Sources of Resistance to Powdery Mildew in Wild Barley (Hordeum vulgare subsp. spontaneum) Collected in Jordan, Lebanon, and Libya
Abstract
:1. Introduction
2. Materials and Method
2.1. Plant Material
2.2. Pathogen
2.3. Populations and Single Plant Lines Resistance Tests
2.4. Postulation of Resistance Alleles
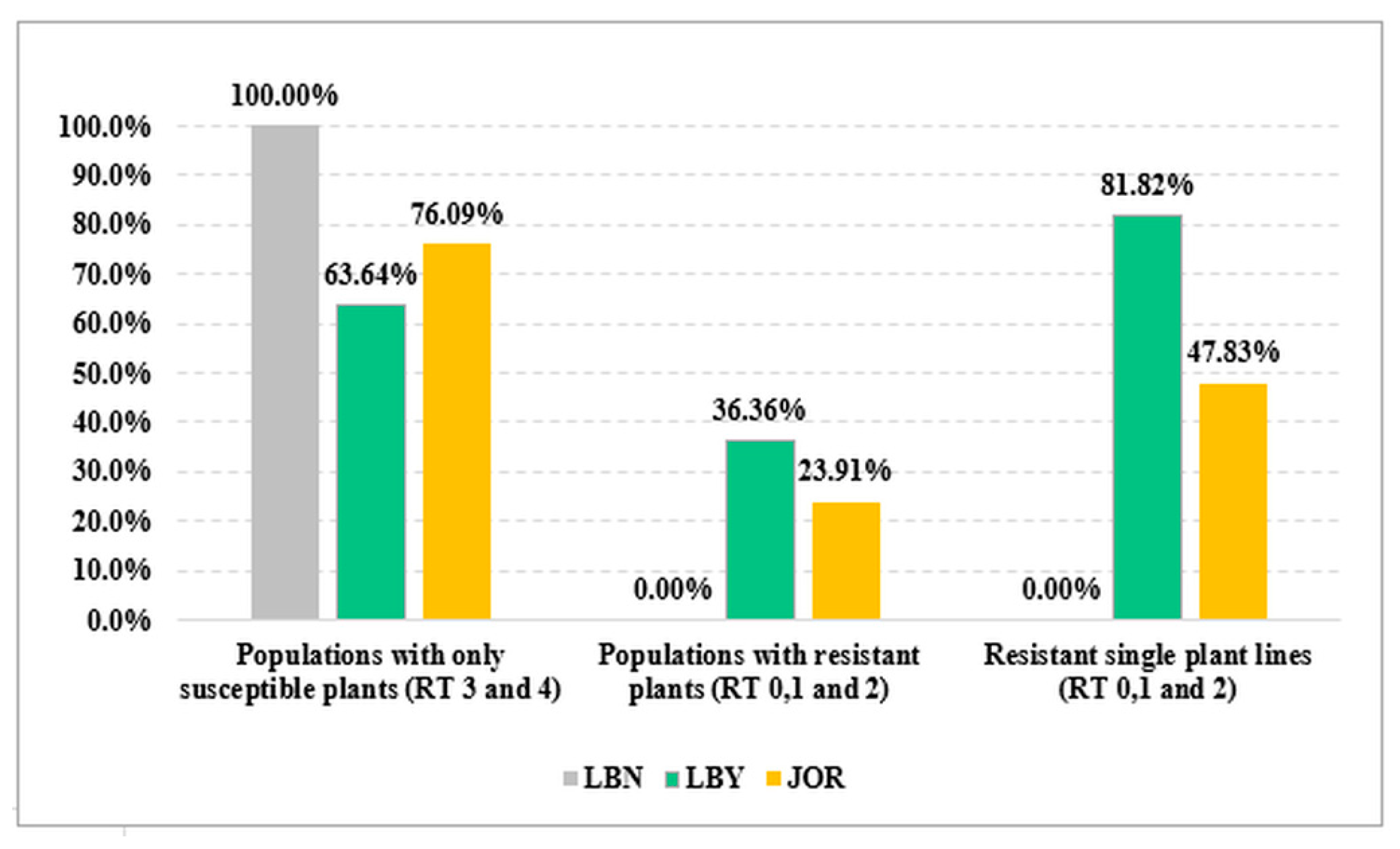
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dai, F.; Zhang, G. Domestication and improvement of cultivated barley. In Exploration, Identification and Utilization of Barley Germplasm; Zhang, L.C., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–26. [Google Scholar] [CrossRef]
- El-Hashash, E.F.; El-Absy, K.M. Barley (Hordeum vulgare L.) Breeding. In Advances in Plant Breeding Strategies: Cereals; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2019; pp. 1–45. [Google Scholar] [CrossRef]
- Dracatos, P.M.; Lu, J.; Sánchez-Martín, J.; Wulff, B.B. Resistance that stacks up: Engineering rust and mildew disease control in the cereal crops wheat and barley. Plant Biotechnol. J. 2023, 21, 1938–1951. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.; Li, M.; Zhang, G.; Ye, L. Barley: A Potential Cereal for Producing Healthy and Functional Foods. Food Qual. Saf. 2022, 6, fyac012. [Google Scholar] [CrossRef]
- Lukinac, J.; Jukić, M. Barley in the Production of Cereal-Based Products. Plants 2022, 11, 3519. [Google Scholar] [CrossRef]
- Newton, A.C.; Akar, T.; Baresel, J.P.; Bebeli, P.J.; Bettencourt, E.; Bladenopoulos, K.V.; Czembor, J.H.; Fasoula, D.A.; Katsiotis, A.; Koutis, K.; et al. Cereal landraces for sustainable agriculture. A review. Agron. Sustain. Dev. 2010, 30, 237–269. [Google Scholar] [CrossRef]
- Weise, S.; Lohwasser, U.; Oppermann, M. Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants 2020, 9, 1050. [Google Scholar] [CrossRef]
- Kaur, V.; Aravind, J.; Jacob, S.R.; Kumari, J.; Panwar, B.S.; Pal, N.; Rana, J.C.; Pandey, A.; Kumar, A. Phenotypic Characterization, Genetic Diversity Assessment in 6778 Accessions of Barley (Hordeum vulgare L. ssp. vulgare) Germplasm Conserved in National Genebank of India and Development of a Core Set. Front. Plant Sci. 2022, 13, 771920. [Google Scholar] [CrossRef]
- Khoury, C.K.; Brush, S.; Costich, D.E.; Curry, H.A.; Haan, S.D.; Engels, J.M.M.; Guarino, L.; Hoban, S.; Mercer, K.L.; Miller, A.J.; et al. Crop genetic erosion: Understanding and responding to loss of crop diversity. New Phytol. 2022, 233, 84–118. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Villegas, J.; Khoury, C.K.; Achicanoy, H.A.; Diaz, M.V.; Mendez, A.C.; Sosa, C.C.; Kehel, Z.; Guarino, L.; Abberton, M.; Aunario, J.K.; et al. State of ex situ conservation of landrace groups of 25 major crops. Nat. Plants 2022, 8, 491–499. [Google Scholar] [CrossRef]
- Brown, J.K.; Wulff, B.B. Diversifying the menu for crop powdery mildew resistance. Cell 2022, 185, 761–763. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E.; Krystek, M.; Pukacki, J. AgroGenome: Interactive Genomic-Based Web Server Developed Based on Data Collected for Accessions Stored in Polish Genebank. Agriculture 2023, 13, 193. [Google Scholar] [CrossRef]
- Knupfer, R. Triticeae genetic resources in ex situ genebank collections. In Genetic and Genomics of the Triticeae. Plant Genetics and Genomics: Crops and Models; Muelbauer, G.J., Feuillet, C., Eds.; Springer Science: New York, NY, USA, 2009; Volume 7, pp. 31–80. [Google Scholar] [CrossRef]
- Shakhatreh, Y.; Haddad, N.; Alrababah, M.; Grando, S.; Ceccarelli, S. Phenotypic diversity in wild barley (Hordeum vulgare L. ssp. spontaneum (C. Koch) Thell.) accessions collected in Jordan. Genet. Res. Crop Evol. 2010, 57, 131–146. [Google Scholar] [CrossRef]
- Thormann, I.; Reeves, P.; Reilley, A.; Engels, J.M.M.; Lohwasser, U.; Börner, A.; Pillen, K.; Richards, C.M. Geography of genetic structure in barley wild relative Hordeum vulgare subsp. spontaneum in Jordan. PLoS ONE 2016, 11, e0160745. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajaj, N.; Grando, S.; Ababnah, M.; Alomari, N.; Albatianh, A.; Nesir, J.; Migdali, H.; Shakhatreh, Y.; Ceccarelli, S. Phenotypic evolution of the wild progenitor of cultivated barley (Hordeum vulgare L. subsp. spontaneum (K. Koch) Thell.) across bioclimatic regions in Jordan. Genet. Resour. Crop. Evol. 2022, 69, 1485–1507. [Google Scholar] [CrossRef]
- Chang, C.W.; Fridman, E.; Mascher, M.; Himmelbach, A.; Schmid, K. Physical geography, isolation by distance and environmental variables shape genomic variation of wild barley (Hordeum vulgare L. ssp. spontaneum) in the Southern Levant. Heredity 2022, 128, 107–119. [Google Scholar] [CrossRef]
- Rehman, S.; Amouzoune, M.; Hiddar, H.; Aberkane, H.; Benkirane, R.; Filali-Maltouf, A.; Al-Jaboobi, M.; Acqbouch, L.; Tsivelikas, A.; Verma, R.P.S.; et al. Traits discovery in Hordeum vulgare sbsp. spontaneum accessions and in lines derived from interspecific crosses with wild Hordeum species for enhancing barley breeding efforts. Crop Sci. 2020, 61, 219–233. [Google Scholar] [CrossRef]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef]
- Capasso, G.; Santini, G.; Petraretti, M.; Esposito, S.; Landi, S. Wild and Traditional Barley Genomic Resources as a Tool for Abiotic Stress Tolerance and Biotic Relations. Agriculture 2021, 11, 1102. [Google Scholar] [CrossRef]
- Clare, S.J.; Oğuz, A.Ç.; Effertz, K.; Poudel, R.S.; See, D.R.; Karakaya, A.; Brueggeman, R.S. Genome-wide association mapping of Pyrenophora teres f. maculata and Pyrenophora teres f. teres resistance loci utilizing natural Turkish wild and landrace barley populations. G3: Genes|Genomes|Genetics, 11. G3 (Bethesda) 2021, 11, jkab269. [Google Scholar] [CrossRef]
- Henningsen, E.C.; Sallam, A.H.; Matny, O.N.; Szinyei, T.; Figueroa, M.; Steffenson, B.J. Rpg7: A New Gene for Stem Rust Resistance from Hordeum vulgare ssp. spontaneum. Phytopathology 2021, 111, 548–558. [Google Scholar] [CrossRef]
- Hovmoller, M.S.; Caffier, V.; Jalli, M.; Andersen, O.; Besenhofer, G.; Czembor, J.H.; Dreiseitl, A.; Flath, K.; Fleck, A.; Heinrics, F.; et al. The Eurpean barley powdery mildew virulence survey and disease nursery 1993–1999. Agonomie 2000, 20, 729–744. [Google Scholar] [CrossRef]
- Mehnaz, M.; Dracatos, P.M.; Park, R.F.; Singh, D. Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust. Agronomy 2021, 11, 2146. [Google Scholar] [CrossRef]
- Mehnaz, M.; Dracatos, P.M.; Pham, A.; March, T.J.; Maurer, A.; Pillen, K.; Forrest, K.L.; Kulkarni, T.V.; Pourkheirandish, M.; Park, R.F.; et al. Discovery and fine mapping of Rph28: A new gene conferring resistance to Puccinia hordei from wild barley. Theor. Appl. Genet. 2021, 134, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Sayed, M.A.; Tarawneh, R.A.; Youssef, H.M.; Pillen, K.; Börner, A. Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines. Plants 2021, 10, 2246. [Google Scholar] [CrossRef]
- Jørgensen, J.H.; Wolfe, M. Genetics of Powdery Mildew Resistance in Barley. Crit. Rev. Plant Sci. 1994, 13, 97–119. [Google Scholar] [CrossRef]
- Weibull, J.; Walther, U.; Sato, K.; Habekuß, A.; Kopahnke, D.; Proeseler, G. Diversity in resistance to biotic stresses. In Diversity in Barley (Hordeum Vulgare); Von Bothmer, R., van Hintum, T., Knüpfer, H., Sato, K., Eds.; Elsevier Science, B.V.: Amsterdam, The Netherlands, 2003; Chapter 8; pp. 143–178. [Google Scholar]
- Dreiseitl, A. The Hordeum vulgare subsp. spontaneum-Blumeria graminis f. sp. hordei pathosystem: Its position in resistance research and breeding applications. Eur. J. Plant Pathol. 2014, 138, 561–568. [Google Scholar] [CrossRef]
- Dreiseitl, A. Great pathotype diversity and reduced virulence complexity in a Central European population of Blumeria graminis f. sp. hordei in 2015–2017. Eur. J. Plant Pathol. 2019, 153, 801–811. [Google Scholar] [CrossRef]
- Kusch, S.; Qian, J.; Loos, A.; Kümmel, F.; Spanu, P.D.; Panstruga, R. Long-term and rapid evolution in powdery mildew fungi. Mol. Ecol. 2023. [Google Scholar] [CrossRef]
- Dreiseitl, A. Specific resistance of barley to powdery mildew, its use and beyond: A concise critical review. Genes 2020, 11, 971. [Google Scholar] [CrossRef]
- Dreiseitl, A. Powdery mildew resistance genes in European barley cultivars registered in the Czech Republic from 2016 to 2020. Genes 2022, 13, 1274. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, Y.; Shang, Y.; Hua, W.; Wang, J.; Jia, Q.; Liu, M.; Yang, J. Genetic evidence of local adaption and long distance migration in Blumeria graminis f. sp. hordei populations from China. J. Gen. Plant Pathol. 2016, 82, 69–81. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Wang, F.; Lang, X.; Zhao, X.; Zhu, J.; Hu, C.; Hu, J.; Zhang, Y.; Yao, X.; et al. Virulence Variability and Genetic Diversity in Blumeria graminis f. sp. hordei in Southeastern and Southwestern China. Plant Dis. 2023, 107, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhuoma, Q.; Xu, Z.; Peng, Y.; Wang, M. Virulence and Genetic Types of Blumeria graminis f. sp. hordei in Tibet and Surrounding Areas. J. Fungi 2023, 9, 363. [Google Scholar] [CrossRef]
- Murray, G.M.; Brennan, J.P. Estimating Disease Losses to the Australian Barley Industry. Australas. Plant Pathol. 2010, 39, 85–96. [Google Scholar] [CrossRef]
- Kauppi, K.; Rajala, A.; Huusela, E.; Kaseva, J.; Ruuttunen, P.; Jalli, H.; Alakukku, L.; Jalli, M. Impact of Pests on Cereal Grain and Nutrient Yield in Boreal Growing Conditions. Agronomy 2021, 11, 592. [Google Scholar] [CrossRef]
- Hysing, S.C.; Rosenqvist, H.; Wiik, L. Agronomic and economic effect of host resistance vs. fungicide control of barley powdery mildew in southern Sweden. Crop. Prot. 2012, 41, 122–127. [Google Scholar] [CrossRef]
- Hollomon, D.W. Fungicide resistance: Facing the challenge. Plant Prot. Sci. 2015, 51, 170–176. [Google Scholar] [CrossRef]
- Vielba-Fernández, A.; Polonio, Á.; Ruiz-Jiménez, L.; de Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Fungicide Resistance in Powdery Mildew Fungi. Microorganisms 2020, 8, 1431. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef]
- Burdon, J.J.; Barrett, L.G.; Rebetzke, G.; Thrall, P.H. Guiding deployment of resistance in cereals using evolutionary principles. Evol. Appl. 2014, 7, 609–624. [Google Scholar] [CrossRef]
- Mundt, C.H. Pyramiding for resistance durability: Theory and practise. Phytopathology 2018, 108, 792–802. [Google Scholar] [CrossRef]
- Walters, D.R.; Avrova, A.; Bingham, I.J.; Burnett, F.J.; Fountaine, J.; Havis, N.D.; Hoad, S.P.; Hughes, G.; Looseley, M.; Oxley, S.J.P.; et al. Control of foliar diseases in barley: Towards an integrated approach. Eur. J. Plant Pathol. 2012, 133, 33–73. [Google Scholar] [CrossRef]
- Jørgensen, L.N.; Hovmoller, M.; Hansen, J.G.; Lassen, P.; Clark, B.; Bayles, R.; Rodemann, B.; Flath, K.; Jahn, M.; Goral, T.; et al. IPM Strategies and Their Dilemmas Including an Introduction to www.eurowheat.org. J. Integr. Agric. 2014, 13, 265–281. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Arseniuk, E.; Boonekamp, P.; Czembor, J.H.; Decroocq, V.; Enjalbert, J.; Finckh, M.R.; Korbin, M.; Koppel, M.M.; Kudsk, P.; et al. Advocating a need for suitable breeding approaches to boost Integrated Pest Management: A European perspective. Pest Manag. Sci. 2018, 74, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Stetkiewicz, S.; Bruce, A.; Burnett, F.J.; Ennos, R.A.; Topp, C.F.E. An interdisciplinary method for assessing IPM potential: Case study in Scottish spring barley. CABI Agric. Biosci. 2022, 3, 23. [Google Scholar] [CrossRef]
- Biffen, R.H. Studies in the inheritance of disease resistance. J. Agric. Sci. 1907, 2, 109–128. [Google Scholar] [CrossRef]
- Czembor, J.H.; Johnston, M.R. Resistance to powdery mildew in selections from Tunisian barley landraces. Plant Breed. 1999, 118, 503–509. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, H.J. Resistance to powdery mildew in barley landraces collected from Jordan. Plant Breed. Seed Sci. 1999, 43, 65–80. [Google Scholar]
- Czembor, J.H. Resistance to powdery mildew in barley (Hordeum vulgare L.) landraces from Egypt. Plant Genet. Resour. Newsl. 2000, 123, 52–60. [Google Scholar]
- Czembor, J.H. Resistance to powdery mildew in populations of barley landraces from Morocco. Australas. Plant Pathol. 2000, 29, 137–148. [Google Scholar] [CrossRef]
- Czembor, J.H. Powdery mildew resistance in selections from barley landraces collected in Jordan, Egypt, Libya and Tunisia. Sci. Agric. Bohem. 2001, 32, 43–65. [Google Scholar]
- Czembor, J.H.; Czembor, H.J. Inheritance of resistance to powdery mildew (Blumeria graminis f.sp. hordei) in selections from Moroccan landraces of barley. Cereal Res. Comm. 2001, 29, 281–288. [Google Scholar] [CrossRef]
- Czembor, J.H. Sources of resistance to powdery mildew (Blumeria graminis f. sp. hordei) in Moroccan barley landraces. Can. J Plant Path Rev. Can. De Phytopathol. 2001, 23, 260–269. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, H.J. Selections from barley landrace collected in Libya as new sources of effective resistance to powdery mildew (Blumeria graminis f.sp. hordei). Rostl. Vyrob. 2002, 48, 217–223. [Google Scholar] [CrossRef]
- Czembor, J.H. Resistance to powdery mildew in selections from Moroccan barley landraces. Euphytica 2002, 125, 397–409. [Google Scholar] [CrossRef]
- Czembor, J.H.; Frese, L. Powdery mildew resistance in selections from barley landraces collected from Turkey. Die Bodenkult. 2003, 54, 35–40. [Google Scholar]
- Silvar, C.; Flath, K.; Kopahnke, D.; Gracia, M.P.; Lasa, J.M.; Casas, A.M.; Igartua, E.; Ordon, F. Analysis of powdery mildew resistance in the Spanish barley core collection. Plant Breed. 2011, 130, 195–202. [Google Scholar] [CrossRef]
- Surlan-Momirovic, G.; Flath, K.; Silvar, C.; Brankovic, G.; Kopahnke, D.; Knezevic, D.; Schliephake, E.; Ordon, F.; Perovic, D. Exploring the Serbian GenBank barley (Hordeum vulgare L. subsp. vulgare) collection for powdery mildew resistance. Genet. Resour. Crop Evol. 2016, 63, 275–287. [Google Scholar] [CrossRef]
- Piechota, U.; Czembor, P.C.; Słowacki, P.; Czembor, J.H. Identifying a novel powdery mildew resistance gene in a barley landrace from Morocco. J. Appl. Genet. 2019, 60, 243–254. [Google Scholar] [CrossRef]
- Piechota, U.; Czembor, P.C.; Czembor, J.H. Evaluating barley landraces collected in North Africa and the Middle East for powdery mildew infection at seedling and adult plant stages. Cereal Res. Commun. 2020, 48, 179–185. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E. Mlo resistance to powdery mildew (Blumeria graminis f. sp. hordei) in barley landraces collected in Yemen. Agronomy 2021, 11, 1582. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E. Sources of Resistance to Powdery Mildew in Barley Landraces from Turkey. Agriculture 2021, 11, 1017. [Google Scholar] [CrossRef]
- Fischbeck, G.; Schwarzbach, E.; Sobel, Z.; Wahl, I. Mildew resistance in Israeli populations of 2-rowed wild barley (Hordeum spontaneum). Z. Pflanz. 1976, 76, 163–166. [Google Scholar]
- Moseman, J.G.; Nevo, E.; Zohary, D. Resistance of Hordeum spontaneum collected in Israel to infection with Erysiphe graminis hordei. Crop Sci. 1983, 23, 1115–1119. [Google Scholar] [CrossRef]
- Jahoor, A.; Fischbeck, G. Identification of new genes for mildew resistance of barley at the Mla locus in lines derived from Hordeum spontaneum. Plant Breed. 1993, 110, 116–122. [Google Scholar] [CrossRef]
- Kintzios, S.; Jahoor, A.; Fischbeck, G. Powdery-mildew-resistance genes Mla29 and Mla32 in H. spontaneum derived winter-barley lines. Plant Breed. 1995, 114, 265–266. [Google Scholar] [CrossRef]
- Kintzios, S.; Fischbeck, G. Identification of new sources for resistance to powdery mildew in H. spontaneum derived winter barley lines. Genet. Resour. Crop Evol. 1996, 43, 25–31. [Google Scholar] [CrossRef]
- Mastebroek, H.D.; Balkema-Bomstra, A.G.; Gaj, M. Genetic analysis of powdery mildew (Erysiphe graminis f. sp. hordei) resistance derived from wild barley (Hordeum vulgare ssp. spontaneum). Plant Breed. 1995, 114, 121–125. [Google Scholar] [CrossRef]
- Fetch, T.G.; Steffenson, B.J.; Nevok, E. Diversity and sources of multiple disease resistance in Hordeum spontaneum. Plant Dis. 2003, 87, 1439–1448. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Bockelman, H.E. Sources of powdery mildew resistance in a wild barley collection. Genet. Resour. Crop Evol. 2003, 50, 345–350. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Dinoor, A. Phenotypic diversity of barley powdery mildew resistance sources. Genet. Resour. Crop Evol. 2004, 51, 251–258. [Google Scholar] [CrossRef]
- Ames, N.; Dreiseitl, A.; Steffenson, B.J.; Muehlbauer, G.J. Mining wild barley for powdery mildew resistance. Plant Pathol. 2015, 64, 1396–1406. [Google Scholar] [CrossRef]
- Dreiseitl, A. High diversity of powdery mildew resistance in the ICARDA wild barley collection. Crop Pasture Sci. 2017, 68, 134–139. [Google Scholar] [CrossRef]
- Dreiseitl, A. Heterogeneity of powdery mildew resistance revealed in accessions of the ICARDA wild barley collection. Front. Plant Sci. 2017, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Czembor, J.H.; Czembor, E. Mlo resistance of barley to powdery mildew (Blumeria graminis f.sp. hordei) Part II. Sources of resistance and their use in barley breeding. Biul. IHAR 2003, 230, 355–374. [Google Scholar]
- Czembor, J.H.; Czembor, E. Mlo resistance of barley to powdery mildew (Blumeria graminis f.sp. hordei) Part III. Durability of resistance. Biul. IHAR 2003, 230, 375–386. [Google Scholar]
- Reinstädler, A.; Müller, J.; Czembor, J.H.; Piffanelli, P.; Panstruga, R. Novel induced mlo mutant alleles in 1 combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol. 2010, 10, 31. [Google Scholar] [CrossRef]
- Kølster, P.; Munk, L.; Stølen, O.; Løhde, J. Near-Isogenic Barley Lines with Genes for Resistance to Powdery Mildew. Crop. Sci. 1986, 26, 903–907. [Google Scholar] [CrossRef]
- Mains, E.B.; Dietz, S.M. Physiologic forms of barley mildew, Erysiphe graminis hordei Marchal. Phytopathology 1930, 20, 229–239. [Google Scholar]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Spies, A.; Korzun, V.; Bayles, R.; Rajaraman, J.; Himmelbach, A.; Hedley, P.E.; Schweizer, P. Allele mining in barley genetic resources reveals genes of race-non-specific powdery mildew resistance. Front. Plant Sci. 2012, 2, 113. [Google Scholar] [CrossRef]
- Hickey, L.T.; Lawson, W.; Platz, G.J.; Fowler, R.A.; Arief, V.; Dieters, M.; German, S.; Fletcher, S.; Park, R.F.; Singh, D.; et al. Mapping quantitative trait loci for partial resistance to powdery mildew in an Australian barley population. Crop Sci. 2012, 52, 1021–1032. [Google Scholar] [CrossRef]
- Bengtsson, T.; Åhman, I.; Manninen, O.; Reitan, L.; Christerson, T.; Due Jensen, J.; Krusell, L.; Jahoor, A.; Orabi, J.A. Novel QTL for Powdery Mildew Resistance in Nordic Spring Barley (Hordeum vulgare L. ssp. vulgare) Revealed by Genome-Wide Association Study. Front Plant Sci. 2017, 8, 1954. [Google Scholar] [CrossRef] [PubMed]
- Cowger, C.; Brown, J.K. Durability of quantitative resistance in crops: Greater than we know? Annu. Rev. Phytopathol. 2019, 57, 253–277. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; D’Antuono, M.; Bradley, J.; Li, C.; Loughman, R. Identification and expression of adult plant resistance in barley to powdery mildew (Blumeria graminis f. sp. hordei) in Australia. Euphytica 2015, 203, 595–605. [Google Scholar] [CrossRef]
- Arabi, M.I.; Jawhar, M.; Al-shehadah, E. Identification of Barley Lines with Resistance to Powdery Mildew Based on Seedling and Adult Plant Responses. Acta Phytopathol. Entomol. Hung. 2020, 55, 3–10. [Google Scholar] [CrossRef]
- Ge, C.; Wentzel, E.; Ellwood, S.R.; Souza, N.D.; Chen, K.; Oliver, R.P. Adult resistance genes to barley powdery mildew confer basal penetration resistance associated with broad-spectrum resistance. Plant Genome 2021, 14, e20129. [Google Scholar] [CrossRef] [PubMed]
- Genievskaya, Y.; Zatybekov, A.; Abugalieva, S.; Turuspekov, Y. Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan. Plants 2023, 12, 2375. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E.; Suchecki, R.; Watson-Haigh, N.S. Genome-Wide Association Study for Powdery Mildew and Rusts Adult Plant Resistance in European Spring Barley from Polish Gene Bank. Agronomy 2022, 12, 7. [Google Scholar] [CrossRef]
- Singh, B.; Mehta, S.; Aggarwal, S.K.; Tiwari, M.; Bhuyan, S.I.; Bhatia, S.; Islam, A. Barley, Disease Resistance, and Molecular Breeding Approaches. In Disease Resistance in Crop Plants; Wani, S.H., Ed.; Springer Nature Switzerland AG: Basel, Switzerland, 2019; Chapter 11; pp. 261–299. [Google Scholar] [CrossRef]
- Riaz, A.; Kanwal, F.; Börner, A.; Pillen, K.; Dai, F.; Alqudah, A.M. Advances in Genomics-Based Breeding of Barley: Molecular Tools and Genomic Databases. Agronomy 2021, 11, 894. [Google Scholar] [CrossRef]

| No. | Near-Isogenic Lines/Cultivars | Resistance Genes | Isolates | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |||
| Bgh 1 | Bgh 2 | Bgh 3 | Bgh 4 | Bgh 8 | Bgh 9 | Bgh 11 | Bgh 13 | Bgh 14 | Bgh 24 | Bgh 28 | Bgh 29 | Bgh 31 | Bgh 33 | Bgh 36 | Bgh 40 | Bgh 48 | Bgh 51 | Bgh 57 | Bgh 63 | |||
| 1 | Pallas | Mla8 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 2 | P1 | Mla1 | 0 | 0 | 4 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 |
| 3 | P2 | Mla3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 4 | 0 | 0 | 0 | 0 |
| 4 | P3 | Mla6, Mla14 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 4 | 0 | 4 | 0 | 0 | 0 | 4 | 4 | 4 | 4 | 4 | 4 |
| 5 | P4A | Mla7, Mlk, +? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 4 | 0 | 2 | 2 | 2 | 4 | 4 | 2 |
| 6 | P4B | Mla7, +? | 4 | 4 | 4 | 1 | 0 | 2 | 2 | 4 | 4 | 0 | 2 | 4 | 4 | 1 | 4 | 4 | 1 | 4 | 4 | 4 |
| 7 | P6 | Mla7, MlLG2 | 4 | 4 | - * | 0 | 0 | 2 | 1 | 2 | 4 | 0 | 2 | 2 | 4 | 0 | 4 | 2 | 0 | 4 | 4 | 4 |
| 8 | P7 | Mla9, Mlk | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| 9 | P8A | Mla9, Mlk | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| 10 | P8B | Mla9 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| 11 | P9 | Mla10, MlDu2 | 4 | 4 | 4 | 0 | 1 | 4 | 0 | 4 | 0 | 2 | 0 | 4 | 4 | 4 | 4 | 0 | 0 | 4 | 4 | 4 |
| 12 | P10 | Mla12 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 4 | 0 | 4 | 4 | 0 | 4 | 0 | 4 |
| 13 | P11 | Mla13, MlRu3 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 4 |
| 14 | P12 | Mla22 | 4 | 4 | 0 | 4 | 4 | 0 | 4 | 0 | 4 | 4 | 4 | 4 | 4 | 0 | 0 | 4 | 4 | 4 | 0 | 0 |
| 15 | P13 | Mla23 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 16 | P14 | Mlra | 4 | 4 | 4 | 0 | 4 | 4 | 4 | 4 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 17 | P15 | Ml(Ru2) | 4 | 4 | 4 | 4 | 3 | 4 | 2 | 4 | 4 | 2 | 0 | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 4 | 4 |
| 18 | P17 | Mlk | 4 | 4 | 4 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 0 | 4 | 4 | 2 | 4 | 2 | 2 | 4 | 4 | 4 |
| 19 | P18 | Mlnn | 4 | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 4 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 |
| 20 | P19 | Mlp | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 21 | P20 | Mlat | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 2 |
| 22 | P21 | Mlg, Ml(CP) | 4 | 4 | 4 | 0 | 0 | 0 | 4 | 0 | 4 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 0 | 4 | 0 | 4 |
| 23 | P22 | mlo5 | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 3 | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) | 0(4) |
| 24 | P23 | Ml(La) | 4 | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 25 | P24 | Mlh | 4 | 4 | 4 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 0 | 4 | 4 | 4 | 4 | 4 |
| 26 | Benedicte | Mla9, Ml(IM9) | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 4 | 0 | 4 | 4 | 0 | 4 | 0 | 4 |
| 27 | Lenka | Mla13, Ml(Ab) | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 4 |
| 28 | Gunnar | Mla3, Ml(Tu2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 29 | Steffi | Ml(St1), Ml(St2) | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 3 | 0 | 4 | 2 | 0 | 2 | 0 | 4 |
| 30 | Kredit | Ml(Kr) | 4 | 2 | 4 | 0 | 2 | 0 | 0 | 2 | 4 | 4 | 4 | 2 | 4 | 0 | 4 | 2 | 2 | 4 | 4 | 4 |
| 31 | Jarek | Ml(Kr), +? | 4 | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 2 | 4 |
| 32 | Trumph | Mla7, Ml(Ab) | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 33 | Borwina | Ml(Bw) | 4 | 3 | 3 | 0 | 4 | 0 | 4 | 4 | 4 | 2 | 2 | 3 | 4 | 4 | 4 | 3 | 4 | 4 | 2 | 2 |
| 34 | Manchurian | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |
| No. | ICARDA | OTHERNUMB *** (IHAR No.) | Isolate Bh Bgh 33 | No. | ICARDA | OTHERNUMB (IHAR No.) | Bgh 33 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IG * | Crop No | ORI ID ** | IG | Crop Nr | ORI ID | ||||||
| 1 | 38616 | 180007 | JOR | 1075 | 4 | 41 | 40177 | 181568 | LBN | 1116 | 4 |
| 2 | 38617 | 180008 | JOR | 1076 | 4 | 42 | 40178 | 181569 | LBN | 1117 | 4 |
| 3 | 38618 | 180009 | JOR | 1077 | 4 | 43 | 40179 | 181570 | LBN | 1118 | 4 |
| 4 | 38619 | 180010 | JOR | 1078 | 4 | 44 | 40180 | 181571 | LBN | 1119 | 4 |
| 5 | 38620 | 180011 | JOR | 1079 | 4 | 45 | 40181 | 181572 | LBN | 1120 | 4 |
| 6 | 38621 | 180012 | JOR | 1080 | 4 | 46 | 40182 | 181573 | LBN | 1121 | 4 |
| 7 | 38622 | 180013 | JOR | 1081 | 4 | 47 | 40183 | 181574 | LBN | 1122 | 4 |
| 8 | 38623 | 180014 | JOR | 1082 | 4 | 48 | 40184 | 181575 | LBN | 1124 | 4 |
| 9 | 38624 | 180015 | JOR | 1083 | 0, 4 | 49 | 40185 | 181576 | LBN | 1125 | 4 |
| 10 | 38625 | 180016 | JOR | 1084 | 0, 4 | 50 | 40186 | 181577 | LBN | 1126 | 4 |
| 11 | 38626 | 180017 | JOR | 1085 | 4 | 51 | 40187 | 181578 | LBN | 1127 | 4 |
| 12 | 38627 | 180018 | JOR | 1086 | 0 | 52 | 40188 | 181579 | LBN | 1128 | 4 |
| 13 | 38628 | 180019 | JOR | 1087 | 4 | 53 | 40189 | 181580 | LBN | 1129 | 4 |
| 14 | 38629 | 180020 | JOR | 1088 | 4 | 54 | 40190 | 181581 | LBN | 1130 | 4 |
| 15 | 38630 | 180021 | JOR | 1089 | 0, 4 | 55 | 40191 | 181582 | LBN | 1131 | 4 |
| 16 | 38631 | 180022 | JOR | 1090 | 4 | 56 | 40193 | 181584 | LBN | 1132 | 4 |
| 17 | 38632 | 180023 | JOR | 1091 | 4 | 57 | 40194 | 181585 | LBN | 1133 | 4 |
| 18 | 38633 | 180024 | JOR | 1092 | 4 | 58 | 112846 | 181657 | LBY | 1134 | 0, 2, 4 |
| 19 | 39398 | 180789 | JOR | 1093 | 4 | 59 | 112847 | 181658 | LBY | 1135 | 4 |
| 20 | 39821 | 181212 | JOR | 1094 | 2, 4 | 60 | 110816 | 181628 | LBN | 1136 | 4 |
| 21 | 39822 | 181213 | JOR | 1095 | 4 | 61 | 110819 | 181629 | LBN | 1137 | 4 |
| 22 | 39823 | 181214 | JOR | 1096 | 4 | 62 | 110823 | 181630 | LBN | 1138 | 4 |
| 23 | 39824 | 181215 | JOR | 1097 | 4 | 63 | 110831 | 181631 | LBN | 1139 | 4 |
| 24 | 39825 | 181216 | JOR | 1098 | 4 | 64 | 110833 | 181632 | LBN | 1140 | 4 |
| 25 | 39826 | 181217 | JOR | 1099 | 4 | 65 | 116004 | 181639 | LBY | 1141 | 4 |
| 26 | 39827 | 181218 | JOR | 1100 | 4 | 66 | 116005 | 181640 | LBY | 1142 | 0, 4 |
| 27 | 39828 | 181219 | JOR | 1101 | 2, 4 | 67 | 115780 | 181660 | JOR | 1143 | 4 |
| 28 | 39829 | 181220 | JOR | 1102 | 4 | 68 | 115781 | 181661 | JOR | 1144 | 0, 4 |
| 29 | 39850 | 181241 | JOR | 1103 | 4 | 69 | 115782 | 181662 | JOR | 1145 | 4 |
| 30 | 39851 | 181242 | JOR | 1104 | 4 | 70 | 115784 | 181664 | JOR | 1146 | 0 |
| 31 | 39877 | 181268 | JOR | 1105 | 4 | 71 | 115785 | 181665 | JOR | 1147 | 4 |
| 32 | 39933 | 181324 | LBY | 1107 | 4 | 72 | 115786 | 181666 | JOR | 1148 | 2, 4 |
| 33 | 39934 | 181325 | LBY | 1108 | 3 | 73 | 115787 | 181667 | JOR | 1149 | 4 |
| 34 | 39935 | 181326 | LBY | 1109 | 0, 2 | 74 | 115788 | 181668 | JOR | 1150 | 4 |
| 35 | 39936 | 181327 | LBY | 1110 | 4 | 75 | 115789 | 181669 | JOR | 1151 | 4 |
| 36 | 39937 | 181328 | LBY | 1111 | 0, 4 | 76 | 115790 | 181670 | JOR | 1152 | 0 |
| 37 | 39938 | 181329 | LBY | 1112 | 4 | 77 | 115791 | 181671 | JOR | 1153 | 4 |
| 38 | 39939 | 181330 | LBY | 1113 | 4 | 78 | 115792 | 181672 | JOR | 1154 | 4 |
| 39 | 40156 | 181547 | LBN | 1114 | 4 | 79 | 115793 | 181673 | JOR | 1155 | 0, 2 |
| 40 | 40168 | 181559 | LBN | 1115 | 4 | 80 | 115795 | 181674 | JOR | 1156 | 4 |
| 81 | 115796 | 181675 | JOR | 1157 | 4 | ||||||
| No. | ICARDA | Othernumb (IHAR Project No.-Line) | Isolates | Postulated Resistance Alleles | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICARDA-IG | IG Line No. | Crop No | ORI ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |||
| Bgh 1 | Bgh 2 | Bgh 3 | Bgh 4 | Bgh 8 | Bgh 9 | Bgh 11 | Bgh 13 | Bgh 14 | Bgh 24 | Bgh 28 | Bgh 29 | Bgh 31 | Bgh 33 | Bgh 36 | Bgh 40 | Bgh 48 | Bgh 51 | Bgh 57 | Bgh 63 | |||||||
| 1 | 38624 | 1 | 180015 | JOR | 1083-1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un * |
| 2 | 38625 | 1 | 180016 | JOR | 1084-1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | un |
| 3 | 38625 | 2 | 180016 | JOR | 1084-3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | un |
| 4 | 38625 | 3 | 180016 | JOR | 1084-4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 5 | 38627 | 1 | 180018 | JOR | 1086-2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 6 | 38630 | 1 | 180021 | JOR | 1089-1 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | un |
| 7 | 39821 | 1 | 181212 | JOR | 1094-1 | 4 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 2 | 4 | 0 | 2 | 0 | 0 | 0 | 2 | 2 | 0 | un |
| 8 | 39821 | 2 | 181212 | JOR | 1094-2 | 2 | 0 | 4 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | 4 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 2 | un |
| 9 | 39828 | 1 | 181219 | JOR | 1101-1 | 2 | 0 | 2 | - ** | 0 | - | 2 | 0 | 0 | 0 | - | 2 | 0 | - | 0 | 0 | 0 | - | - | - | un |
| 10 | 39828 | 2 | 181219 | JOR | 1101-2 | 0 | 0 | 0 | - | 0 | 2 | 2 | 0 | 0 | 2 | 0 | 2 | 0 | - | 0 | 0 | 0 | - | - | 0 | un |
| 11 | 39828 | 3 | 181219 | JOR | 1101-3 | 2 | 0 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 12 | 39935 | 1 | 181326 | LBY | 1109-1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | un |
| 13 | 39935 | 2 | 181326 | LBY | 1109-2 | 0 | 0 | 0 | - | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | - | 0 | 0 | 0 | - | - | 0 | un |
| 14 | 39935 | 3 | 181326 | LBY | 1109-3 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | un |
| 15 | 39935 | 4 | 181326 | LBY | 1109-4 | 0 | 0 | 2 | 0 | 2 | 0 | 2 | 2 | 0 | 2 | 0 | 2 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | un |
| 16 | 39937 | 1 | 181328 | LBY | 1111-1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 4 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 2 | 0 | un |
| 17 | 39937 | 2 | 181328 | LBY | 1111-2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | un |
| 18 | 112846 | 1 | 181657 | LBY | 1134-1 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 19 | 112846 | 2 | 181657 | LBY | 1134-2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | un |
| 20 | 116005 | 1 | 181640 | LBY | 1142-1 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 2 | 0 | un |
| 21 | 115781 | 1 | 181661 | JOR | 1144-1 | 4 | 2 | 4 | - | 0 | 0 | 2 | 2 | 0 | 2 | 0 | 2 | 0 | - | 2 | 4 | 0 | - | - | 2 | un |
| 22 | 115781 | 2 | 181661 | JOR | 1144-2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | un |
| 23 | 115781 | 3 | 181661 | JOR | 1144-3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | un |
| 24 | 115784 | 1 | 181664 | JOR | 1146-1 | 0 | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | un |
| 25 | 115786 | 1 | 181666 | JOR | 1148-2 | 0 | 0 | 0 | 2 | 0 | - | 2 | 2 | 0 | 2 | - | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | - | un |
| 26 | 115790 | 1 | 181670 | JOR | 1152-1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 27 | 115790 | 2 | 181670 | JOR | 1152-2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 28 | 115793 | 1 | 181673 | JOR | 1155-1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | un |
| 29 | 115793 | 2 | 181673 | JOR | 1155-3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | un |
| 30 | 115793 | 3 | 181673 | JOR | 1155-3 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | un |
| 31 | 115793 | 2 | 181673 | JOR | 1155-2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 4 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | un |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czembor, J.H.; Czembor, E. Sources of Resistance to Powdery Mildew in Wild Barley (Hordeum vulgare subsp. spontaneum) Collected in Jordan, Lebanon, and Libya. Agronomy 2023, 13, 2462. https://doi.org/10.3390/agronomy13102462
Czembor JH, Czembor E. Sources of Resistance to Powdery Mildew in Wild Barley (Hordeum vulgare subsp. spontaneum) Collected in Jordan, Lebanon, and Libya. Agronomy. 2023; 13(10):2462. https://doi.org/10.3390/agronomy13102462
Chicago/Turabian StyleCzembor, Jerzy H., and Elzbieta Czembor. 2023. "Sources of Resistance to Powdery Mildew in Wild Barley (Hordeum vulgare subsp. spontaneum) Collected in Jordan, Lebanon, and Libya" Agronomy 13, no. 10: 2462. https://doi.org/10.3390/agronomy13102462
APA StyleCzembor, J. H., & Czembor, E. (2023). Sources of Resistance to Powdery Mildew in Wild Barley (Hordeum vulgare subsp. spontaneum) Collected in Jordan, Lebanon, and Libya. Agronomy, 13(10), 2462. https://doi.org/10.3390/agronomy13102462







