A Review of the Diverse Genes and Molecules Involved in Sucrose Metabolism and Innovative Approaches to Improve Sucrose Content in Sugarcane
Abstract
:1. Introduction
2. Sugar and Other By-Products
3. Tissue Culture and Mutation
4. Sucrose Content Improvement
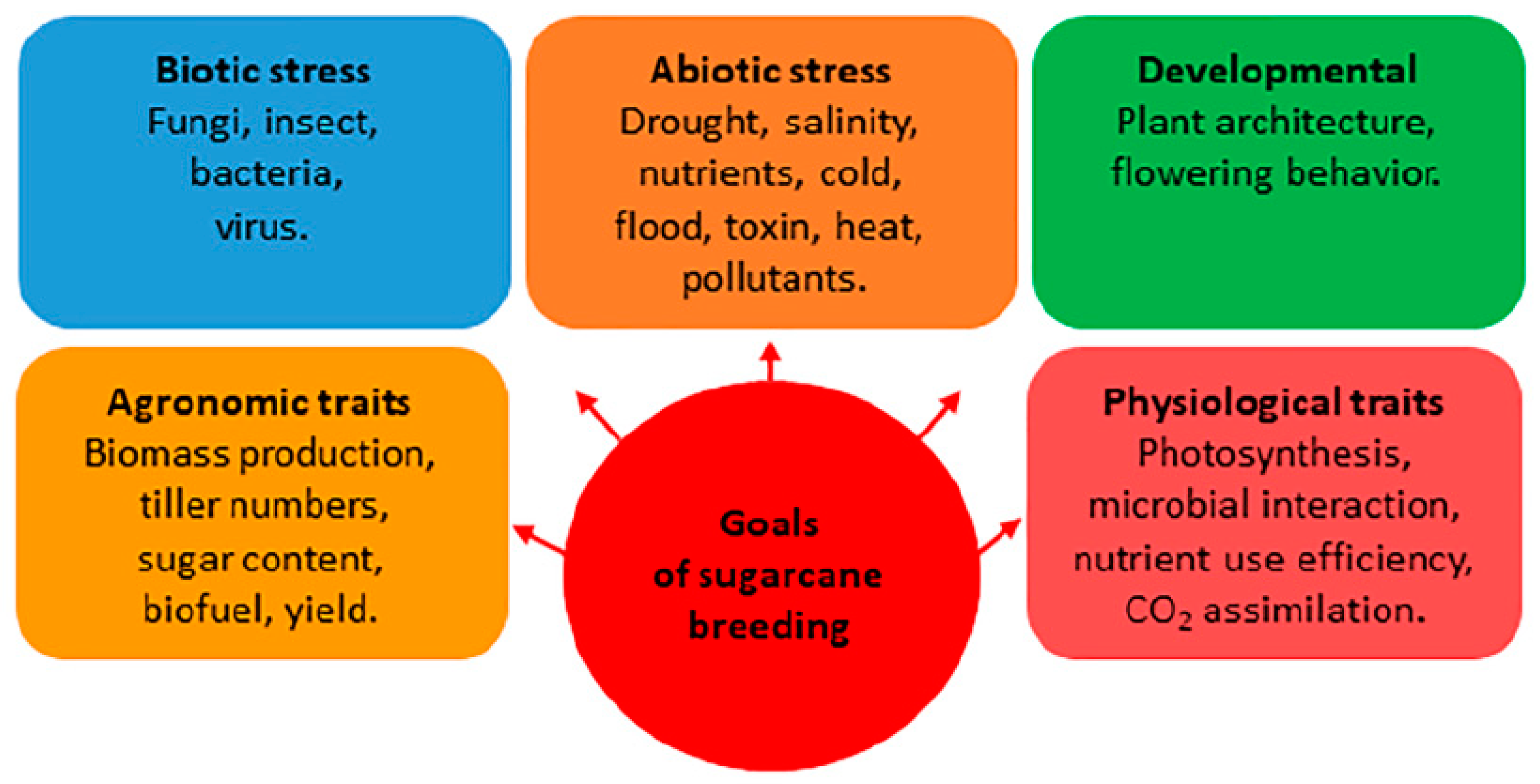
5. Sugarcane Breeding
6. Genetic Engineering
7. Application of Transcriptomic Tools
8. Application of Proteomic Approaches
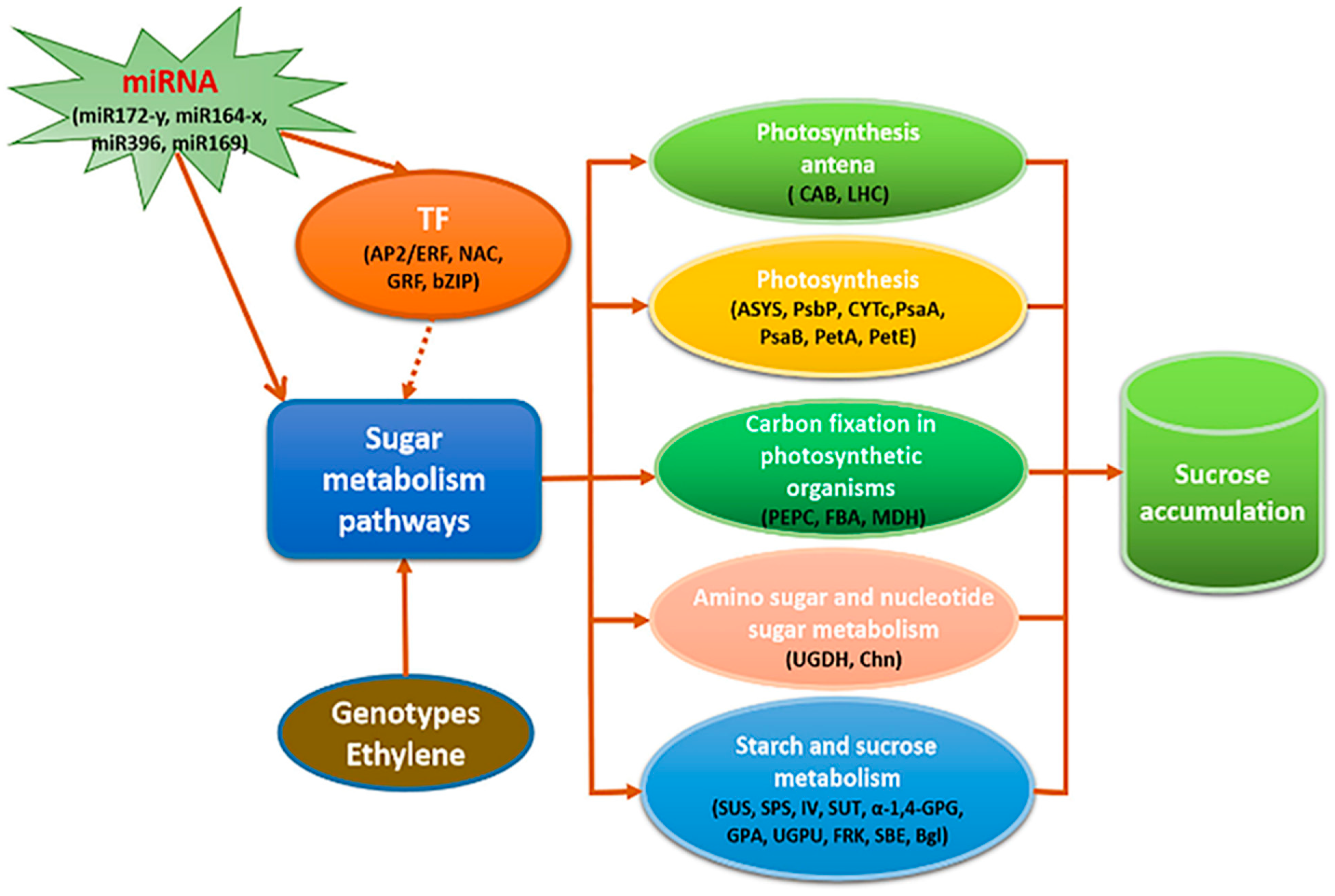
9. Sucrose Synthesis
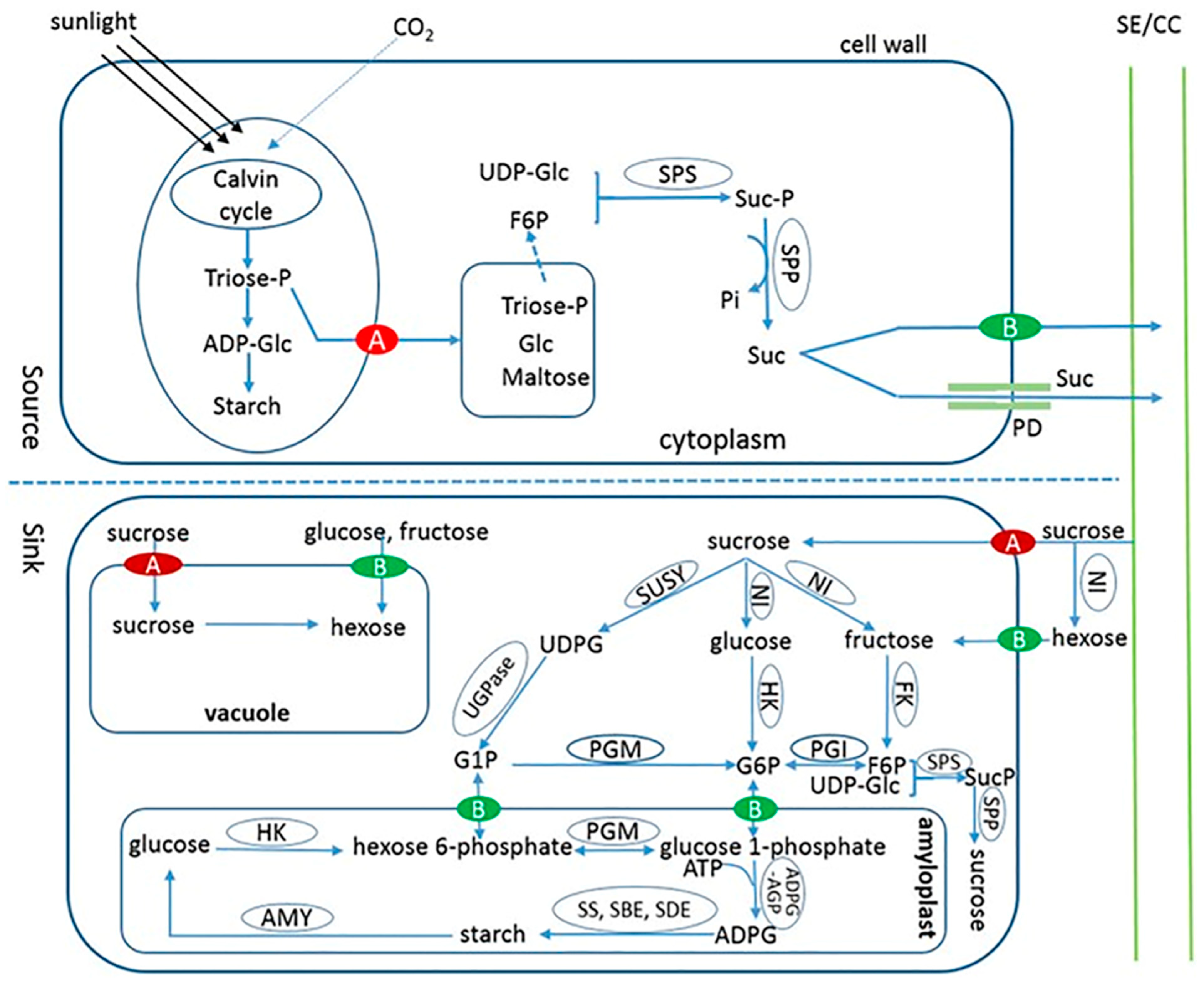
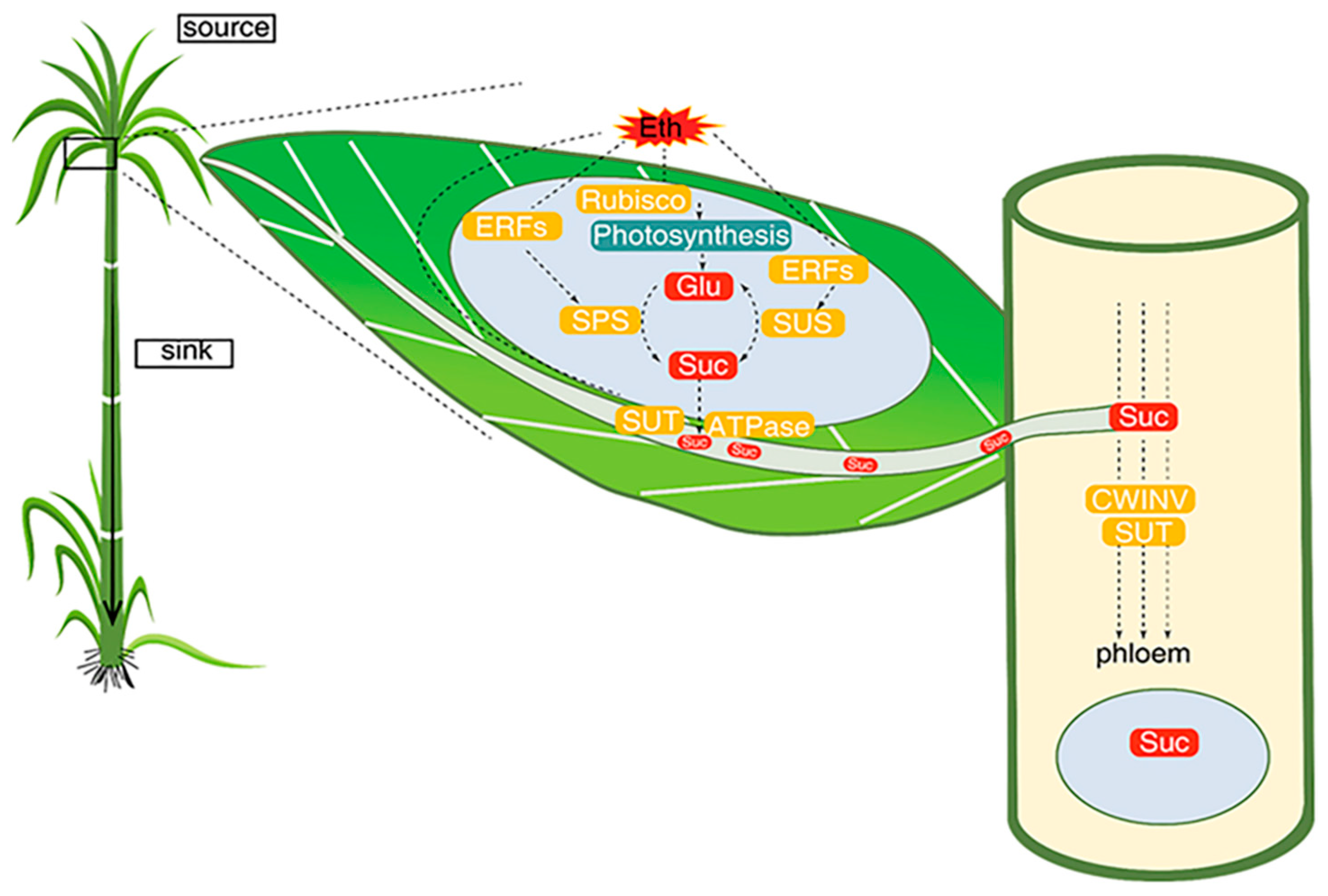
10. Relationship between Source and Sink
11. Sucrose Phosphate Synthase (SPS)
12. Sucrose Synthase (SuSy)
13. Invertase
14. Cellulose Synthase (CeS)
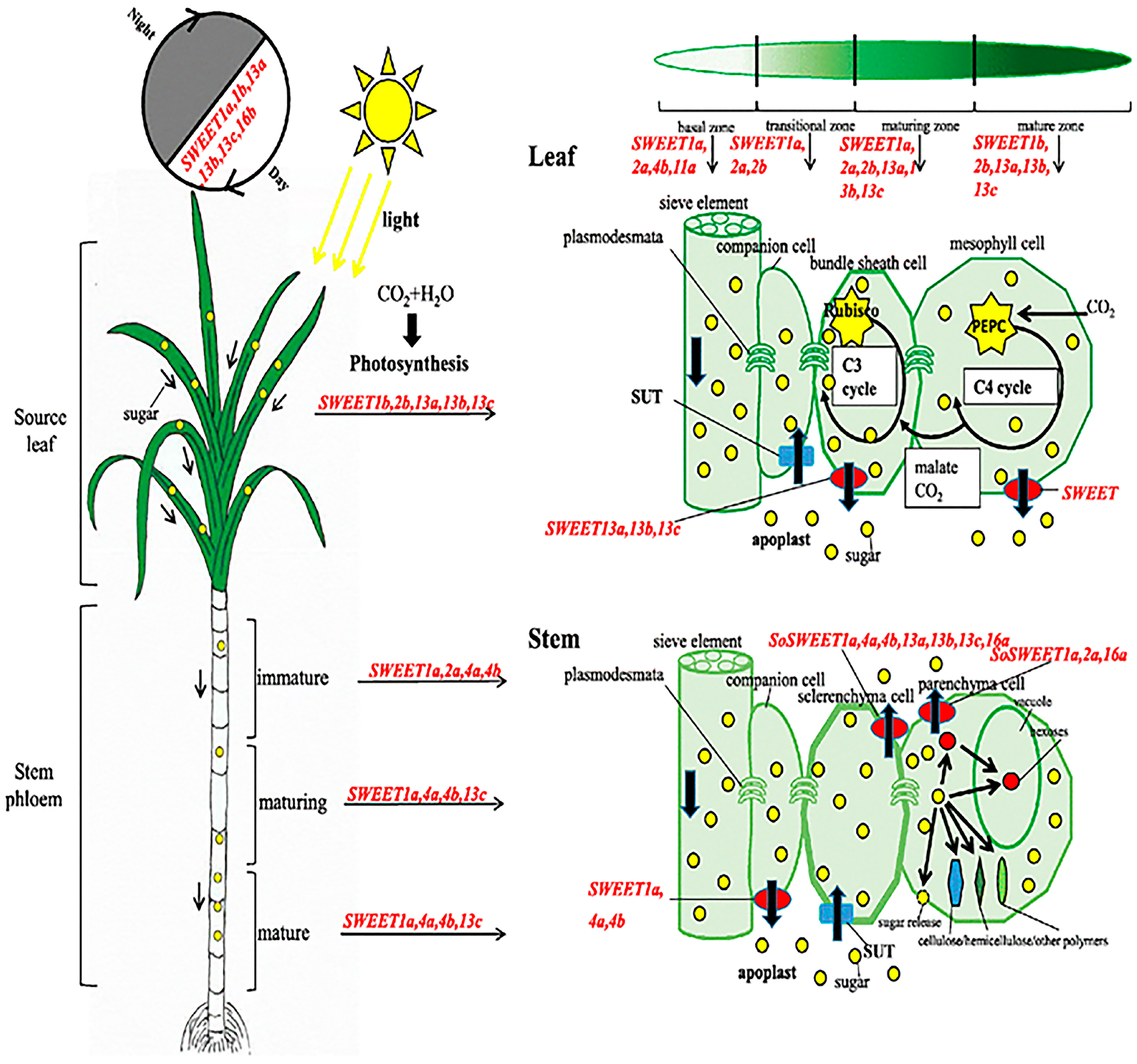
15. Sucrose Transporter (SUT) and SWEET
16. Trehalose
17. Transcription Factor (TF)
18. Protein Kinase (PTK)
19. Phytohormones
19.1. IAA
19.2. ETH
19.3. GA
19.4. ABA
20. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Watson, L.; Clifford, H.T.; Dallwitz, M.J. The classification of Poaceae: Subfamilies and supertribes. Aust. J. Bot. 1985, 33, 433–484. [Google Scholar] [CrossRef]
- Bonnett, G.D.; Henry, R.J. Saccharum. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 165–177. [Google Scholar]
- Cheavegatti-Gianotto, A.; de Abreu, H.M.C.; Arruda, P.; Bespalhok Filho, J.C.; Burnquist, W.L.; Creste, S.; di Ciero, L.; Ferro, J.A.; de Oliveira Figueira, A.V.; de Sousa Filgueiras, T. Sugarcane (Saccharum X officinarum): A reference study for the regulation of genetically modified cultivars in Brazil. Trop. Plant Biol. 2011, 4, 62–89. [Google Scholar] [CrossRef]
- Moore, P.H. Temporal and spatial regulation of sucrose accumulation in the sugarcane stem. Funct. Plant Biol. 1995, 22, 661–679. [Google Scholar] [CrossRef]
- Bihmidine, S.; Hunter, C.T., III; Johns, C.E.; Koch, K.E.; Braun, D.M. Regulation of assimilate import into sink organs: Update on molecular drivers of sink strength. Front. Plant Sci. 2013, 4, 177. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.M.; Wang, L.; Ruan, Y.-L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 2014, 65, 1713–1735. [Google Scholar] [CrossRef]
- Yadav, U.P.; Ayre, B.G.; Bush, D.R. Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: Assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 2015, 6, 275. [Google Scholar] [CrossRef]
- McCormick, A.; Watt, D.; Cramer, M. Supply and demand: Sink regulation of sugar accumulation in sugarcane. J. Exp. Bot. 2009, 60, 357–364. [Google Scholar] [CrossRef]
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef]
- Hoang, N.V.; Furtado, A.; Botha, F.C.; Simmons, B.A.; Henry, R.J. Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front. Bioeng. Biotechnol. 2015, 3, 182. [Google Scholar] [CrossRef] [PubMed]
- Inman-Bamber, G.; Jackson, P.; Bonnett, G.; Morgan, T. Have we reached peak CCS? Int. Sugar J. 2011, 113, 798–803. [Google Scholar]
- Dahlquist, E. Biomass as Energy Source: Resources, Systems and Applications; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Zhang, J.; Zhou, M.; Walsh, J.; Zhu, L.; Chen, Y.; Ming, R. Sugarcane genetics and genomics. In Sugarcane: Physiology, Biochemistry, and Functional Biology; Wiley: Hoboken, NJ, USA, 2013; pp. 623–643. [Google Scholar]
- Li, Y.-R.; Yang, L.-T. Sugarcane agriculture and sugar industry in China. Sugar Tech 2015, 17, 1–8. [Google Scholar] [CrossRef]
- Luo, J.; Pan, Y.-B.; Xu, L.; Grisham, M.P.; Zhang, H.; Que, Y. Rational regional distribution of sugarcane cultivars in China. Sci. Rep. 2015, 5, 15721. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Solomon, S. Potential of developing sugarcane by-product based industries in India. Sugar Tech 2006, 8, 104–111. [Google Scholar] [CrossRef]
- Barnabas, L.; Ramadass, A.; Amalraj, R.S.; Palaniyandi, M.; Rasappa, V. Sugarcane proteomics: An update on current status, challenges, and future prospects. Proteomics 2015, 15, 1658–1670. [Google Scholar] [CrossRef] [PubMed]
- Formann, S.; Hahn, A.; Janke, L.; Stinner, W.; Sträuber, H.; Logroño, W.; Nikolausz, M. Beyond sugar and ethanol production: Value generation opportunities through sugarcane residues. Front. Energy Res. 2020, 8, 579577. [Google Scholar] [CrossRef]
- Kumar, S.; Prakasha, G.; Hanumanthapa, D.; Kuri, S. Diversified use of byproducts of sugarcane and cotton-a review. Int. J. Curr. Microbiol. Appl. Sci 2018, 7, 1616–1634. [Google Scholar] [CrossRef]
- Guilherme, A.; Dantas, P.; Santos, E.; Fernandes, F.A.; Macedo, G.R. Evaluation of composition, characterization and enzymatic hydrolysis of pretreated sugar cane bagasse. Braz. J. Chem. Eng. 2015, 32, 23–33. [Google Scholar] [CrossRef]
- Hofsetz, K.; Silva, M.A. Brazilian sugarcane bagasse: Energy and non-energy consumption. Biomass Bioenergy 2012, 46, 564–573. [Google Scholar] [CrossRef]
- Ghulam, S.; Khan, M.; Usman, K.; Ullah, S. Effect of different rates of press mud on plant growth and yield of lentil in calcareous soil. Sarhad J. Agric. 2012, 28, 249–252. [Google Scholar]
- Cantarino, C.; Vogt, C. Seqüestro de Carbono, Cana-de-Açúcar e o Efeito Cinderela; ComCiência: Sao Paolo, Brazil, 2010. [Google Scholar]
- Sardar, S.; Ilyas, S.U.; Malik, S.R.; Javaid, K.; Sardar, S.; Ilyas, S.U.; Malik, S.R. Compost fertilizer production from sugar press mud (SPM). Int. J. Chem. Environ. Eng. 2012, 3, 39–43. [Google Scholar]
- Dussán, K.J.; Justo, O.R.; Perez, V.H.; David, G.F.; Junior, E.G.S.; da Silva, S.S. Bioethanol production from sugarcane bagasse hemicellulose hydrolysate by immobilized S. shehatae in a fluidized bed fermenter under magnetic field. BioEnergy Res. 2019, 12, 338–346. [Google Scholar] [CrossRef]
- Devi, G.K.; Vignesh, K.; Chozhavendhan, S. Effective utilization of sugarcane trash for energy production. In Refining Biomass Residues for Sustainable Energy and Bioproducts; Elsevier: Amsterdam, The Netherlands, 2020; pp. 259–273. [Google Scholar]
- Farzad, S.; Mandegari, M.A.; Guo, M.; Haigh, K.F.; Shah, N.; Görgens, J.F. Multi-product biorefineries from lignocelluloses: A pathway to revitalisation of the sugar industry? Biotechnol. Biofuels 2017, 10, 87. [Google Scholar] [CrossRef]
- Dewanti, P.; Widuri, L.I.; Alfian, F.N.; Addy, H.S.; Okviandari, P.; Sugiharto, B. Rapid propagation of virus-free sugarcane (Saccharum officinarum) by somatic embryogenesis. Agric. Agric. Sci. Procedia 2016, 9, 456–461. [Google Scholar] [CrossRef]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Rani, V.; Raina, S. Genetic fidelity of organized meristem-derived micropropagated plants: A critical reappraisal. In Vitro Cell. Dev. Biol. Plant 2000, 36, 319–330. [Google Scholar] [CrossRef]
- Devarumath, R.; Nandy, S.; Rani, V.; Marimuthu, S.; Muraleedharan, N.; Raina, S. RAPD, ISSR and RFLP fingerprints as useful markers to evaluate genetic integrity of micropropagated plants of three diploid and triploid elite tea clones representing Camellia sinensis (China type) and C. assamica ssp. assamica (Assam-India type). Plant Cell Rep. 2002, 21, 166–173. [Google Scholar]
- Guasmi, F.; Elfalleh, W.; Hannachi, H.; Feres, K.; Touil, L.; Marzougui, N.; Triki, T.; Ferchichi, A. The use of ISSR and RAPD markers for genetic diversity among south tunisian barley. Int. Sch. Res. Not. 2012, 2012, 952196. [Google Scholar] [CrossRef]
- Rajpal, V.R.; Sharma, S.; Devarumath, R.M.; Chaudhary, M.; Kumar, A.; Khare, N.; Raina, S. Nuclear and Organelle DNA Fingerprints as the Most Useful Markers to Evaluate Genetic Integrity of Micropropagated Plants. In Tree Biotechnology; CRC Press: Boca Raton, FL, USA, 2014; p. 303. [Google Scholar]
- Sugiyama, A.; Saida, Y.; Yoshimizu, M.; Takanashi, K.; Sosso, D.; Frommer, W.B.; Yazaki, K. Molecular characterization of LjSWEET3, a sugar transporter in nodules of Lotus japonicus. Plant Cell Physiol. 2017, 58, 298–306. [Google Scholar]
- Carothers, A.M.; Rizvi, H.; Hasson, R.M.; Heit, Y.I.; Davids, J.S.; Bertagnolli, M.M.; Cho, N.L. Mesenchymal Stromal Cell Mutations and Wound Healing Contribute to the Etiology of Desmoid TumorsAPC-Deficient MSCs in Desmoid Tumors. Cancer Res. 2012, 72, 346–355. [Google Scholar] [CrossRef]
- Dumont, J.; Euwart, D.; Mei, B.; Estes, S.; Kshirsagar, R. Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Crit. Rev. Biotechnol. 2016, 36, 1110–1122. [Google Scholar] [CrossRef]
- Long, S.P.; ZHU, X.G.; Naidu, S.L.; Ort, D.R. Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 2006, 29, 315–330. [Google Scholar] [CrossRef]
- Tao, Y.; George-Jaeggli, B.; Bouteillé-Pallas, M.; Tai, S.; Cruickshank, A.; Jordan, D.; Mace, E. Genetic diversity of C4 photosynthesis pathway genes in Sorghum bicolor (L.). Genes 2020, 11, 806. [Google Scholar] [CrossRef]
- von Caemmerer, S.; Furbank, R.T. Strategies for improving C4 photosynthesis. Curr. Opin. Plant Biol. 2016, 31, 125–134. [Google Scholar] [CrossRef] [PubMed]
- McCormack, J.E.; Hird, S.M.; Zellmer, A.J.; Carstens, B.C.; Brumfield, R.T. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenetics Evol. 2013, 66, 526–538. [Google Scholar] [CrossRef] [PubMed]
- Steele, P.R.; Hertweck, K.L.; Mayfield, D.; McKain, M.R.; Leebens-Mack, J.; Pires, J.C. Quality and quantity of data recovered from massively parallel sequencing: Examples in Asparagales and Poaceae. Am. J. Bot. 2012, 99, 330–348. [Google Scholar] [CrossRef] [PubMed]
- Arthan, W.; McKain, M.R.; Traiperm, P.; Welker, C.A.; Teisher, J.K.; Kellogg, E.A. Phylogenomics of Andropogoneae (Panicoideae: Poaceae) of mainland Southeast Asia. Syst. Bot. 2017, 42, 418–431. [Google Scholar] [CrossRef]
- Estep, M.C.; McKain, M.R.; Vela Diaz, D.; Zhong, J.; Hodge, J.G.; Hodkinson, T.R.; Layton, D.J.; Malcomber, S.T.; Pasquet, R.; Kellogg, E.A. Allopolyploidy, diversification, and the Miocene grassland expansion. Proc. Natl. Acad. Sci. USA 2014, 111, 15149–15154. [Google Scholar] [CrossRef]
- Welker, C.A.; Souza-Chies, T.T.; Longhi-Wagner, H.M.; Peichoto, M.C.; McKain, M.R.; Kellogg, E.A. Phylogenetic analysis of Saccharum sl (Poaceae; Andropogoneae), with emphasis on the circumscription of the South American species. Am. J. Bot. 2015, 102, 248–263. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.A. Breeding for improved sugar content in sugarcane. Field Crops Res. 2005, 92, 277–290. [Google Scholar] [CrossRef]
- Morais, L.K.d.; Aguiar, M.S.d.; Albuquerque e Silva, P.d.; Câmara, T.M.M.; Cursi, D.E.; Júnior, A.R.F.; Chapola, R.G.; Carneiro, M.S.; Bespalhok Filho, J.C. Breeding of sugarcane. In Industrial Crops; Springer: Berlin/Heidelberg, Germany, 2015; pp. 29–42. [Google Scholar]
- Todd, J.; Glaz, B.; Burner, D.; Kimbeng, C. Historical use of cultivars as parents in Florida and Louisiana sugarcane breeding programs. Int. Sch. Res. Not. 2015, 2015, 257417. [Google Scholar] [CrossRef]
- Cursi, D.E.; Hoffmann, H.P.; Barbosa, G.; Bressiani, J.A.; Gazaffi, R.; Chapola, R.G.; Fernandes Junior, A.; Balsalobre, T.W.A.; Diniz, C.A.; Santos, J.M. History and current status of sugarcane breeding, germplasm development and molecular genetics in Brazil. Sugar Tech 2021, 24, 112–133. [Google Scholar] [CrossRef]
- Piperidis, G.; Piperidis, N.; D’Hont, A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genom. 2010, 284, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, A.; Tejedor, M.T.; Erazzu, L.E.; Cabada, S.; Sopena, R. Pedigree comparison highlights genetic similarities and potential industrial values of sugarcane cultivars. Euphytica 2017, 213, 121. [Google Scholar] [CrossRef]
- Lima, M.; Garcia, A.A.F.; Oliveira, K.; Matsuoka, S.; Arizono, H.; de Souza, A. Analysis of genetic similarity detected by AFLP and coefficient of parentage among genotypes of sugar cane (Saccharum spp.). Theor. Appl. Genet. 2002, 104, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Deren, C. Genetic base of US mainland sugarcane. Crop Sci. 1995, 35, 1195–1199. [Google Scholar] [CrossRef]
- Liu, H.; Yang, X.; You, Q.; Song, J.; Wang, L.; Zhang, J.; Deng, Z.; Ming, R.; Wang, J. Pedigree, marker recruitment, and genetic diversity of modern sugarcane cultivars in China and the United States. Euphytica 2018, 214, 48. [Google Scholar] [CrossRef]
- You, Q.; Xu, L.; Zheng, Y.; Que, Y. Genetic diversity analysis of sugarcane parents in Chinese breeding programmes using gSSR markers. Sci. World J. 2013, 2013, 613062. [Google Scholar] [CrossRef]
- Zan, F.; Zhang, Y.; Wu, Z.; Zhao, J.; Wu, C.; Zhao, Y.; Chen, X.; Zhao, L.; Qin, W.; Yao, L. Genetic analysis of agronomic traits in elite sugarcane (Saccharum spp.) germplasm. PLoS ONE 2020, 15, e0233752. [Google Scholar] [CrossRef]
- Basnayake, S.W.; Morgan, T.C.; Wu, L.; Birch, R.G. Field performance of transgenic sugarcane expressing isomaltulose synthase. Plant Biotechnol. J. 2012, 10, 217–225. [Google Scholar] [CrossRef]
- Casu, R.E.; Rae, A.L.; Nielsen, J.M.; Perroux, J.M.; Bonnett, G.D.; Manners, J.M. Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families. Plant Mol. Biol. 2015, 89, 607–628. [Google Scholar] [CrossRef] [PubMed]
- Van Emon, J.M. The omics revolution in agricultural research. J. Agric. Food Chem. 2016, 64, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Khan, Q.; Kashif, M.; Shah, S.J. Comprehensive analysis of the mechanism underlying plastic microbiome and plants interaction, with future perspectives. J. Soil Plant Environ. 2022, 1, 31–43. [Google Scholar] [CrossRef]
- Cardoso-Silva, C.B.; Costa, E.A.; Mancini, M.C.; Balsalobre, T.W.A.; Canesin, L.E.C.; Pinto, L.R.; Carneiro, M.S.; Garcia, A.A.F.; de Souza, A.P.; Vicentini, R. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE 2014, 9, e88462. [Google Scholar] [CrossRef]
- Ma, H.-M.; Schulze, S.; Lee, S.; Yang, M.; Mirkov, E.; Irvine, J.; Moore, P.; Paterson, A. An EST survey of the sugarcane transcriptome. Theor. Appl. Genet. 2004, 108, 851–863. [Google Scholar] [CrossRef]
- Vettore, A.L.; da Silva, F.R.; Kemper, E.L.; Souza, G.M.; da Silva, A.M.; Ferro, M.I.T.; Henrique-Silva, F.; Giglioti, É.A.; Lemos, M.V.; Coutinho, L.L. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003, 13, 2725–2735. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wang, J.; Shang, H.; Huang, Y.; Yao, W.; Chen, B.; Zhang, M. Transcriptomic characterization and potential marker development of contrasting sugarcane cultivars. Sci. Rep. 2018, 8, 1683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Q.; Li, L.; Tang, H.; Zhang, Q.; Chen, Y.; Arrow, J.; Zhang, X.; Wang, A.; Miao, C. Recent polyploidization events in three Saccharum founding species. Plant Biotechnol. J. 2019, 17, 264–274. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef]
- Khan, Q.; Chen, J.Y.; Zeng, X.P.; Qin, Y.; Guo, D.J.; Mahmood, A.; Yang, L.T.; Liang, Q.; Song, X.P.; Xing, Y.X. Transcriptomic exploration of a high sucrose mutant in comparison with the low sucrose mother genotype in sugarcane during sugar accumulating stage. GCB Bioenergy 2021, 13, 1448–1465. [Google Scholar] [CrossRef]
- Mutz, K.-O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.H.; Chen, F.; Torres-Jerez, I.; Tang, Y.; Wang, M.; Du, Q.; Cheng, X.; Wen, J.; Dixon, R. Transcriptome analysis of secondary cell wall development in Medicago truncatula. BMC Genom. 2016, 17, 23. [Google Scholar] [CrossRef]
- Hoang, N.V.; Furtado, A.; Mason, P.J.; Marquardt, A.; Kasirajan, L.; Thirugnanasambandam, P.P.; Botha, F.C.; Henry, R.J. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genom. 2017, 18, 395. [Google Scholar] [CrossRef]
- Thirugnanasambandam, P.P.; Mason, P.J.; Hoang, N.V.; Furtado, A.; Botha, F.C.; Henry, R.J. Analysis of the diversity and tissue specificity of sucrose synthase genes in the long read transcriptome of sugarcane. BMC Plant Biol. 2019, 19, 160. [Google Scholar] [CrossRef]
- Singh, N.P.; Shami, V. Abiotic stress tolerance in sugarcane using genomics and proteomics techniques. In Biotechnology to Enhance Sugarcane Productivity and Stress Tolerance; CRC Press: Boca Raton, FL, USA, 2018; pp. 97–114. [Google Scholar]
- Fan, Y.-G.; Chen, R.-F.; Qiu, L.-H.; Zhou, Z.-F.; Zhou, H.-W.; Wei, J.-G.; Song, X.-P.; Luo, T.; Lakshmanan, P.; Huang, X. Quantitative proteomics analysis of sugarcane Ratoon crop Chlorosis. Sugar Tech 2021, 23, 673–681. [Google Scholar] [CrossRef]
- Vélez-Bermúdez, I.C.; Wen, T.-N.; Lan, P.; Schmidt, W. Isobaric tag for relative and absolute quantitation (iTRAQ)-based protein profiling in plants. In Plant Proteostasis; Springer: Berlin/Heidelberg, Germany, 2016; pp. 213–221. [Google Scholar]
- Ali, A.; Pan, Y.-B.; Wang, Q.-N.; Wang, J.-D.; Chen, J.-L.; Gao, S.-J. Genetic diversity and population structure analysis of Saccharum and Erianthus genera using microsatellite (SSR) markers. Sci. Rep. 2019, 9, 395. [Google Scholar] [CrossRef]
- Calderan-Rodrigues, M.J.; Jamet, E.; Douché, T.; Bonassi, M.B.R.; Cataldi, T.R.; Fonseca, J.G.; San Clemente, H.; Pont-Lezica, R.; Labate, C.A. Cell wall proteome of sugarcane stems: Comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol. 2016, 16, 14. [Google Scholar] [CrossRef]
- Calderan-Rodrigues, M.J.; Jamet, E.; Bonassi, M.B.C.R.; Guidetti-Gonzalez, S.; Begossi, A.C.; Setem, L.V.; Franceschini, L.M.; Fonseca, J.G.; Labate, C.A. Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 2014, 14, 738–749. [Google Scholar] [CrossRef]
- Ma, P.; Zhang, X.; Chen, L.; Zhao, Q.; Zhang, Q.; Hua, X.; Wang, Z.; Tang, H.; Yu, Q.; Zhang, M. Comparative analysis of sucrose phosphate synthase (SPS) gene family between Saccharum officinarum and Saccharum spontaneum. BMC Plant Biol. 2020, 20, 422. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; de Moura Vale, E.; Heringer, A.S.; Santa-Catarina, C.; Silveira, V. Putrescine induces somatic embryo development and proteomic changes in embryogenic callus of sugarcane. J. Proteom. 2016, 130, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Heringer, A.S.; Reis, R.S.; Passamani, L.Z.; de Souza-Filho, G.A.; Santa-Catarina, C.; Silveira, V. Comparative proteomics analysis of the effect of combined red and blue lights on sugarcane somatic embryogenesis. Acta Physiol. Plant. 2017, 39, 52. [Google Scholar] [CrossRef]
- Fonseca, J.G.; Calderan-Rodrigues, M.J.; de Moraes, F.E.; Cataldi, T.R.; Jamet, E.; Labate, C.A. Cell wall proteome of sugarcane young and mature leaves and stems. Proteomics 2018, 18, 1700129. [Google Scholar] [CrossRef]
- Salvato, F.; Wilson, R.; Portilla Llerena, J.P.; Kiyota, E.; Lima Reis, K.; Boaretto, L.F.; Balbuena, T.S.; Azevedo, R.A.; Thelen, J.J.; Mazzafera, P. Luxurious nitrogen fertilization of two sugar cane genotypes contrasting for lignin composition causes changes in the stem proteome related to carbon, nitrogen, and oxidant metabolism but does not alter lignin content. J. Proteome Res. 2017, 16, 3688–3703. [Google Scholar] [CrossRef]
- Suresha, G.; Mahadevaiah, C.; Appunu, C. Biotechnological interventions for improving sucrose accumulation in sugarcane. In Sugarcane Biotechnology: Challenges and Prospects; Springer: Berlin/Heidelberg, Germany, 2017; pp. 111–122. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- D’Hont, A.; Grivet, L.; Feldmann, P.; Glaszmann, J.; Rao, S.; Berding, N. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol. Gen. Genet. MGG 1996, 250, 405–413. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Dileep, A.; Lebonah, D.E.; Pramoda Kumari, J. A short review on proteomics and its applications. Int. Lett. Nat. Sci. 2014, 12, 77–84. [Google Scholar] [CrossRef]
- Anderson, N.G.; Anderson, N.L. Twenty years of two-dimensional electrophoresis: Past, present and future. Electrophoresis 1996, 17, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Poljak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Blackstock, W.P.; Weir, M.P. Proteomics: Quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999, 17, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Kersten, B.; Agrawal, G.K.; Durek, P.; Neigenfind, J.; Schulze, W.; Walther, D.; Rakwal, R. Plant phosphoproteomics: An update. Proteomics 2009, 9, 964–988. [Google Scholar] [CrossRef] [PubMed]
- Sangha, J.S.; Yolanda, H.C.; Kaur, J.; Khan, W.; Abduljaleel, Z.; Alanazi, M.S.; Mills, A.; Adalla, C.B.; Bennett, J.; Prithiviraj, B. Proteome analysis of rice (Oryza sativa L.) mutants reveals differentially induced proteins during brown planthopper (Nilaparvata lugens) infestation. Int. J. Mol. Sci. 2013, 14, 3921–3945. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Rampitsch, C.; Bykova, N.V. Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front. Plant Sci. 2015, 6, 209. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Zieske, L.R. A perspective on the use of iTRAQ™ reagent technology for protein complex and profiling studies. J. Exp. Bot. 2006, 57, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, L.; Wang, Z.; Peng, Q.; Yang, Y.; Chen, Y.; Que, Y. Comparative proteomics reveals that central metabolism changes are associated with resistance against Sporisorium scitamineum in sugarcane. BMC Genom. 2016, 17, 800. [Google Scholar] [CrossRef]
- Khan, Q.; Qin, Y.; Guo, D.-J.; Lu, Z.; Xie, X.-Q.; Yang, L.-T.; Liang, Q.; Song, X.-P.; Xing, Y.-X.; Li, Y.-R. Proteome Based Comparative Investigation of a High Sucrose Sugarcane Mutant in Contrast to the Low Sucrose Mother Variety by Using TMT Quantitative Proteomics. Sugar Tech 2022, 24, 1246–1259. [Google Scholar] [CrossRef]
- Ma, S.; Li, Y.; Li, X.; Sui, X.; Zhang, Z. Phloem unloading strategies and mechanisms in crop fruits. J. Plant Growth Regul. 2019, 38, 494–500. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef]
- Wang, M.; Li, A.M.; Liao, F.; Qin, C.X.; Chen, Z.L.; Zhou, L.; Li, Y.R.; Li, X.F.; Lakshmanan, P.; Huang, D.L. Control of sucrose accumulation in sugarcane (Saccharum spp. hybrids) involves miRNA-mediated regulation of genes and transcription factors associated with sugar metabolism. GCB Bioenergy 2022, 14, 173–191. [Google Scholar] [CrossRef]
- Whittaker, A.; Botha, F.C. Carbon partitioning during sucrose accumulation in sugarcane internodal tissue. Plant Physiol. 1997, 115, 1651–1659. [Google Scholar] [CrossRef]
- Rae, A.L.; Perroux, J.M.; Grof, C.P. Sucrose partitioning between vascular bundles and storage parenchyma in the sugarcane stem: A potential role for the ShSUT1 sucrose transporter. Planta 2005, 220, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Uys, L.; Botha, F.C.; Hofmeyr, J.-H.S.; Rohwer, J.M. Kinetic model of sucrose accumulation in maturing sugarcane culm tissue. Phytochemistry 2007, 68, 2375–2392. [Google Scholar] [CrossRef] [PubMed]
- Welbaum, G.E.; Meinzer, F.C. Compartmentation of solutes and water in developing sugarcane stalk tissue. Plant Physiol. 1990, 93, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nayak, S.; Koch, K.; Ming, R. Carbon partitioning in sugarcane (Saccharum species). Front. Plant Sci. 2013, 4, 201. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhou, H.; Ma, B.; Owiti, A.; Korban, S.S.; Han, Y. Divergent evolutionary pattern of sugar transporter genes is associated with the difference in sugar accumulation between grasses and eudicots. Sci. Rep. 2016, 6, 29153. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Stitt, M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007, 30, 1126–1149. [Google Scholar] [CrossRef] [PubMed]
- McCormick, A.J.; Cramer, M.D.; Watt, D.A. Regulation of photosynthesis by sugars in sugarcane leaves. J. Plant Physiol. 2008, 165, 1817–1829. [Google Scholar] [CrossRef]
- McCarthy, R.L.; Zhong, R.; Ye, Z.-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef]
- Watt, D.; McCormick, A.; Govender, C.; Carson, D.; Cramer, M.; Huckett, B.; Botha, F. Increasing the utility of genomics in unravelling sucrose accumulation. Field Crops Res. 2005, 92, 149–158. [Google Scholar] [CrossRef]
- Grof, C.; So, C.T.E.; Perroux, J.; Bonnett, G.; Forrester, R. The five families of sucrose-phosphate synthase genes in Saccharum spp. are differentially expressed in leaves and stem. Funct. Plant Biol. 2006, 33, 605–610. [Google Scholar] [CrossRef]
- Ishimaru, K.; Ono, K.; Kashiwagi, T. Identification of a new gene controlling plant height in rice using the candidate-gene strategy. Planta 2004, 218, 388–395. [Google Scholar] [CrossRef]
- Sarquís, J.I.; Gonzalez, H.; de Jiménez, E.S.; Dunlap, J.R. Physiological traits associated with mass selection for improved yield in a maize population. Field Crops Res. 1998, 56, 239–246. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Komor, E.; Moore, P.H. Sucrose accumulation in the sugarcane stem is regulated by the difference between the activities of soluble acid invertase and sucrose phosphate synthase. Plant Physiol. 1997, 115, 609–616. [Google Scholar] [CrossRef] [PubMed]
- LF, L.; CE, C. The biosynthesis of sucrose phosphate. J. Biol. Chem. 1955, 214, 157–165. [Google Scholar]
- Lunn, J.E. Sucrose-phosphatase gene families in plants. Gene 2003, 303, 187–196. [Google Scholar] [CrossRef]
- Lutfiyya, L.L.; Xu, N.; Robert, L.; Morrell, J.A.; Miller, P.W.; Duff, S.M. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 2007, 164, 923–933. [Google Scholar] [CrossRef]
- Langenkämper, G.; Fung, R.W.; Newcomb, R.D.; Atkinson, R.G.; Gardner, R.C.; MacRae, E.A. Sucrose phosphate synthase genes in plants belong to three different families. J. Mol. Evol. 2002, 54, 322–332. [Google Scholar] [CrossRef]
- Galtier, N.; Foyer, C.H.; Huber, J.; Voelker, T.A.; Huber, S.C. Effects of elevated sucrose-phosphate synthase activity on photosynthesis, assimilate partitioning, and growth in tomato (Lycopersicon esculentum var UC82B). Plant Physiol. 1993, 101, 535–543. [Google Scholar] [CrossRef]
- Micallef, B.J.; Haskins, K.A.; Vanderveer, P.J.; Roh, K.-S.; Shewmaker, C.K.; Sharkey, T.D. Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta 1995, 196, 327–334. [Google Scholar] [CrossRef]
- Seger, M.; Gebril, S.; Tabilona, J.; Peel, A.; Sengupta-Gopalan, C. Impact of concurrent overexpression of cytosolic glutamine synthetase (GS1) and sucrose phosphate synthase (SPS) on growth and development in transgenic tobacco. Planta 2015, 241, 69–81. [Google Scholar] [CrossRef]
- Verma, A.K.; Upadhyay, S.; Verma, P.C.; Solomon, S.; Singh, S.B. Functional analysis of sucrose phosphate synthase (SPS) and sucrose synthase (SS) in sugarcane (Saccharum) cultivars. Plant Biol. 2011, 13, 325–332. [Google Scholar] [CrossRef]
- Buczynski, S.; Thom, M.; Chourey, P.; Maretzki, A. Tissue distribution and characterization of sucrose synthase isozymes in sugarcane. J. Plant Physiol. 1993, 142, 641–646. [Google Scholar] [CrossRef]
- Lingle, S.E.; Smith, R.C. Sucrose metabolism related to growth and ripening in sugarcane internodes. Crop Sci. 1991, 31, 172–177. [Google Scholar] [CrossRef]
- Barratt, D.P.; Barber, L.; Kruger, N.J.; Smith, A.M.; Wang, T.L.; Martin, C. Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol. 2001, 127, 655–664. [Google Scholar] [CrossRef]
- Carson, D.L.; Huckett, B.I.; Botha, F.C. Sugarcane ESTs differentially expressed in immature and maturing internodal tissue. Plant Sci. 2002, 162, 289–300. [Google Scholar] [CrossRef]
- Chourey, P.; Taliercio, E.; Carlson, S.; Ruan, Y.-L. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. MGG 1998, 259, 88–96. [Google Scholar] [CrossRef]
- Huang, J.-W.; Chen, J.-T.; Yu, W.-P.; Shyur, L.-F.; Wang, A.-Y.; Sung, H.-Y.; Lee, P.-D.; Su, J.-C. Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Biosci. Biotechnol. Biochem. 1996, 60, 233–239. [Google Scholar] [CrossRef]
- Botha, F.C.; Black, K.G. Sucrose phosphate synthase and sucrose synthase activity during maturation of internodal tissue in sugarcane. Funct. Plant Biol. 2000, 27, 81–85. [Google Scholar] [CrossRef]
- Tomlinson, P.T.; Duke, E.R.; Nolte, K.D.; Koch, K.E. Sucrose synthase and invertase in isolated vascular bundles. Plant Physiol. 1991, 97, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Russell, D. Maize sucrose synthase-1 promoter directs phloem cell-specific expression of Gus gene in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 1990, 87, 4144–4148. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Van den Ende, W.; Van Laere, A.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-H.; Huang, Y.-C.; Yang, C.-S.; Yang, C.-C.; Wang, A.-Y.; Sung, H.-Y. Insights into the catalytic properties of bamboo vacuolar invertase through mutational analysis of active site residues. Phytochemistry 2009, 70, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Venkateshwar, M.; Chaitanya, K.; Altaf, M.; Mahammad, E.; Bee, H.; Reddy, G. Influence of micronutrients on yeast growth and β-D-fructofuranosidase production. Indian J. Microbiol. 2010, 50, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, H.; Tanase, K.; Shiratake, K.; Mori, H.; Yamaki, S. Purification and characterization of two soluble acid invertase isozymes from Japanese pear fruit. Phytochemistry 2003, 63, 125–129. [Google Scholar] [CrossRef]
- Kim, D.; Lee, G.; Chang, M.; Park, J.; Chung, Y.; Lee, S.; Lee, T.-K. Purification and biochemical characterization of insoluble acid invertase (INAC-INV) from pea seedlings. J. Agric. Food Chem. 2011, 59, 11228–11233. [Google Scholar] [CrossRef]
- Hatch, M.; Glasziou, K. Sugar accumulation cycle in sugar cane. II. Relationship of invertase activity to sugar content & growth rate in storage tissue of plants grown in controlled environments. Plant Physiol. 1963, 38, 344. [Google Scholar] [PubMed]
- Ma, H.; Albert, H.H.; Paull, R.; Moore, P.H. Metabolic engineering of invertase activities in different subcellular compartments affects sucrose accumulation in sugarcane cells. Funct. Plant Biol. 2000, 27, 1021–1030. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Shivalingamurthy, S.G.; Anangi, R.; Kalaipandian, S.; Glassop, D.; King, G.F.; Rae, A.L. Identification and functional characterization of sugarcane invertase inhibitor (ShINH1): A potential candidate for reducing pre-and post-harvest loss of sucrose in sugarcane. Front. Plant Sci. 2018, 9, 598. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. Biochemistry and Molecular Biology of Plants; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Anterola, A.M.; Jeon, J.-H.; Davin, L.B.; Lewis, N.G. Transcriptional control of monolignol biosynthesis in Pinus taeda: Factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J. Biol. Chem. 2002, 277, 18272–18280. [Google Scholar] [CrossRef]
- Kimura, S.; Laosinchai, W.; Itoh, T.; Cui, X.; Linder, C.R.; Brown, R.M., Jr. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 1999, 11, 2075–2085. [Google Scholar] [CrossRef]
- Wendler, R.; Veith, R.; Dancer, J.; Stitt, M.; Komor, E. Sucrose storage in cell suspension cultures of Saccharum sp. (sugarcane) is regulated by a cycle of synthesis and degradation. Planta 1991, 183, 31–39. [Google Scholar] [CrossRef]
- Bindon, K.A.; Botha, F.C. Carbon allocation to the insoluble fraction, respiration and triose-phosphate cycling in the sugarcane culm. Physiol. Plant. 2002, 116, 12–19. [Google Scholar] [CrossRef]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Slewinski, T.L.; Meeley, R.; Braun, D.M. Sucrose transporter1 functions in phloem loading in maize leaves. J. Exp. Bot. 2009, 60, 881–892. [Google Scholar] [CrossRef]
- Srivastava, A.C.; Ganesan, S.; Ismail, I.O.; Ayre, B.G. Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 2008, 148, 200–211. [Google Scholar] [CrossRef]
- Chincinska, I.; Gier, K.; Krügel, U.; Liesche, J.; He, H.; Grimm, B.; Harren, F.J.; Cristescu, S.M.; Kühn, C. Photoperiodic regulation of the sucrose transporter StSUT4 affects the expression of circadian-regulated genes and ethylene production. Front. Plant Sci. 2013, 4, 26. [Google Scholar] [CrossRef]
- Payyavula, R.S.; Tay, K.H.; Tsai, C.J.; Harding, S.A. The sucrose transporter family in Populus: The importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 2011, 65, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Hua, X.; Zhang, Q.; Wang, J.; Shen, Q.; Zhang, X.; Wang, K.; Yu, Q.; Lin, Y.-R.; Ming, R. New insights into the evolution and functional divergence of the SWEET family in Saccharum based on comparative genomics. BMC Plant Biol. 2018, 18, 270. [Google Scholar] [CrossRef]
- ElSayed, A.I.; Lehrer, A.; Ebrahim, M.; Mohamed, A.H.; Komor, E. Assessment of sucrose transporters, metabolites and sucrose phosphate synthase in different sugarcane tissues. Physiol. Mol. Biol. Plants 2017, 23, 703–712. [Google Scholar] [CrossRef]
- Rae, A.L.; Grof, C.P.; Casu, R.E.; Bonnett, G.D. Sucrose accumulation in the sugarcane stem: Pathways and control points for transport and compartmentation. Field Crops Res. 2005, 92, 159–168. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, M.; Liang, Y.; Xing, Y.; Yang, L.; Chen, M.; Comstock, J.C.; Li, Y.; Yang, L. Morphological and physiological responses of sugarcane to Leifsonia xyli subsp. xyli infection. Plant Dis. 2016, 100, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Schulz, A.; Beyhl, D.; Marten, I.; Wormit, A.; Neuhaus, E.; Poschet, G.; Büttner, M.; Schneider, S.; Sauer, N.; Hedrich, R. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 2011, 68, 129–136. [Google Scholar] [CrossRef]
- Jeena, G.S.; Kumar, S.; Shukla, R.K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 2019, 100, 351–365. [Google Scholar] [CrossRef]
- Cabib, E.; Leloir, L.F. The biosynthesis of trehalose phosphate. J. Biol. Chem. 1958, 231, 259–275. [Google Scholar] [CrossRef]
- Elbein, A.D.; Pan, Y.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Gonzalez-Uriarte, A.; Griffiths, C.A.; Hassani-Pak, K. The role of trehalose 6-phosphate in crop yield and resilience. Plant Physiol. 2018, 177, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Watson, A.; Griffiths, C.A. Trehalose 6-phosphate signalling and impact on crop yield. Biochem. Soc. Trans. 2020, 48, 2127–2137. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.; Feng, Y.; Tu, M.; Wittich, P.E.; Bate, N.J.; Messing, J. Transcriptome and metabolome reveal distinct carbon allocation patterns during internode sugar accumulation in different sorghum genotypes. Plant Biotechnol. J. 2019, 17, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Junior, N.N.; Nicolau, M.S.A.P.; Mantovanini, L.J.; Zingaretti, S.M. Expression analysis of two genes coding for trehalose-6-phosphate synthase (TPS), in sugarcane (Saccharum spp.) under water stress. Am. J. Plant Sci. 2013, 2013, 91–99. [Google Scholar] [CrossRef]
- Hu, X.; Wu, Z.-D.; Luo, Z.-Y.; Burner, D.M.; Pan, Y.-B.; Wu, C.-W. Genome-wide analysis of the trehalose-6-phosphate synthase (TPS) gene family and expression profiling of ScTPS genes in sugarcane. Agronomy 2020, 10, 969. [Google Scholar] [CrossRef]
- Bosch, S. Trehalose and Carbon Partitioning in Sugarcane. Ph.D. Thesis, University of Stellenbosch, Stellenbosch, South Africa, 2005. [Google Scholar]
- Eastmond, P.J.; Van Dijken, A.J.; Spielman, M.; Kerr, A.; Tissier, A.F.; Dickinson, H.G.; Jones, J.D.; Smeekens, S.C.; Graham, I.A. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 2002, 29, 225–235. [Google Scholar] [CrossRef]
- Schluepmann, H.; Pellny, T.; van Dijken, A.; Smeekens, S.; Paul, M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6849–6854. [Google Scholar] [CrossRef]
- Gómez, L.D.; Gilday, A.; Feil, R.; Lunn, J.E.; Graham, I.A. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010, 64, 1–13. [Google Scholar] [CrossRef]
- Schluepmann, H.; Berke, L.; Sanchez-Perez, G.F. Metabolism control over growth: A case for trehalose-6-phosphate in plants. J. Exp. Bot. 2012, 63, 3379–3390. [Google Scholar] [CrossRef]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef]
- Wu, L.; Birch, R.G. Physiological basis for enhanced sucrose accumulation in an engineered sugarcane cell line. Funct. Plant Biol. 2010, 37, 1161–1174. [Google Scholar] [CrossRef]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant Sci. 2011, 2, 70. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Bevan, M. The regulation of transcription factor activity in plants. Trends Plant Sci. 1998, 3, 378–383. [Google Scholar] [CrossRef]
- Pugh, B.F. Mechanisms of transcription complex assembly. Curr. Opin. Cell Biol. 1996, 8, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Ranish, J.A.; Hahn, S. Transcription: Basal factors and activation. Curr. Opin. Genet. Dev. 1996, 6, 151–158. [Google Scholar] [CrossRef]
- Cowell, I.G. Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 1994, 19, 38–42. [Google Scholar] [CrossRef]
- Alessio, V.M.; Cavaçana, N.; Dantas, L.L.d.B.; Lee, N.; Hotta, C.T.; Imaizumi, T.; Menossi, M. The FBH family of bHLH transcription factors controls ACC synthase expression in sugarcane. J. Exp. Bot. 2018, 69, 2511–2525. [Google Scholar] [CrossRef]
- Figueiredo, R.; Portilla Llerena, J.P.; Kiyota, E.; Ferreira, S.S.; Cardeli, B.R.; de Souza, S.C.R.; dos Santos Brito, M.; Sodek, L.; Cesarino, I.; Mazzafera, P. The sugarcane ShMYB78 transcription factor activates suberin biosynthesis in Nicotiana benthamiana. Plant Mol. Biol. 2020, 104, 411–427. [Google Scholar] [CrossRef]
- Wang, D.; Wang, L.; Su, W.; Ren, Y.; You, C.; Zhang, C.; Que, Y.; Su, Y. A class III WRKY transcription factor in sugarcane was involved in biotic and abiotic stress responses. Sci. Rep. 2020, 10, 20964. [Google Scholar] [CrossRef]
- Li, Z.; Hua, X.; Zhong, W.; Yuan, Y.; Wang, Y.; Wang, Z.; Ming, R.; Zhang, J. Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol. 2020, 61, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ling, H.; Liu, F.; Huang, N.; Wang, L.; Mao, H.; Li, C.; Tang, H.; Su, W.; Su, Y. Cloning and expression analysis of a II d sub-group WRKY transcription factor gene from sugarcane. Sci. Agric. Sin. 2018, 51, 4409–4423. [Google Scholar]
- Papini-Terzi, F.S.; Rocha, F.R.; Vêncio, R.Z.; Felix, J.M.; Branco, D.S.; Waclawovsky, A.J.; Del Bem, L.E.; Lembke, C.G.; Costa, M.D.; Nishiyama, M.Y. Sugarcane genes associated with sucrose content. BMC Genom. 2009, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sheen, J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, F.; Hummel, M.; Schuurmans, J.; Wiese-Klinkenberg, A.; Smeekens, S.; Hanson, J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009, 150, 1356–1367. [Google Scholar] [CrossRef]
- Zhou, X.; Herbst-Robinson, K.J.; Zhang, J. Visualizing dynamic activities of signaling enzymes using genetically encodable FRET-based biosensors: From designs to applications. Methods Enzymol. 2012, 504, 317–340. [Google Scholar]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-activated protein kinase cascades in plant hormone signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar] [CrossRef]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef]
- Sugden, C.; Donaghy, P.G.; Halford, N.G.; Hardie, D.G. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999, 120, 257–274. [Google Scholar] [CrossRef]
- Vicentini, R.; de Maria Felix, J.; Dornelas, M.C.; Menossi, M. Characterization of a sugarcane (Saccharum spp.) gene homolog to the brassinosteroid insensitive1-associated receptor kinase 1 that is associated to sugar content. Plant Cell Rep. 2009, 28, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Hardtke, C.S. Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 2011, 21, R365–R373. [Google Scholar] [CrossRef]
- Botha, F.C.; Lakshmanan, P.; O’Connell, A.; Moore, P.H. Hormones and growth regulators. In Sugarcane: Physiology, Biochemistry, and Functional Biology; Wiley: Hoboken, NJ, USA, 2013; pp. 331–377. [Google Scholar]
- Gray, W.M. Hormonal regulation of plant growth and development. PLoS Biol. 2004, 2, e311. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E. Communal benefits of transgenic corn. Science 2010, 330, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Chory, J. Crosstalk in cellular signaling: Background noise or the real thing? Dev. Cell 2011, 21, 985–991. [Google Scholar] [CrossRef]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 2016, 7, 1240. [Google Scholar] [CrossRef]
- Frick, E.M.; Strader, L.C. Roles for IBA-derived auxin in plant development. J. Exp. Bot. 2018, 69, 169–177. [Google Scholar] [CrossRef]
- Zhu, F.; Wai, C.M.; Zhang, J.; Jones, T.C.; Nagai, C.; Ming, R. Differential expression of hormone related genes between extreme segregants of a Saccharum interspecific F2 population. Euphytica 2018, 214, 55. [Google Scholar] [CrossRef]
- Liscum, E.; Reed, J. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]
- Ntambo, M.S.; Meng, J.Y.; Rott, P.; Royer, M.; Lin, L.H.; Zhang, H.L.; Gao, S.J. Identification and characterization of Xanthomonas albilineans causing sugarcane leaf scald in China using multilocus sequence analysis. Plant Pathol. 2019, 68, 269–277. [Google Scholar] [CrossRef]
- Masood, A.; Iqbal, N.; Khan, N.A. Role of ethylene in alleviation of cadmium-induced photosynthetic capacity inhibition by sulphur in mustard. Plant Cell Environ. 2012, 35, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Nazar, R.; Khan, M.I.R.; Iqbal, N.; Masood, A.; Khan, N.A. Involvement of ethylene in reversal of salt-inhibited photosynthesis by sulfur in mustard. Physiol. Plant. 2014, 152, 331–344. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Fukao, T.; Ronald, P.; Ismail, A.; Heuer, S.; Mackill, D. Submergence tolerant rice: SUB1′s journey from landrace to modern cultivar. Rice 2010, 3, 138–147. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Etheridge, N.; Schaller, G.E. Ethylene signal transduction. Ann. Bot. 2005, 95, 901–915. [Google Scholar] [CrossRef]
- Chang, C. Q&A: How do plants respond to ethylene and what is its importance? BMC Biol. 2016, 14, 7. [Google Scholar]
- Chen, Z.; Qin, C.; Wang, M.; Liao, F.; Liao, Q.; Liu, X.; Li, Y.; Lakshmanan, P.; Long, M.; Huang, D. Ethylene-mediated improvement in sucrose accumulation in ripening sugarcane involves increased sink strength. BMC Plant Biol. 2019, 19, 285. [Google Scholar] [CrossRef] [PubMed]
- Cunha, C.P.; Roberto, G.G.; Vicentini, R.; Lembke, C.G.; Souza, G.M.; Ribeiro, R.V.; Machado, E.C.; Lagôa, A.M.; Menossi, M. Ethylene-induced transcriptional and hormonal responses at the onset of sugarcane ripening. Sci. Rep. 2017, 7, 43364. [Google Scholar] [CrossRef]
- de Almeida Silva, M.; Caputo, M.M. Ripening and the use of ripeners for better sugarcane management. In Crop Management—Cases and Tools for Higher Yield and Sustainability; InTech: London, UK, 2012; Chapter 1. [Google Scholar]
- Gao, X.X.; Fan, X.; Dao, J.M.; Deng, J.; Li, R.D.; Zhang, Y.B.; Guo, J.W.; Liu, S.C. Relationship between endogenous ethylene production and natural defoliation traits during the maturation of sugarcane. Bragantia 2015, 74, 189–195. [Google Scholar] [CrossRef]
- Yang, C.; Lu, X.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol. Plant 2015, 8, 495–505. [Google Scholar] [CrossRef]
- Jan, A.; Komatsu, S. Functional characterization of gibberellin-regulated genes in rice using microarray system. Genom. Proteom. Bioinform. 2006, 4, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, J.; Weinstain, R.; Shani, E. Gibberellin localization and transport in plants. Trends Plant Sci. 2018, 23, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A century of gibberellin research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 2001, 20, 387–442. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.; Osgood, R.; Carr, J.; Ginoza, H. Sugarcane studies with gibberellin. V. Plot harvests vs. stalk harvests to assess the effect of applied GA3 on sucrose yield. J. Plant Growth Regul. 1982, 1, 205–210. [Google Scholar]
- Gupta, R.; Chakrabarty, S. Gibberellic acid in plant: Still a mystery unresolved. Plant Signal. Behav. 2013, 8, e25504. [Google Scholar] [CrossRef]
- Resende, P.; Soares, J.; Hudetz, M. Moddus, a plant growth regulator and management tool for sugarcane production in Brazil. Sugar Cane Int. 2000, 103, 5–9. [Google Scholar]
- Claeys, H.; De Bodt, S.; Inzé, D. Gibberellins and DELLAs: Central nodes in growth regulatory networks. Trends Plant Sci. 2014, 19, 231–239. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, F.; Zhao, X.; Ge, H.; Chai, L.; Chen, S.; Perl, A.; Ma, H. Proteomic analysis of berry-sizing effect of GA3 on seedless Vitis vinifera L. Proteomics 2012, 12, 86–94. [Google Scholar] [CrossRef]
- Pribil, M.; Hermann, S.; Dun, G.; Karno, X.; Ngo, C.; O’neill, S.; Wang, L.; Bonnett, G.; Chandler, P.; Beveridge, C. Altering sugarcane shoot architecture through genetic engineering: Prospects for increasing cane and sugar yield. In Proceedings of the Australian Society of Sugar Cane Technologists, Cairns, Australia, 8–11 May 2007; pp. 251–257. [Google Scholar]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef]
- Moore, P.H.; Ginoza, H. Gibberellin Studies with Sugarcane. III. Effects of Rate and Frequency of Gibberellic Acid Applications on Stalk Length and Fresh Weight 1. Crop Sci. 1980, 20, 78–82. [Google Scholar] [CrossRef]
- Qiu, L.-H.; Chen, R.-F.; Luo, H.-M.; Fan, Y.-G.; Huang, X.; Liu, J.-X.; Xiong, F.-Q.; Zhou, H.-W.; Gan, C.-K.; Wu, J.-M. Effects of exogenous GA3 and DPC treatments on levels of endogenous hormone and expression of key gibberellin biosynthesis pathway genes during stem elongation in sugarcane. Sugar Tech 2019, 21, 936–948. [Google Scholar] [CrossRef]
- Kuhnle, J.; Moore, P.; Haddon, W.; Fitch, M. Identification of gibberellins from sugarcane plants. J. Plant Growth Regul. 1983, 2, 59–71. [Google Scholar] [CrossRef]
- Roopendra, K.; Sharma, A.; Chandra, A.; Saxena, S. Gibberellin-induced perturbation of source–sink communication promotes sucrose accumulation in sugarcane. 3 Biotech 2018, 8, 418. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Laby, R.J.; Kincaid, M.S.; Kim, D.; Gibson, S.I. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000, 23, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Assmann, S.M. An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell 1996, 8, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Tsz-fung, F.C. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA-binding START proteins. Science 2009, 324, 1068. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.-J.; Wan, Y.; Liu, W.; Xie, S. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 2018, 23, 3340–3351.e3345. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Kwak, J.M.; Allen, G.J. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 2001, 410, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef]
- Li, C.-N.; Yang, L.-T.; Srivastava, M.K.; Li, Y.-R. Foliar application of abscisic acid improves drought tolerance of sugarcane plant under severe water stress. Int. J. Agric. Innov. Res. 2014, 3, 101–107. [Google Scholar]
- Hayamichi, Y. Effects of abscisic acid treatment on the growth and sugar accumulation of sugarcane plant. Jpn. J. Trop. Agric. 1997, 41, 22–26. [Google Scholar]

| Sucrose phosphate synthase (SPS); SPS1, SPS2, SPS4, SPS5 |
| Sucrose synthase (SuSy); SuSy1, SuSy2, SuSy4 |
| Soluble acid invertase (SAI) |
| Cell wall invertase (CWI) |
| Neutral invertase (NI) |
| Sucrose transporter; SWEET1b, SWEET13c, SWEET4a/4b, SUT1, SUT4, SUT5, SUT6, ShPST2a, ShPST2b, ShSUT4. |
| SNF1-like kinases |
| Trehalose-phosphate synthase |
| Cellulose synthase (CeS); CesA1, CesA7, CesA9, bk2l3,CesA10, CesA11, CesA12 |
| Trehalose 6-phosphate (T6P) |
| Trehalose-6-phosphate phosphatase (TPP) |
| Transcription factors (TF); WRKY, MYB, NAC, AP2/ERF |
| Basic helix-loop-helix (bHLH) |
| ScFBHs and ScACS2 |
| Mitogen-activated PTK (MAPK) |
| Sucrose-nonfermentation1-related protein kinase1-2 (ScSnRK1-2) |
| BCL2 antagonist/killer 1 (ScBAK1) |
| Phytohormones; Auxin (IAA), AUX/IAA, Gibberellin (GA), Cytokinin (CTK), Abscisic acid (ABA), Ethylene (ETH). |
| Brassinosteroid (BR), jasmonates, salicylic acid, a peptide hormone, and strigolactone |
| Gretchen Hagen3 (GH3), small auxin-up RNAs (SAUR |
| Ethylene receptor (ETR), Ethylene response sensor (ERS), Ethylene insensitive (EIN) ETP, EIN2, Targeting protein genes EIL, EIN3 like, EBF (EIN3-F) |
| GA1, GA3, GA4 and GA7 A19, GA20 and GA29 |
| ABA receptor pyrabactin resistance 1 (PYR1), PYR1-like (PYL) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, Q.; Qin, Y.; Guo, D.-J.; Yang, L.-T.; Song, X.-P.; Xing, Y.-X.; Li, Y.-R. A Review of the Diverse Genes and Molecules Involved in Sucrose Metabolism and Innovative Approaches to Improve Sucrose Content in Sugarcane. Agronomy 2023, 13, 2957. https://doi.org/10.3390/agronomy13122957
Khan Q, Qin Y, Guo D-J, Yang L-T, Song X-P, Xing Y-X, Li Y-R. A Review of the Diverse Genes and Molecules Involved in Sucrose Metabolism and Innovative Approaches to Improve Sucrose Content in Sugarcane. Agronomy. 2023; 13(12):2957. https://doi.org/10.3390/agronomy13122957
Chicago/Turabian StyleKhan, Qaisar, Ying Qin, Dao-Jun Guo, Li-Tao Yang, Xiu-Peng Song, Yong-Xiu Xing, and Yang-Rui Li. 2023. "A Review of the Diverse Genes and Molecules Involved in Sucrose Metabolism and Innovative Approaches to Improve Sucrose Content in Sugarcane" Agronomy 13, no. 12: 2957. https://doi.org/10.3390/agronomy13122957
APA StyleKhan, Q., Qin, Y., Guo, D.-J., Yang, L.-T., Song, X.-P., Xing, Y.-X., & Li, Y.-R. (2023). A Review of the Diverse Genes and Molecules Involved in Sucrose Metabolism and Innovative Approaches to Improve Sucrose Content in Sugarcane. Agronomy, 13(12), 2957. https://doi.org/10.3390/agronomy13122957







