organic carbon decomposition under plant species differing in soil acidification and root exudation. New Phytol. 2016, 211, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Hou, W.; Liu, J.; Malik, K.; Kong, X.; Wang, L.; Chen, X.; Tang, M.; Zhu, R.; Cheng, C.; et al. Effects of different land use types and soil depths on soil mineral elements, soil enzyme activity, and fungal community in karst area of southwest China. Int. J. Environ. Res. Public Health 2022, 19, 3120. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Liu, L.; Zhang, Z.; Ze, S.; Ji, M.; Li, Z.; Yu, J.; Yang, B.; Zhao, N. Do mixed Pinus yunnanensis plantations improve soil’s physicochemical properties and enzyme activities? Diversity 2022, 14, 214. [Google Scholar] [CrossRef]
- Wang, F.; Cui, H.; He, F.; Liu, Q.; Zhu, Q.; Wang, W.; Liao, H.; Yao, D.; Cao, W.; Lu, P. The green manure (Astragalus sinicus L.) improved rice yield and quality and changed soil microbial communities of rice in the karst mountains area. Agronomy 2022, 12, 1851. [Google Scholar] [CrossRef]
- Bian, X.; Yang, X.; Li, Q.; Sun, X. Effects of planting of two common crops, Allium fistulosum and Brassica napus, on soil properties and microbial communities of ginseng cultivation in northeast China. BMC Microbiol. 2022, 22, 182. [Google Scholar] [CrossRef]
- Gupta, R.S.; Mok, A. Phylogenomics and signature proteins for the alpha Proteobacteria and its main groups. BMC Microbiol. 2007, 7, 106. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Sun, X.R.; Feng, J.; Zhu, S.F.; Shui, W. Metagenomic analysis reveals the different characteristics of microbial communities inside and outside the karst tiankeng. BMC Microbiol. 2022, 22, 115. [Google Scholar] [CrossRef]
- Gao, Z.; Hu, Y.; Han, M.; Xu, J.; Wang, X.; Liu, L.; Tang, Z.; Jiao, W.; Jin, R.; Liu, M.; et al. Effects of continuous cropping of sweet potatoes on the bacterial community structure in rhizospheric soil. BMC Microbiol. 2021, 21, 102. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, J.; Baumgartner, V.; Birkhofer, K.; Boeddinghaus, R.S.; Bunk, B.; Fischer, M.; Fösel, B.U.; Friedrich, M.W.; Göker, M.; Hölzel, N.; et al. The evolution of ecological diversity in Acidobacteria. Front. Microbiol. 2022, 13, 715637. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Wu, J.; Hayes, R.B.; Ahn, J.Y. The oral fungal mycobiome: Characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 2017, 17, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badotti, F.; de Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Goes-Neto, A. Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 17, 42. [Google Scholar] [CrossRef] [Green Version]
- Khalil, A.M.A.; Hassan, S.E.-D.; Alsharif, S.M.; Eid, A.M.; Ewais, E.E.-D.; Azab, E.; Gobouri, A.A.; Elkelish, A.; Fouda, A. Isolation and characterization of fungal endophytes isolated from medicinal plant ephedra pachyclada as plant growth-promoting. Biomolecules 2021, 11, 140. [Google Scholar] [CrossRef]
- Coelho, M.A.; Bakkeren, G.; Sun, S.; Hood, M.E.; Giraud, T. Fungal sex: The Basidiomycota. Microbiol. Spectr. 2017, 10, 1128. [Google Scholar]
- Marler, T.E. Soil from Serianthes rhizosphere influences growth and leaf nutrient content of Serianthes plants. Agronomy 2022, 12, 1938. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Yuan, H.; Lv, G. Differed growth stage dynamics of root-associated bacterial and fungal community structure associated with halophytic plant Lycium ruthenicum. Microorganisms 2022, 10, 1644. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, X.; Wu, T.; Dai, W.; Liu, H.; Yang, R. Analysis of unculturable bacterial communities in tea orchard soils based on nested PCR-DGGE. World J. Microbiol. Biotechnol. 2012, 28, 1967–1979. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Jiang, Y.; Weng, B.; Lin, W. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef]
- An, S.; Wei, Y.; Li, H.; Zhao, Z.; Hu, J.; Philp, J.; Ryder, M.; Toh, R.; Li, J.; Zhou, Y.; et al. Long-term monocultures of American ginseng change the rhizosphere microbiome by reducing phenolic acids in soil. Agriculture 2022, 12, 640. [Google Scholar] [CrossRef]
- Manici, L.M.; Kelderer, M.; Franke-Whittle, I.H.; Rühmer, T.; Baab, G.; Nicoletti, F.; Caputo, F.; Topp, A.; Insam, H.; Naef, A. Relationship between root-endophytic microbial communities and replant disease in specialized apple growing areas in Europe. Appl. Soil Ecol. 2013, 72, 207–214. [Google Scholar] [CrossRef]
- Cui, J.; Zhang, E.; Zhang, X.; Wang, Q.; Liu, Q. Effects of 2,4-di-tert-butylphenol at different concentrations on soil functionality and microbial community structure in the Lanzhou lily rhizosphere. Appl. Soil Ecol. 2022, 172, 104367. [Google Scholar] [CrossRef]
- Pang, Z.; Tayyab, M.; Kong, C.; Liu, Q.; Liu, Y.; Hu, C.; Huang, J.; Weng, P.; Islam, W.; Lin, W.; et al. Continuous sugarcane planting negatively impacts soil microbial community structure, soil fertility, and sugarcane agronomic parameters. Microorganisms 2021, 9, 2008. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Cui, Y.; Li, H.; Kuang, A.; Li, X.; Wei, Y.; Ji, X. Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol. Res. 2017, 194, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Xiong, W.; Xing, Y.; Sun, Y.; Lin, X.; Dong, Y. Long-term coffee monoculture alters soil chemical properties and microbial communities. Sci. Rep. 2018, 8, 6116. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, C.; Zhang, T.; Wang, X. Soil biology & biochemistry fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar]

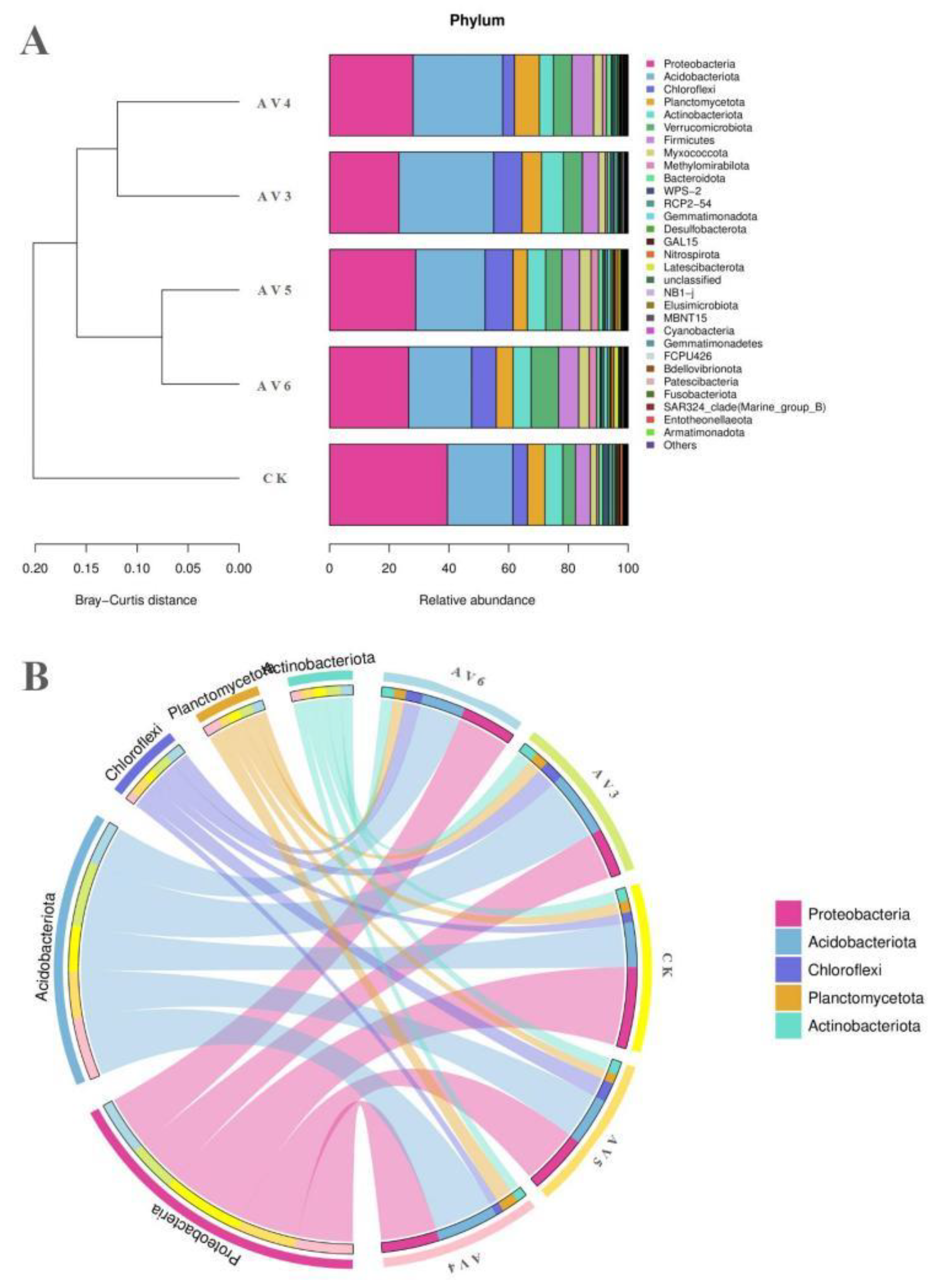
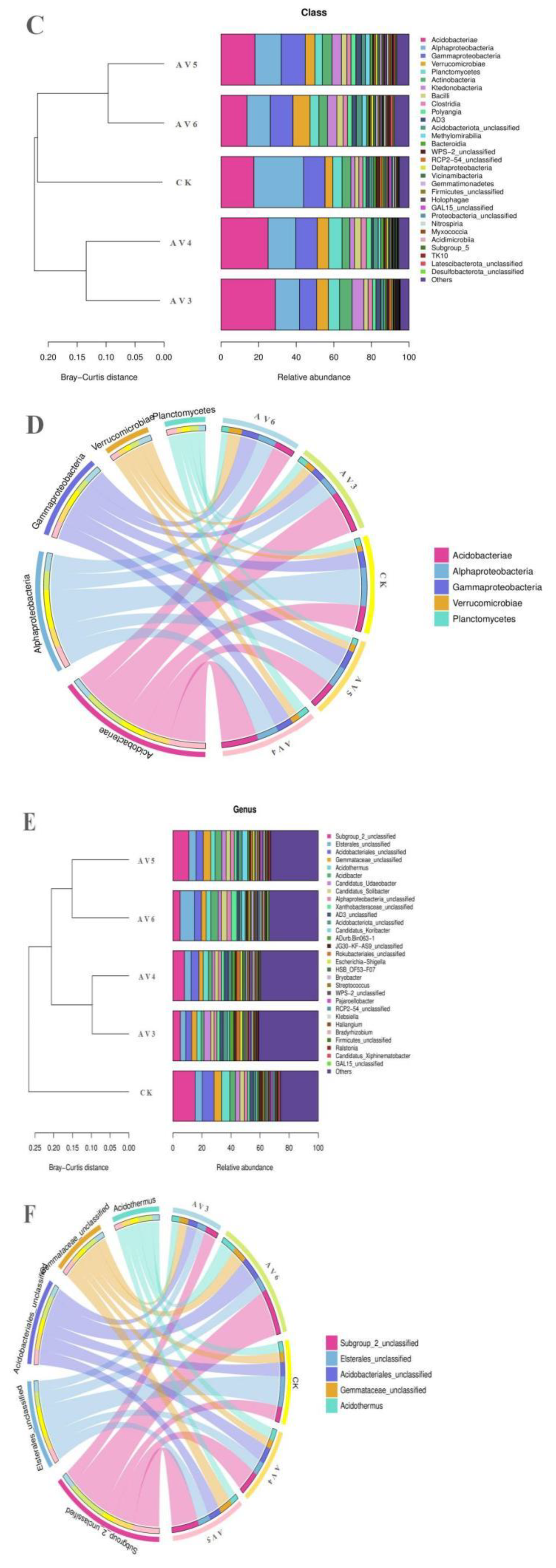
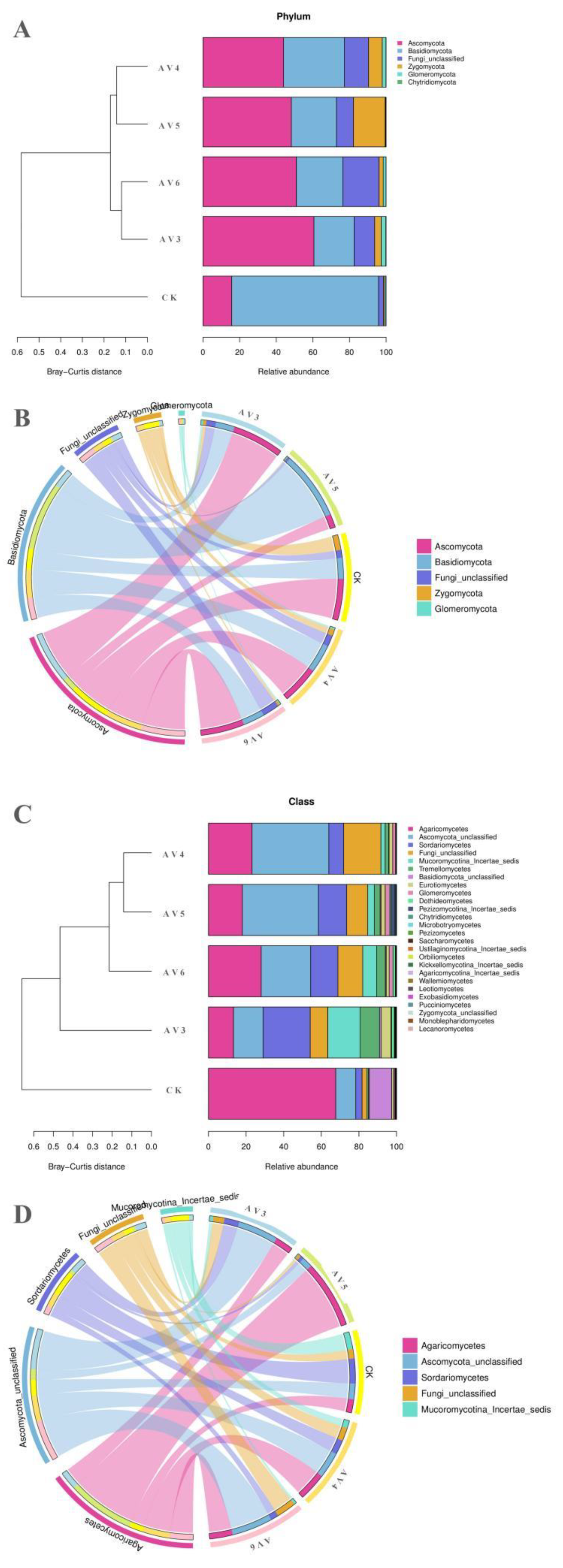
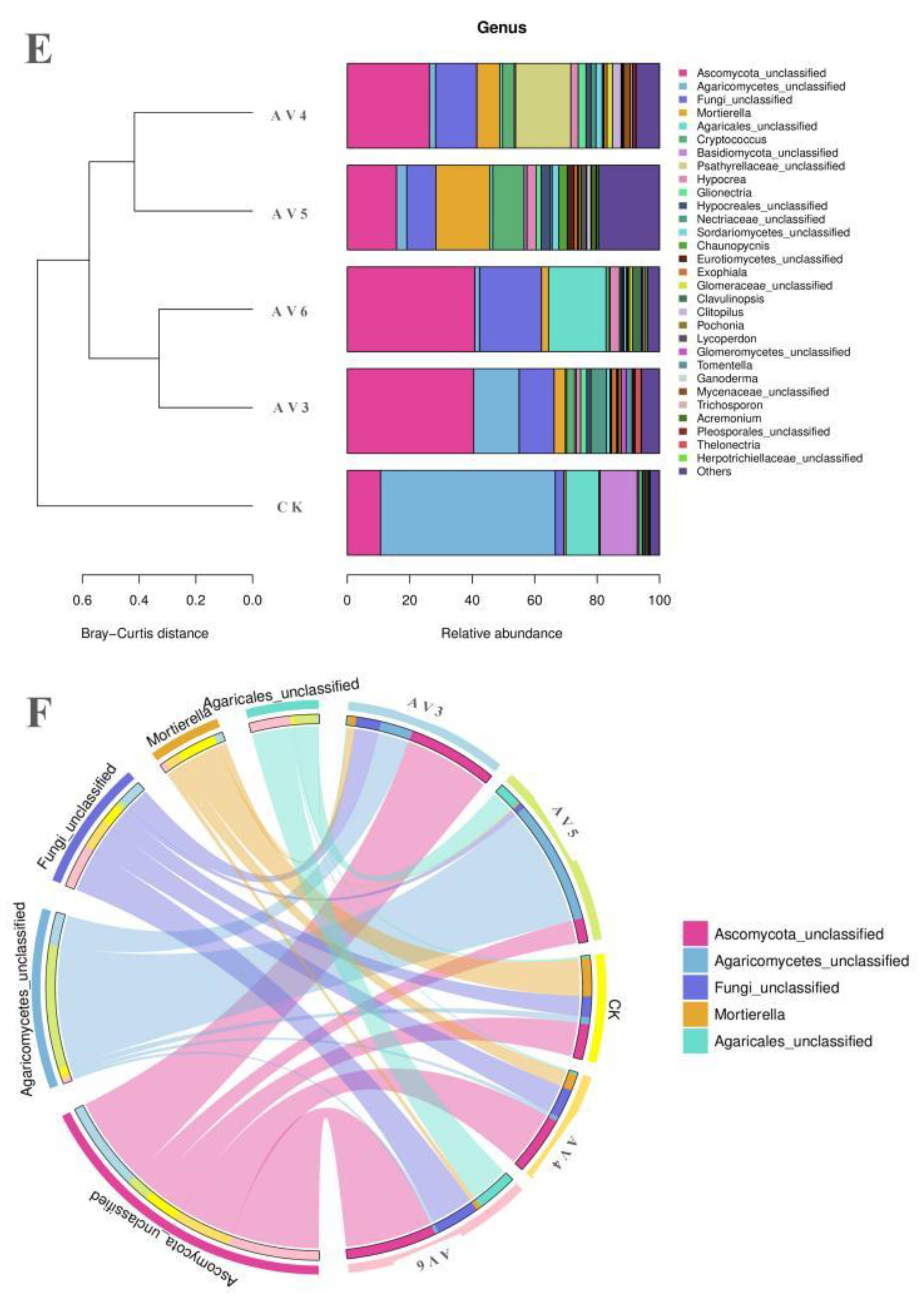
| Samples | pH | OM (g/kg−1) | TN (g/kg−1) | AN (g/kg−1) | TP (g/kg−1) | AP (g/kg−1) | TK (g/kg−1) | AK (g/kg−1) |
|---|---|---|---|---|---|---|---|---|
| AV3 | 4.42 ± 0.12a | 23.50 ± 1.10a | 0.70 ± 0.01a | 107.51 ± 0.55a | 0.70 ± 0.07a | 4.30 ± 0.29a | 4.42 ± 0.06a | 84.02 ± 2.18a |
| AV4 | 4.25 ± 0.01b | 23.50 ± 0.42a | 0.95 ± 0.03b | 142.48 ± 3.13b | 0.78 ± 0.05b | 4.17 ± 0.43b | 4.25 ± 0.11b | 81.96 ± 3.99a |
| AV5 | 4.18 ± 0.03c | 32.33 ± 1.77b | 1.35 ± 0.05c | 177.82 ± 0.68c | 0.89 ± 0.07c | 3.65 ± 0.16c | 3.98 ± 0.12c | 79.62 ± 4.56a |
| AV6 | 4.10 ± 0.02d | 33.43 ± 1.06b | 1.58 ± 0.11d | 341.62 ± 10.72d | 0.94 ± 0.05d | 3.60 ± 0.13c | 3.94 ± 0.25c | 78.98 ± 3.38a |
| CK | 4.36 ± 0.06e | 32.70 ± 1.72b | 0.57 ± 0.01e | 72.38 ± 1.58e | 0.63 ± 0.07e | 3.59 ± 0.11c | 4.02 ± 0.27c | 60.87 ± 4.54b |
| Samples | Urease Activities (mg/d/g) | Acid Phosphatase Activities (μmol/d/g) | Catalase Activities (μmol/d/g) | Sucrase Activities (U/g) |
|---|---|---|---|---|
| AV3 | 0.42 ± 0.03a | 4.36 ± 0.22a | 53.27 ± 1.80a | 235.21 ± 16.00ab |
| AV4 | 0.47 ± 0.02ab | 4.52 ± 0.45b | 52.24 ± 0.73b | 221.63 ± 10.83a |
| AV5 | 0.50 ± 0.02b | 4.84 ± 0.14c | 54.42 ± 0.67c | 252.89 ± 6.29b |
| AV6 | 0.52 ± 0.02b | 5.12 ± 0.32d | 52.70 ± 0.46d | 210.14 ± 9.79a |
| CK | 0.40 ± 0.02a | 3.94 ± 0.19e | 49.97 ± 0.67b | 187.83 ± 7.31c |
| Samples | Observed_OTUs | Shannon | Simpson | Chao1 | Goods_coverage | Pielou_e |
|---|---|---|---|---|---|---|
| AV3 | 1524.67 ± 78.02a | 9.55 ± 0.13a | 1.00 ± 0.00a | 1532.47 ± 77.92a | 1.00 ± 0.00a | 0.90 ± 0.01a |
| AV4 | 1460.00 ± 60.00b | 9.59 ± 0.09a | 1.00 ± 0.00a | 1465.07 ± 58.46b | 1.00 ± 0.00a | 0.91 ± 0.00b |
| AV5 | 1448.50 ± 94.50b | 9.45 ± 0.10a | 1.00 ± 0.00a | 1459.23 ± 87.73b | 1.00 ± 0.00a | 0.90 ± 0.00a |
| AV6 | 1428.33 ± 19.33b | 9.46 ± 0.07a | 1.00 ± 0.00a | 1439.18 ± 25.85b | 1.00 ± 0.00a | 0.90 ± 0.01a |
| CK | 1681.00 ± 39.00c | 9.91 ± 0.05b | 1.00 ± 0.00a | 1686.32 ± 37.63c | 1.00 ± 0.00a | 0.92 ± 0.00c |
| Samples | Observed_OTUs | Shannon | Simpson | Chao1 | Goods_coverage | Pielou_e |
|---|---|---|---|---|---|---|
| AV3 | 969.67 ± 3.03a | 6.98 ± 0.63a | 0.96 ± 0.02a | 971.03 ± 7.85a | 1.00 ± 0.00a | 0.71 ± 0.03a |
| AV4 | 576.00 ± 0.00b | 6.06 ± 0.06b | 0.95 ± 0.00a | 577.04 ± 0.62b | 1.00 ± 0.00a | 0.66 ± 0.01ab |
| AV5 | 439.50 ± 6.50b | 4.95 ± 0.16c | 0.87 ± 0.02b | 439.62 ± 6.47b | 1.00 ± 0.00a | 0.56 ± 0.02b |
| AV6 | 329.33 ± 5.79b | 3.35 ± 0.85d | 0.75 ± 0.07c | 329.98 ± 5.26b | 1.00 ± 0.00a | 0.40 ± 0.02c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Lu, Y.; Li, W.; He, S.; Lin, R.; Qu, P.; Chen, H.; Zhang, F.; Zhao, M.; Shi, X.; et al. Effects of the Continuous Cropping of Amomum villosum on Rhizosphere Soil Physicochemical Properties, Enzyme Activities, and Microbial Communities. Agronomy 2022, 12, 2548. https://doi.org/10.3390/agronomy12102548
Wang B, Lu Y, Li W, He S, Lin R, Qu P, Chen H, Zhang F, Zhao M, Shi X, et al. Effects of the Continuous Cropping of Amomum villosum on Rhizosphere Soil Physicochemical Properties, Enzyme Activities, and Microbial Communities. Agronomy. 2022; 12(10):2548. https://doi.org/10.3390/agronomy12102548
Chicago/Turabian StyleWang, Butian, Yunfeng Lu, Weifeng Li, Suming He, Rong Lin, Peng Qu, Hongmei Chen, Fengying Zhang, Meng Zhao, Xuedong Shi, and et al. 2022. "Effects of the Continuous Cropping of Amomum villosum on Rhizosphere Soil Physicochemical Properties, Enzyme Activities, and Microbial Communities" Agronomy 12, no. 10: 2548. https://doi.org/10.3390/agronomy12102548






