Alternative Starter Fertilization Strategies in Maize (Zea mays L.) Cultivation: Agronomic Potential of Microgranular Fertilizer and Plant Growth-Promoting Microorganisms and Their Impact on the Soil Native Microbial Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Weather Data
2.2. Preparation of Combined Plant-Beneficial Microorganism Inoculum
2.3. Experiment Setup and Field Treatments
2.4. Soil and Root Sampling
2.5. Measurement of Soil Nutrient Concentrations
2.6. Measurement of Mycorrhizal Colonisation Rate
2.7. Measurement of Dry Mass Content and DNA Extraction
2.8. qPCR Analysis
2.9. Statistical Analysis
3. Results
3.1. The Effect of Starter Fertilization and Inoculation with Plant-Beneficial Microorganisms on Concentrations of Soil-Available Nutrients and Maize Yield
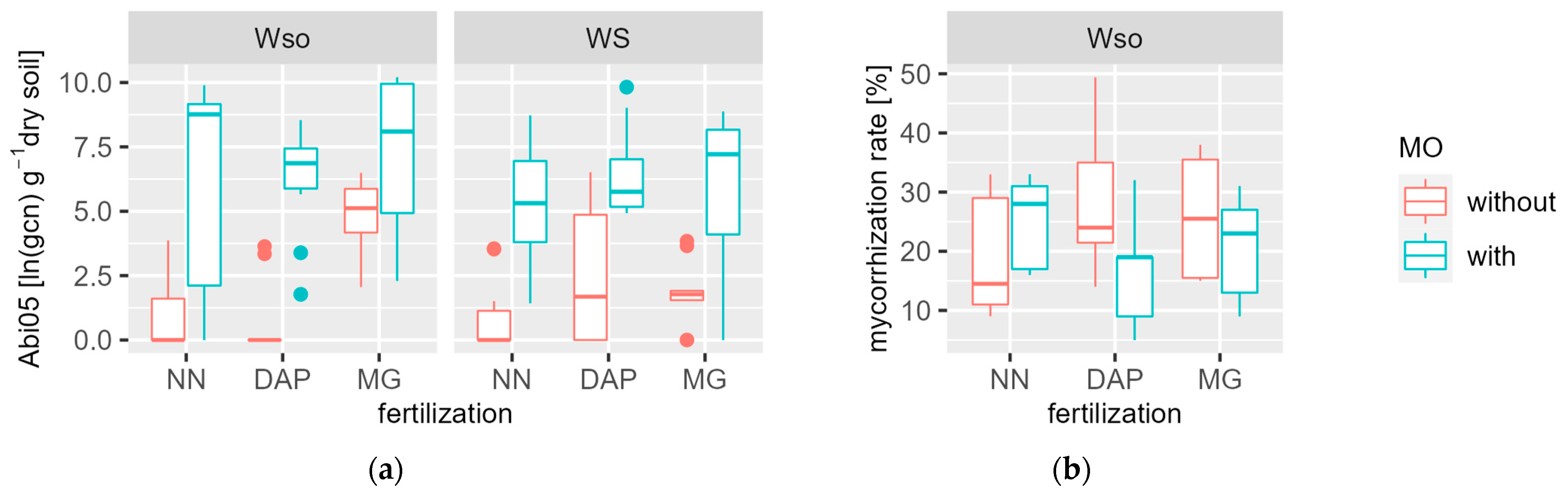
3.2. Root Mycorrhizal Colonisation Rate and Recovery of Inocluated Bacteria
3.3. Treatment Effects on the Soil Microbial Community
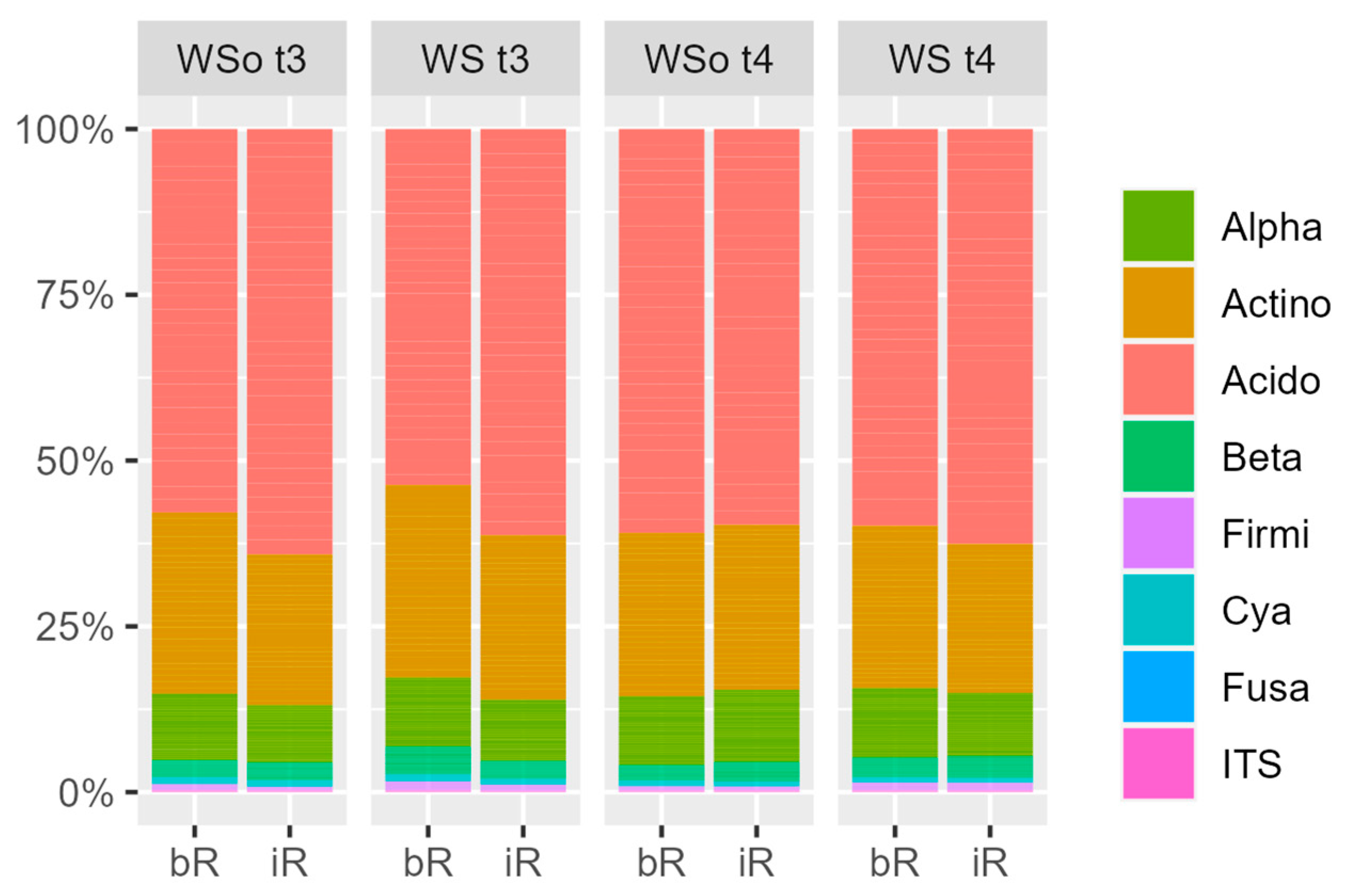
3.4. Description of the Soil Microbial Community and Its Spacial and Temporal Differences
3.4.1. Soil Microbial Community Description
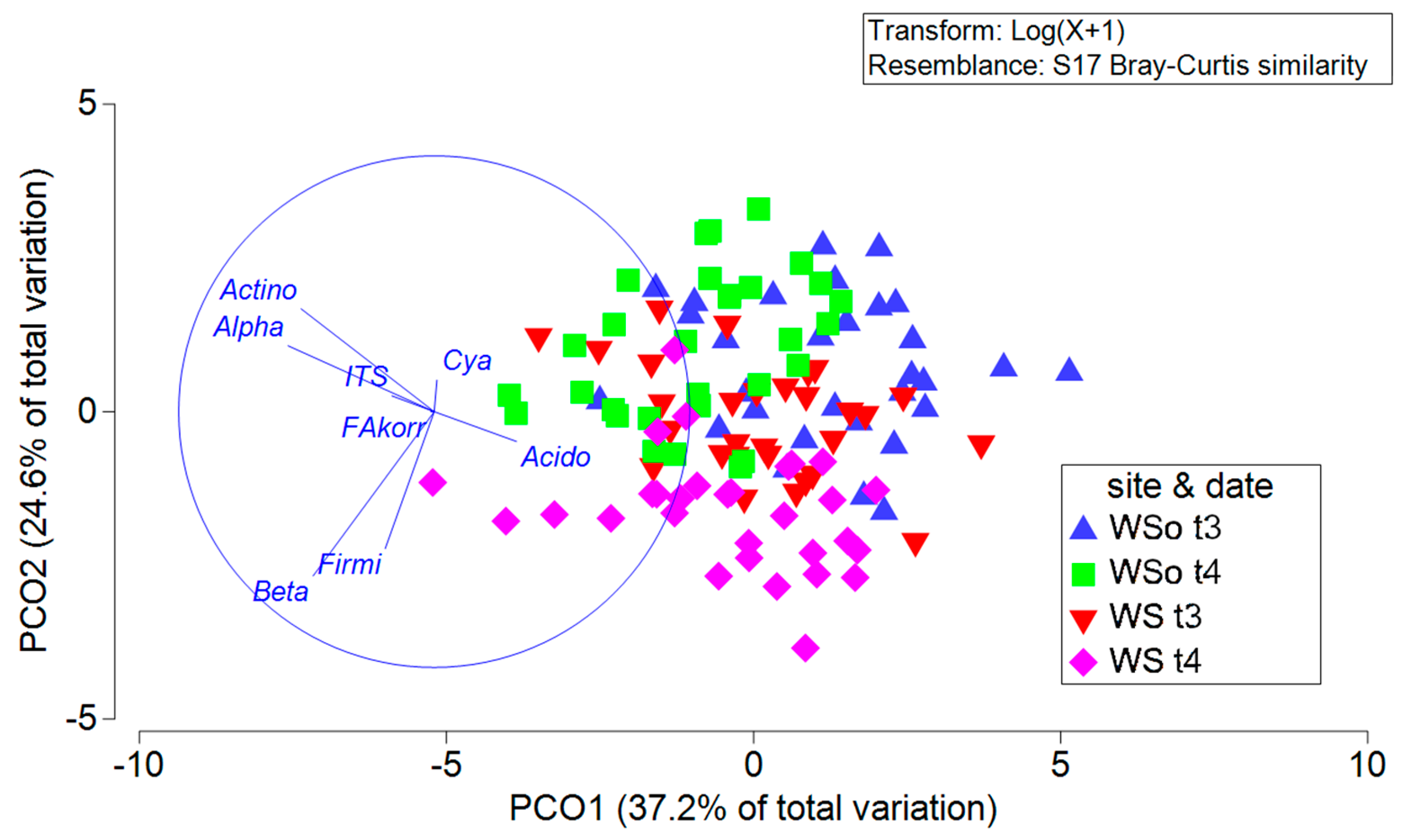
3.4.2. Spatial Distribution of Soil Microbial Communities
3.4.3. Effects of Site and Sampling Date
3.5. Correlations of the Maize Yield with Soil Nutrient Concentrations, Dry Mass Content, and Microbial Relative Abundances
4. Discussion
4.1. Drivers of Yield and Plant-Available Nutrient Concentrations
4.2. Recovery of Inoculated Bacterial Strain and Mycorrhizal Colonisation Rate
4.3. Treatment Effects on the Soil Microbial Community
4.4. Soil Microbial Community: Influence of Site, Sampling Date and Sampling Area
4.5. Correlations of Maize Yield with Soil’s Physical and Microbial Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Assay | Forward Primer | Reverse Primer | Ref. | AL [bp] | Target Region | DNA | Annealing | Cyc | Standard Strain | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidobact. | Acido31 | Eub518 | [45] | 500 | 16S-rRNA | 1 µL, 1:10 | 61 °C, 45 s | 35 | Acidobacterium capsulatum | ||
| Actinobact. | S-P-Acti-1154-a-S-19 | S-P-Acti-1339-a-A-18 | [101] | 166 | 16S-rRNA | 1 µL, 1:10 | 62 °C, 30 s | 35 | Microbacterium foliorum | ||
| α-Proteobact. | S-C-aProt-0528-a-S-19 | S-C-aProt-0689-a-A-21 | [101] | 142 | 16S-rRNA | 1 µL, 1:25 | 58 °C, 25 s | 35 | Sphingomonas paucimobilis | ||
| Β-Proteobact. | S-C-bProt-0972-a-S-18 | S-C-bProt-1221-a-A-17 | [101] | 231 | 16S-rRNA | 1 µL, 1:10 | 57 °C, 45 s | 35 | Variovorax paradoxus | ||
| Firmicutes | S-P-Firm-0525-a-S-18 | 1040FirmR | [101] | 501 | 16S-rRNA | 1 µL, 1:25 | 64 °C, 30 s | 35 | Bacillus subtilis | ||
| Cyanobact. | CYA361f | CYA785r | [102] | 440 | 16S-rRNA | 1 µL, 1:25 | 62.2 °C, 30 s | 40 | Chroococcidiopsis cubana | ||
| Fungi (ITS) | ITS F | 5,8S-R | [45] | 300 | ITS1 | 1 µL, 1:10 | 59 °C, 30 s | 35 | A. tenuissima GH50t | ||
| Inhibition | T7 F | M13 rev | [56] | 210 | - | 1 µL, und. | 58 °C, 30 s | 30 | Artificial DNA-Fragment | ||
| Forward Primer | Reverse Primer | Probe | |||||||||
| Fusarium | FA pl3 forward | Fus pl reverse | S FUS pl | [103] | TEF1 | 2 µL, und | 67 °C, 90 s | 45 | F. graminearum 486 | ||
References
- Panagos, P.; Köningner, J.; Ballabio, C.; Liakos, L.; Muntwyler, A.; Borrelli, P.; Lugato, E. Improving the phosphorus budget of European agricultural soils. Sci. Total Environ. 2022, 853, 158706. [Google Scholar] [CrossRef]
- MacDonald, G.K.; Bennett, E.M.; Potter, P.A.; Ramankutty, N. Agronomic phosphorus imbalances across the world’s croplands. Proc. Natl. Acad. Sci. USA 2011, 108, 3086–3091. [Google Scholar] [CrossRef]
- Einarsson, R.; Pitulia, D.; Cederberg, C. Subnational nutrient budgets to monitor environmental risks in EU agriculture: Calculating phosphorus budgets for 243 EU28 regions using public data. Nutr. Cycl. Agroecosystems 2020, 117, 199–213. [Google Scholar] [CrossRef]
- Bennett, E.M.; Carpenter, S.R.; Caraco, N.F. Human Impact on Erodable Phosphorus and Eutrophication: A Global Perspective. BioScience 2001, 51, 227. [Google Scholar] [CrossRef]
- Cordell, D.; White, S. Life’s Bottleneck: Sustaining the World’s Phosphorus for a Food Secure Future. Annu. Rev. Environ. Resour. 2014, 39, 161–188. [Google Scholar] [CrossRef]
- Van Vuuren, D.P.; Bouwman, A.F.; Beusen, A. Phosphorus demand for the 1970–2100 period: A scenario analysis of resource depletion. Glob. Environ. Change 2010, 20, 428–439. [Google Scholar] [CrossRef]
- Blandino, M.; Battisti, M.; Vanara, F.; Reyneri, A. The synergistic effect of nitrogen and phosphorus starter fertilization sub-surface banded at sowing on the early vigor, grain yield and quality of maize. Eur. J. Agron. 2022, 137, 126509. [Google Scholar] [CrossRef]
- Wortmann, C.S.; Xerinda, S.A.; Mamo, M.; Shapiro, C.A. No-Till Row Crop Response to Starter Fertilizer in Eastern Nebraska: I. Irrigated and Rainfed Corn. Agron. J. 2006, 98, 156–162. [Google Scholar] [CrossRef]
- Kaiser, D.E.; Mallarino, A.P.; Bermudez, M. Corn Grain Yield, Early Growth, and Early Nutrient Uptake as Affected by Broadcast and In-Furrow Starter Fertilization. Agron. J. 2005, 97, 620–626. [Google Scholar] [CrossRef]
- Roller, S.; Weiß, T.M.; Li, D.; Liu, W.; Schipprack, W.; Melchinger, A.E.; Hahn, V.; Leiser, W.L.; Würschum, T. Can we abandon phosphorus starter fertilizer in maize? Results from a diverse panel of elite and doubled haploid landrace lines of maize (Zea mays L.). Front. Plant Sci. 2022, 13, 1005931. [Google Scholar] [CrossRef]
- Quinn, D.J.; Lee, C.D.; Poffenbarger, H.J. Corn yield response to sub-surface banded starter fertilizer in the U.S.: A meta-analysis. Field Crops Res. 2020, 254, 107834. [Google Scholar] [CrossRef]
- Buczko, U.; van Laak, M.; Eichler-Löbermann, B.; Gans, W.; Merbach, I.; Panten, K.; Peiter, E.; Reitz, T.; Spiegel, H.; von Tucher, S. Re-evaluation of the yield response to phosphorus fertilization based on meta-analyses of long-term field experiments. Ambio 2018, 47, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Balawejder, M.; Szostek, M.; Gorzelany, J.; Antos, P.; Witek, G.; Matłok, N. A Study on the Potential Fertilization Effects of Microgranule Fertilizer Based on the Protein and Calcined Bones in Maize Cultivation. Sustainability 2020, 12, 1343. [Google Scholar] [CrossRef]
- Olbrycht, M.; Kołodziej, M.; Bochenek, R.; Przywara, M.; Balawejder, M.; Matłok, N.; Antos, P.; Piątkowski, W.; Antos, D. Mechanism of nutrition activity of a microgranule fertilizer fortified with proteins. BMC Plant Biol. 2020, 20, 126. [Google Scholar] [CrossRef] [PubMed]
- Thielicke, M.; Ahlborn, J.; Eichler-Löbermann, B.; Eulenstein, F. On the Negative Impact of Mycorrhiza Application on Maize Plants (Zea mays) Amended with Mineral and Organic Fertilizer. Microorganisms 2023, 11, 1663. [Google Scholar] [CrossRef]
- Thielicke, M.; Ahlborn, J.; Životić, L.; Saljnikov, E.; Eulenstein, F. Microgranular fertilizer and biostimulants as alternatives to diammonium phosphate fertilizer in maize production on marshland soils in northwest Germany. Zemljište Biljka 2022, 71, 53–66. [Google Scholar] [CrossRef]
- Zhang, S.; Lehmann, A.; Zheng, W.; You, Z.; Rillig, M.C. Arbuscular mycorrhizal fungi increase grain yields: A meta-analysis. New Phytol. 2019, 222, 543–555. [Google Scholar] [CrossRef]
- Schütz, L.; Gattinger, A.; Meier, M.; Müller, A.; Boller, T.; Mäder, P.; Mathimaran, N. Improving Crop Yield and Nutrient Use Efficiency via Biofertilization—A Global Meta-analysis. Front. Plant Sci. 2018, 8, 2204. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef]
- Singh, R.; Goodwin, S.B. Exploring the Corn Microbiome: A Detailed Review on Current Knowledge, Techniques, and Future Directions. PhytoFrontiers™ 2022, 2, 158–175. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Bossuyt, S.; Engelen, K.; Marchal, K.; Vanderleyden, J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014, 201, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.; Wu, G.; Veronican Njeri, K.; Shen, Q.; Zhang, N.; Zhang, R. Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 2016, 158, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Djonovic, S.; Vargas, W.A.; Kolomiets, M.V.; Horndeski, M.; Wiest, A.; Kenerley, C.M. A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 2007, 145, 875–889. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Lin, Y.; Ye, G.; Kuzyakov, Y.; Liu, D.; Fan, J.; Ding, W. Long-term manure application increases soil organic matter and aggregation, and alters microbial community structure and keystone taxa. Soil Biol. Biochem. 2019, 134, 187–196. [Google Scholar] [CrossRef]
- Sapp, M.; Harrison, M.; Hany, U.; Charlton, A.; Thwaites, R. Comparing the effect of digestate and chemical fertiliser on soil bacteria. Appl. Soil Ecol. 2015, 86, 1–9. [Google Scholar] [CrossRef]
- Tosi, M.; Deen, W.; Drijber, R.; McPherson, M.; Stengel, A.; Dunfield, K. Long-term N inputs shape microbial communities more strongly than current-year inputs in soils under 10-year continuous corn cropping. Soil Biol. Biochem. 2021, 160, 108361. [Google Scholar] [CrossRef]
- Bakker, M.G.; Chaparro, J.M.; Manter, D.K.; Vivanco, J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil 2015, 392, 115–126. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, S.; Li, R.; Zhang, J.; Liu, Y.; Lv, L.; Zhu, H.; Wu, W.; Li, W. Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 2017, 109, 145–155. [Google Scholar] [CrossRef]
- Baudoin, E.; Benizri, E.; Guckert, A. Impact of growth stage on the bacterial community structure along maize roots, as determined by metabolic and genetic fingerprinting. Appl. Soil Ecol. 2002, 19, 135–145. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Pfeiffer, S.; Mitter, B.; Oswald, A.; Schloter-Hai, B.; Schloter, M.; Declerck, S.; Sessitsch, A. Rhizosphere microbiomes of potato cultivated in the High Andes show stable and dynamic core microbiomes with different responses to plant development. FEMS Microbiol. Ecol. 2017, 93, fiw242. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Garbaye, J.; Tarkka, M. The mycorrhiza helper bacteria revisited. New Phytol. 2007, 176, 22–36. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, J.; George, T.S.; Limpens, E.; Feng, G. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci. 2022, 27, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Cornell, C.; Kokkoris, V.; Richards, A.; Horst, C.; Rosa, D.; Bennett, J.A.; Hart, M.M. Do Bioinoculants Affect Resident Microbial Communities? A Meta-Analysis. Front. Agron. 2021, 3, 753474. [Google Scholar] [CrossRef]
- Vuolo, F.; Novello, G.; Bona, E.; Gorrasi, S.; Gamalero, E. Impact of Plant-Beneficial Bacterial Inocula on the Resident Bacteriome: Current Knowledge and Future Perspectives. Microorganisms 2022, 10, 2462. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Andersson, S.G.E.; Battin, T.J.; Prosser, J.I.; Schimel, J.P.; Whitman, W.B.; Hallin, S. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 2010, 8, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Sanderlin, J.S.; Reeves, J.H.; Jenkins, M.B.; Endale, D.M.; Coleman, D.C.; Whitman, W.B. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol. Biochem. 2008, 40, 2843–2853. [Google Scholar] [CrossRef]
- Von Mering, C.; Hugenholtz, P.; Raes, J.; Tringe, S.G.; Doerks, T.; Jensen, L.J.; Ward, N.; Bork, P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 2007, 315, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Bru, D.; Saby, N.P.A.; Cuhel, J.; Arrouays, D.; Simek, M.; Hallin, S. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 2009, 11, 3096–3104. [Google Scholar] [CrossRef]
- Lanza, G.; Rebensburg, P.; Kern, J.; Lentzsch, P.; Wirth, S. Impact of chars and readily available carbon on soil microbial respiration and microbial community composition in a dynamic incubation experiment. Soil Tillage Res. 2016, 164, 18–24. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Gan, Y.; Hanson, K.G.; Zentner, R.P.; Selles, F.; McDonald, C.L. Response of lentil to microbial inoculation and low rates of fertilization in the semiarid Canadian prairies. Can. J. Plant Sci. 2005, 85, 847–855. [Google Scholar] [CrossRef]
- Chatterjee, A.; Franzen, D.W. Do we need to apply additional phosphorus for corn succeeding sugarbeet? Agron. J. 2020, 112, 4492–4497. [Google Scholar] [CrossRef]
- Köppen, W.; Geiger, R. Handbuch der Klimatologie; Gebrüder Borntraeger: Berlin, Germany, 1930. [Google Scholar]
- Meier, U. Entwicklungsstadien Mono- und Dikotyler Pflanzen: BBCH Monografie; Julius-Kühn-Institut (JKI): Quedlinburg, Germany, 2018. [Google Scholar]
- Vierheilig, H.; Piché, Y. A modified procedure for staining arbuscular mycorrhizal fungi in roots. Z. Pflanzenernaehr. Bodenk. 1998, 161, 601–602. [Google Scholar] [CrossRef]
- Giovannetti, M.; Mosse, B. An Evaluation of Techniques for Measuring Vesicular Arbuscular Mycorrhizal Infection in Roots. New Phytol. 1980, 84, 489–500. [Google Scholar] [CrossRef]
- O’Kelly, B.C. Oven-Drying Characteristics of Soils of Different Origins. Dry. Technol. 2005, 23, 1141–1149. [Google Scholar] [CrossRef]
- Degelmann, D.M.; Borken, W.; Drake, H.L.; Kolb, S. Different Atmospheric Methane-Oxidizing Communities in European Beech and Norway Spruce Soils. Appl. Environ. Microbiol. 2010, 76, 3228–3235. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests, R Package Version 0.7.0; 2021. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 28 September 2023).
- Giani, L.; Makowsky, L.; Mueller, K. Plaggic Anthrosol: Soil of the Year 2013 in Germany: An overview on its formation, distribution, classification, soil function and threats. J. Plant Nutr. Soil Sci. 2014, 177, 320–329. [Google Scholar] [CrossRef]
- Landwirtschaftskammer Niedersachsen. Empfehlungen zur Grunddüngung. Available online: https://www.lwk-niedersachsen.de/services/download.cfm?file=22858 (accessed on 28 September 2023).
- Lanwirtschaftskammer Niedersachsen. Unterfußdüngung zu Mais. Available online: https://www.landwirtschaftskammer.de/landwirtschaft/ackerbau/mais/unterfussduengung-pdf.pdf (accessed on 5 October 2023).
- Anderson, S.F.; Kelley, K.; Maxwell, S.E. Sample-Size Planning for More Accurate Statistical Power: A Method Adjusting Sample Effect Sizes for Publication Bias and Uncertainty. Psychol. Sci. 2017, 28, 1547–1562. [Google Scholar] [CrossRef]
- Von Tucher, S.; Hörndl, D.; Schmidhalter, U. Interaction of soil pH and phosphorus efficacy: Long-term effects of P fertilizer and lime applications on wheat, barley, and sugar beet. Ambio 2018, 47, 41–49. [Google Scholar] [CrossRef]
- Goldmann, K.; Boeddinghaus, R.S.; Klemmer, S.; Regan, K.M.; Heintz-Buschart, A.; Fischer, M.; Prati, D.; Piepho, H.-P.; Berner, D.; Marhan, S.; et al. Unraveling spatiotemporal variability of arbuscular mycorrhizal fungi in a temperate grassland plot. Environ. Microbiol. 2020, 22, 873–888. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Zhang, Z.; Sun, Z.; Chen, Y.; Jiang, J.; Shen, Z. The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 2017, 7, 45134. [Google Scholar] [CrossRef]
- Ziane, S.O.; Talibi, Z.E.A.; Douira, A.; Amir, S.; Meddich, A.; El Modafar, C. Synergistic effects of arbuscular mycorrhizal fungi associated to plant growth-promoting rhizobacteria in suppression of soil-borne Fusarium wilt of date palm. Biocatal. Agric. Biotechnol. 2023, 51, 102753. [Google Scholar] [CrossRef]
- Raklami, A.; Bechtaoui, N.; Tahiri, A.-I.; Anli, M.; Meddich, A.; Oufdou, K. Use of Rhizobacteria and Mycorrhizae Consortium in the Open Field as a Strategy for Improving Crop Nutrition, Productivity and Soil Fertility. Front. Microbiol. 2019, 10, 1106. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.K.; Choi, I.S.; Choi, Y.J. Overexpression and characterization of thermostable chitinase from Bacillus atrophaeus SC081 in Escherichia coli. BMB Rep. 2011, 44, 193–198. [Google Scholar] [CrossRef]
- Sella, S.R.B.R.; Vandenberghe, L.P.S.; Soccol, C.R. Bacillus atrophaeus: Main characteristics and biotechnological applications—A review. Crit. Rev. Biotechnol. 2015, 35, 533–545. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, E.; Yang, X.; Zhang, L.; Yousef, A.E.; Zhong, J. Isolation and characterization of a Bacillus atrophaeus strain and its potential use in food preservation. Food Control 2016, 60, 511–518. [Google Scholar] [CrossRef]
- Rajaofera, M.; Jin, P.F.; Fan, Y.M.; Sun, Q.Q.; Huang, W.K.; Wang, W.B.; Shen, H.Y.; Zhang, S.; Lin, C.H.; Liu, W.B.; et al. Antifungal activity of the bioactive substance from Bacillus atrophaeus strain HAB-5 and its toxicity assessment on Danio rerio. Pestic. Biochem. Physiol. 2018, 147, 153–161. [Google Scholar] [CrossRef]
- Rodríguez, M.; Marín, A.; Torres, M.; Béjar, V.; Campos, M.; Sampedro, I. Aphicidal Activity of Surfactants Produced by Bacillus atrophaeus L193. Front. Microbiol. 2018, 9, 3114. [Google Scholar] [CrossRef]
- Xue, J.; Sun, L.; Xu, H.; Gu, Y.; Lei, P. Bacillus atrophaeus NX-12 Utilizes Exosmotic Glycerol from Fusarium oxysporum f. sp. cucumerinum for Fengycin Production. J. Agric. Food Chem. 2023, 71, 10565–10574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, B.; Wang, Y.; Guo, Q.; Lu, X.; Li, S.; Ma, P. Lipopeptides, a novel protein, and volatile compounds contribute to the antifungal activity of the biocontrol agent Bacillus atrophaeus CAB-1. Appl. Microbiol. Biotechnol. 2013, 97, 9525–9534. [Google Scholar] [CrossRef] [PubMed]
- Köhl, L.; Lukasiewicz, C.E.; van der Heijden, M.G.A. Establishment and effectiveness of inoculated arbuscular mycorrhizal fungi in agricultural soils. Plant Cell Environ. 2016, 39, 136–146. [Google Scholar] [CrossRef]
- Dias, T.; Correia, P.; Carvalho, L.; Melo, J.; de Varennes, A.; Cruz, C. Arbuscular mycorrhizal fungal species differ in their capacity to overrule the soil’s legacy from maize monocropping. Appl. Soil Ecol. 2018, 125, 177–183. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Parke, J.L.; Mueller, S.M.; Senior, L.; Stuber, C.; Tracy, W.F. Variation among Maize Inbred Lines and Detection of Quantitative Trait Loci for Growth at Low Phosphorus and Responsiveness to Arbuscular Mycorrhizal Fungi. Crop Sci. 2000, 40, 358–364. [Google Scholar] [CrossRef]
- Raya-Hernández, A.I.; Jaramillo-López, P.F.; López-Carmona, D.A.; Díaz, T.; Carrera-Valtierra, J.A.; Larsen, J. Field evidence for maize-mycorrhiza interactions in agroecosystems with low and high P soils under mineral and organic fertilization. Appl. Soil Ecol. 2020, 149, 103511. [Google Scholar] [CrossRef]
- Sylvia, D.M.; Neal, L.H. Nitrogen affects the phosphorus response of VA mycorrhiza. New Phytol. 1990, 115, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef]
- Kröber, M.; Wibberg, D.; Grosch, R.; Eikmeyer, F.; Verwaaijen, B.; Chowdhury, S.P.; Hartmann, A.; Pühler, A.; Schlüter, A. Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front. Microbiol. 2014, 5, 252. [Google Scholar] [CrossRef]
- Krnjaja, V.; Nesic, Z.; Stankovic, S.; Radovic, C.; Lukic, M. Nitrogen effects on maize susceptibility to Fusarium ear rot (Fusarium verticillioides). Cereal Res. Commun. 2008, 36, 579–580. [Google Scholar]
- Orr, R.; Dennis, P.G.; Wong, Y.; Browne, D.J.; Cooper, M.; Birt, H.W.G.; Lapis-Gaza, H.R.; Pattison, A.B.; Nelson, P.N. Nitrogen fertilizer rate but not form affects the severity of Fusarium wilt in banana. Front. Plant Sci. 2022, 13, 907819. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Guo, S.; Jousset, A.; Zhao, Q.; Wu, H.; Li, R.; Kowalchuk, G.A.; Shen, Q. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Biochem. 2017, 114, 238–247. [Google Scholar] [CrossRef]
- Li, N.; Wang, C.; Li, X.; Liu, M. Effects of earthworms and arbuscular mycorrhizal fungi on preventing Fusarium oxysporum infection in the strawberry plant. Plant Soil 2019, 443, 139–153. [Google Scholar] [CrossRef]
- Schmidt, R.; Köberl, M.; Mostafa, A.; Ramadan, E.M.; Monschein, M.; Jensen, K.B.; Bauer, R.; Berg, G. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 2014, 5, 64. [Google Scholar] [CrossRef]
- Kalam, S.; Das, S.N.; Basu, A.; Podile, A.R. Population densities of indigenous Acidobacteria change in the presence of plant growth promoting rhizobacteria (PGPR) in rhizosphere. J. Basic Microbiol. 2017, 57, 376–385. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Kuramae, E.E.; de Hollander, M.; Pijl, A.S.; van Veen, J.A.; Tsai, S.M. Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbiol. Ecol. 2013, 83, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Bonk, F.; Popp, D.; Harms, H.; Centler, F. PCR-based quantification of taxa-specific abundances in microbial communities: Quantifying and avoiding common pitfalls. J. Microbiol. Methods 2018, 153, 139–147. [Google Scholar] [CrossRef]
- Starke, R.; Pylro, V.S.; Morais, D.K. 16S rRNA Gene Copy Number Normalization Does Not Provide More Reliable Conclusions in Metataxonomic Surveys. Microb. Ecol. 2021, 81, 535–539. [Google Scholar] [CrossRef]
- Lofgren, L.A.; Uehling, J.K.; Branco, S.; Bruns, T.D.; Martin, F.; Kennedy, P.G. Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 2019, 28, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.K.; Schmidt, T.M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015, 43, D593–D598. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Wu, M.; Eisen, J.A.; Green, J.L. Incorporating 16S Gene Copy Number Information Improves Estimates of Microbial Diversity and Abundance. PLoS Comput. Biol. 2012, 8, e1002743. [Google Scholar] [CrossRef]
- Lopes, L.D.; Schachtman, D.P. Rhizosphere and bulk soil bacterial community succession is influenced more by changes in soil properties than in rhizosphere metabolites across a maize growing season. Appl. Soil Ecol. 2023, 189, 104960. [Google Scholar] [CrossRef]
- Lopes, L.D.; Wang, P.; Futrell, S.L.; Schachtman, D.P. Sugars and Jasmonic Acid Concentration in Root Exudates Affect Maize Rhizosphere Bacterial Communities. Appl. Environ. Microbiol. 2022, 88, e0097122. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.P.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef]
- Panico, S.C.; Esposito, F.; Memoli, V.; Vitale, L.; Polimeno, F.; Magliulo, V.; Maisto, G.; de Marco, A. Variations of agricultural soil quality during the growth stages of sorghum and sunflower. Appl. Soil Ecol. 2020, 152, 103569. [Google Scholar] [CrossRef]
- Nyéki, A.; Kerepesi, C.; Daróczy, B.; Benczúr, A.; Milics, G.; Nagy, J.; Harsányi, E.; Kovács, A.J.; Neményi, M. Application of spatio-temporal data in site-specific maize yield prediction with machine learning methods. Precis. Agric. 2021, 22, 1397–1415. [Google Scholar] [CrossRef]
- Pfeiffer, S.; Pastar, M.; Mitter, B.; Lippert, K.; Hackl, E.; Lojan, P.; Oswald, A.; Sessitsch, A. Improved group-specific primers based on the full SILVA 16S rRNA gene reference database. Environ. Microbiol. 2014, 16, 2389–2407. [Google Scholar] [CrossRef]
- Mühling, M.; Woolven-Allen, J.; Murrell, J.C.; Joint, I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2008, 2, 379–392. [Google Scholar] [CrossRef]
- Müller, T.; Ruppel, S.; Behrendt, U.; Lentzsch, P.; Müller, M.E.H. Antagonistic Potential of Fluorescent Pseudomonads Colonizing Wheat Heads Against Mycotoxin Producing Alternaria and Fusaria. Front. Microbiol. 2018, 9, 2124. [Google Scholar] [CrossRef]

| Site | WS | WSo | ||||
|---|---|---|---|---|---|---|
| Depth [cm] | 0–30 | 30–60 | 60–90 | 0–30 | 30–60 | 60–90 |
| TOC [%] | 3.16 ± 0.30 | 2.19 ± 1.04 | 0.83 ± 0.49 | 2.57 ± 0.29 | 1.21 ± 0.12 | 0.44 ± 0.14 |
| Ntot [%] | 0.21 ± 0.02 | 0.12 ± 0.07 | 0.06 ± 0.01 | 0.23 ± 0.04 | 0.12 ± 0.03 | 0.06 ± 0.02 |
| Nmin [kg ha−1] | 102.3 ± 17.1 | 32.0 ± 9.3 | 12.7 ± 2.1 | 57.4 ± 29.9 | 47.7 ± 19.7 | 2.2 ± 0.84 |
| PDL [mg 100 g−1 soil] | 19.1 ± 1.1 | 10.4 ± 4.8 | 4.7 ± 6.6 | 20.1 ± 1.9 | 20.0 ± 3.2 | 13.3 ± 4.2 |
| KDL [mg 100 g−1 soil] | 22.8 ± 2.1 | 14.4 ± 3.5 | 8.9 ± 3.5 | 7.5 ± 2.5 | 5.0 ± 1.7 | 3.2 ± 0.6 |
| MgDL [mg 100 g−1 soil] | 9.4 ± 1.3 | 5.6 ± 0.6 | 3.5 ± 1.6 | 6.4 ± 2.0 | 5.0 ± 2.0 | 4.5 ± 2.1 |
| pH | 5.16 ± 0.08 | 5.37 ± 0.08 | 5.47 ± 0.08 | 4.61 ± 0.09 | 4.56 ± 0.15 | 4.66 ± 0.08 |
| Treatment | Fertilizer/Inoculum | Application Rate | N Input [kg ha−1] | P Input [kg ha−1] | K Input [kg ha−1] |
|---|---|---|---|---|---|
| DAP | DAP | 100 kg ha−1 | 18.0 | 20.1 | - |
| MG | MG | 25 kg ha−1 | 1.9 | 2.4 | 0.8 |
| MO | MO inoculum in bentonite | 25 kg ha−1 | - | - | - |
| MG + MO | MO inoculum in MG | 25 kg ha−1 | 1.9 | 2.4 | 0.8 |
| DAP + MO | DAP MO inoculum in bentonite | 100 kg ha−1 25 kg ha−1 | 18.0 - | 20.1 - | - - |
| Source of Variation | tot-gcn | Multivariate | Acido | Actino | Firmi | Alpha | Beta | Cya | ITS | Fusa |
|---|---|---|---|---|---|---|---|---|---|---|
| site | <0.001 | <0.001 | 0.915 | 0.632 | <0.001 | 0.195 | 0.047 | 0.123 | 0.004 | 0.003 |
| time | 0.005 | <0.001 | 0.030 | 0.686 | <0.001 | <0.001 | 0.002 | <0.001 | <0.001 | 0.095 |
| fert | 0.394 | 0.619 | 0.757 | 0.895 | 0.514 | 0.516 | 0.672 | 0.565 | 0.718 | 0.167 |
| MO | 0.130 | 0.315 | 0.392 | 0.518 | 0.022 | 0.304 | 0.411 | 0.147 | 0.860 | 0.434 |
| site:time | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.300 | 0.178 | 0.173 | 0.451 |
| site:fert | 0.996 | 0.899 | 0.510 | 0.387 | 0.430 | 0.671 | 0.709 | 0.781 | 0.725 | 0.003 |
| site:MO | 0.027 | 0.085 | 0.107 | 0.299 | 0.027 | 0.294 | 0.056 | 0.404 | 0.426 | 0.823 |
| time:fert | 0.965 | 0.569 | 0.778 | 0.736 | 0.963 | 0.663 | 0.472 | 0.264 | 0.972 | 0.156 |
| time:MO | 0.568 | 0.824 | 0.452 | 0.344 | 0.831 | 0.495 | 0.751 | 0.982 | 0.184 | 0.491 |
| fert:MO | 0.466 | 0.712 | 0.471 | 0.621 | 0.644 | 0.135 | 0.583 | 0.782 | 0.373 | 0.255 |
| site:time:fert | 0.883 | 0.984 | 0.333 | 0.309 | 0.793 | 0.960 | 0.882 | 0.982 | 0.646 | 0.831 |
| site:time:MO | 0.952 | 0.151 | 0.465 | 0.509 | 0.450 | 0.084 | 0.070 | 0.335 | 0.777 | 0.588 |
| site:fert:MO | 0.642 | 0.245 | 0.109 | 0.120 | 0.603 | 0.405 | 0.693 | 0.692 | 0.065 | 0.232 |
| time:fert:MO | 0.525 | 0.274 | 0.044 | 0.108 | 0.567 | 0.207 | 0.261 | 0.316 | 0.403 | 0.278 |
| plot(site:fert:MO) | 0.013 | 0.025 | 0.003 | 0.005 | 0.111 | 0.027 | 0.699 | 0.914 | 0.120 | 0.839 |
| site:time:fert:MO | 0.469 | 0.729 | 0.615 | 0.455 | 0.689 | 0.411 | 0.351 | 0.110 | 0.906 | 0.777 |
| Maize Dry Mass Yield Correlated with: | ||
|---|---|---|
| Time | WSo | WS |
| t2 | Soil DM (−0.27, .) | PDL (0.43, **) |
| t3 | B. atrophaeus Abi05 (0.25, .) | PDL (0.27, .) α-Proteobacteria (0.23, .) |
| t4 | B. atrophaeus Abi05 (0.30, *) tot-gcn (0.25, .) soil DM (−0.25, .) | Firmicutes (0.26, *) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geist, L.; Wolfer, R.; Thiem, R.; Thielicke, M.; Eichler-Löbermann, B.; Eulenstein, F.; Müller, M.E.H. Alternative Starter Fertilization Strategies in Maize (Zea mays L.) Cultivation: Agronomic Potential of Microgranular Fertilizer and Plant Growth-Promoting Microorganisms and Their Impact on the Soil Native Microbial Community. Agronomy 2023, 13, 2900. https://doi.org/10.3390/agronomy13122900
Geist L, Wolfer R, Thiem R, Thielicke M, Eichler-Löbermann B, Eulenstein F, Müller MEH. Alternative Starter Fertilization Strategies in Maize (Zea mays L.) Cultivation: Agronomic Potential of Microgranular Fertilizer and Plant Growth-Promoting Microorganisms and Their Impact on the Soil Native Microbial Community. Agronomy. 2023; 13(12):2900. https://doi.org/10.3390/agronomy13122900
Chicago/Turabian StyleGeist, Lena, Renate Wolfer, Richard Thiem, Matthias Thielicke, Bettina Eichler-Löbermann, Frank Eulenstein, and Marina E. H. Müller. 2023. "Alternative Starter Fertilization Strategies in Maize (Zea mays L.) Cultivation: Agronomic Potential of Microgranular Fertilizer and Plant Growth-Promoting Microorganisms and Their Impact on the Soil Native Microbial Community" Agronomy 13, no. 12: 2900. https://doi.org/10.3390/agronomy13122900
APA StyleGeist, L., Wolfer, R., Thiem, R., Thielicke, M., Eichler-Löbermann, B., Eulenstein, F., & Müller, M. E. H. (2023). Alternative Starter Fertilization Strategies in Maize (Zea mays L.) Cultivation: Agronomic Potential of Microgranular Fertilizer and Plant Growth-Promoting Microorganisms and Their Impact on the Soil Native Microbial Community. Agronomy, 13(12), 2900. https://doi.org/10.3390/agronomy13122900







