BrDMC1, a Recombinase Gene, Is Involved in Seed Germination in Brassica rapa under Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Evolutionary Analysis and Molecular Evolutionary Rate Analysis
2.3. Cis-Acting Elements and Co-Expression Network Analysis
2.4. Subcellular Localization Analysis in N.benthamiana
2.5. Histochemical Estimation of GUS Activity and ROS Activity Determination of BrDMC1
2.6. Analysis of Proline, Malondialdehyde (MDA) and Soluble Sugar Content
2.7. Assays of Antioxidative Enzyme Activity
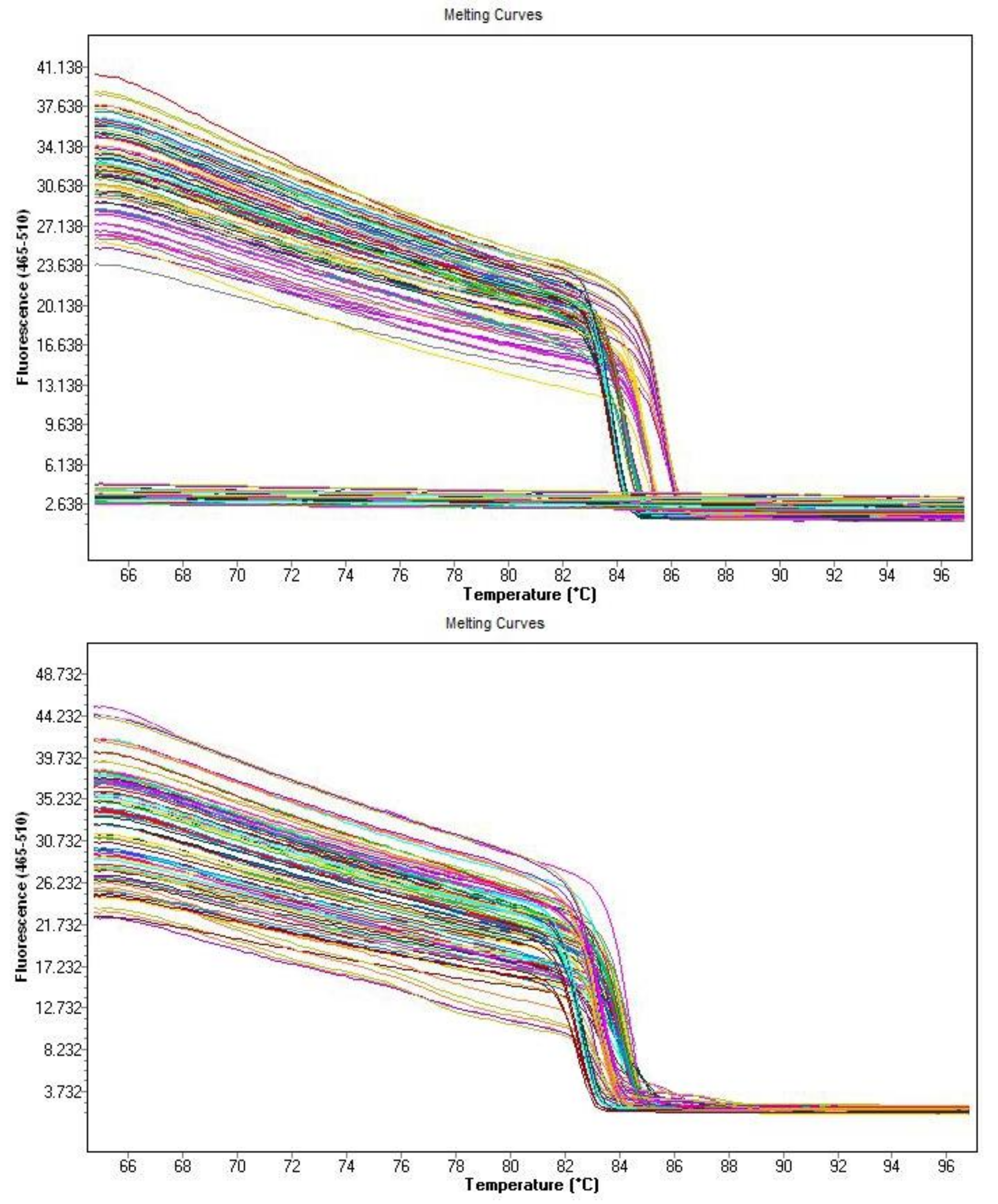
2.8. Quantitative Real-Time PCR Analysis
2.9. Statistical Analyses
3. Results
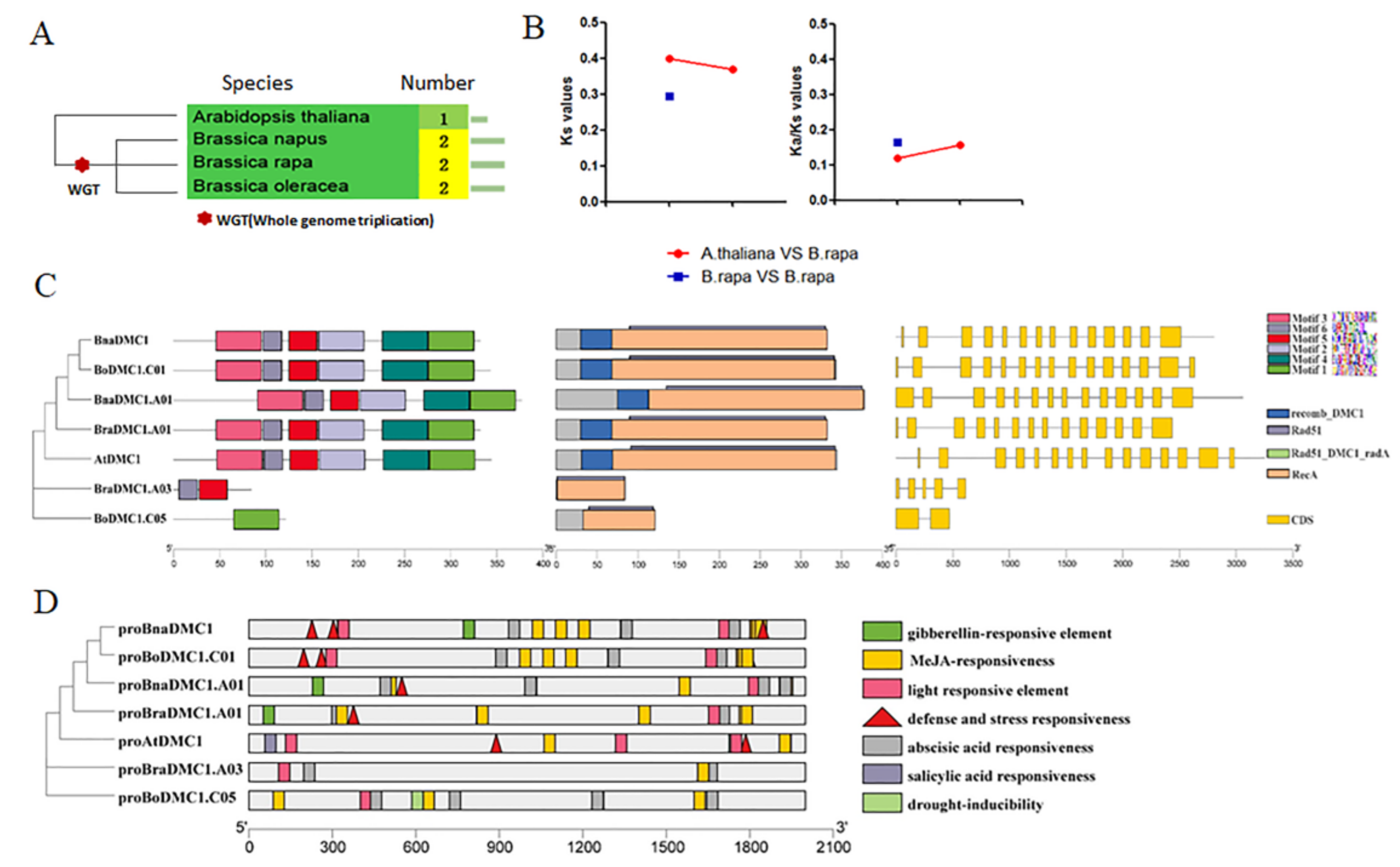
3.1. Analysis of DMC1 Genes following a Whole-Genome Triplication Event in B. rapa
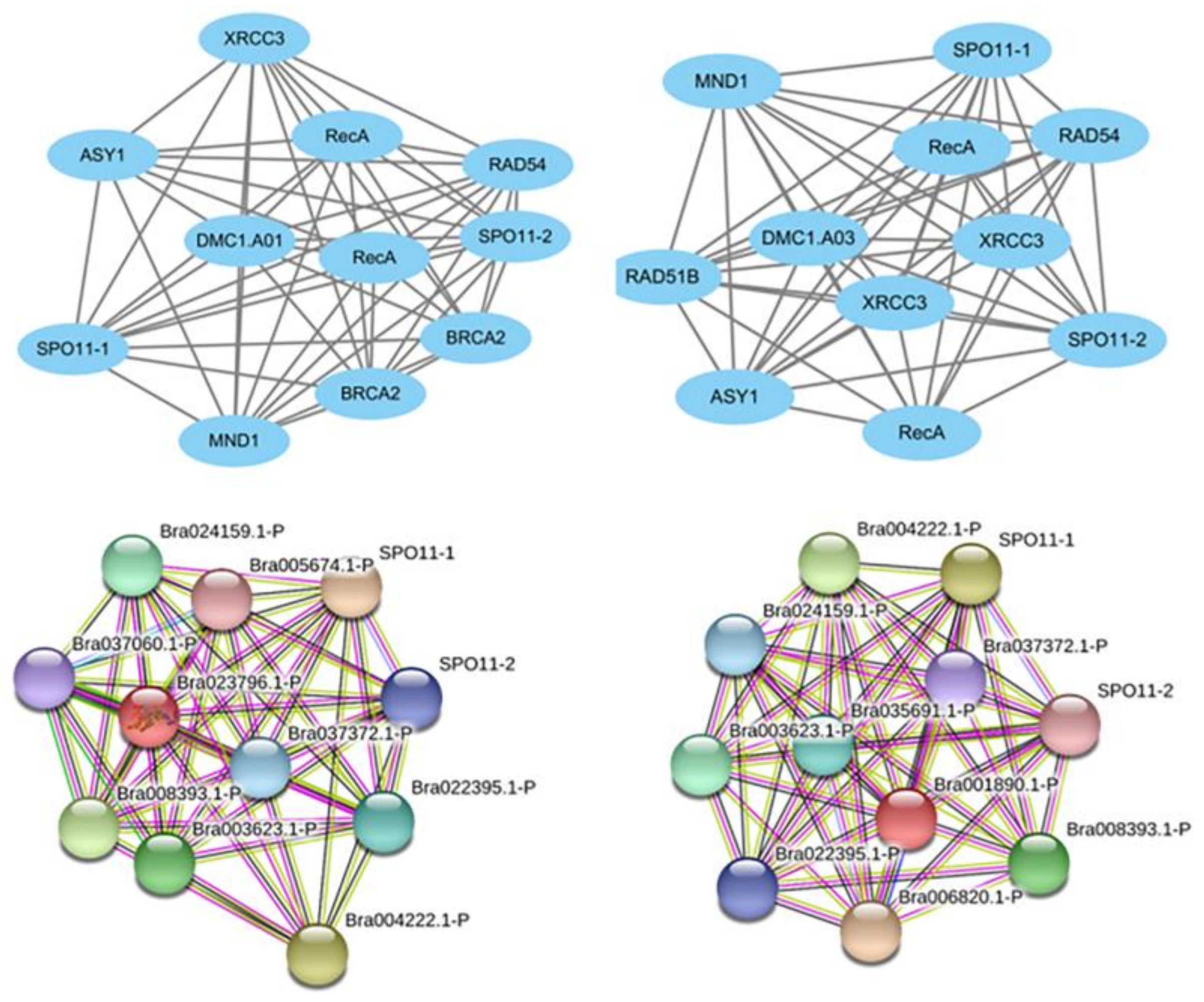
3.2. Interaction Networks of Potential DNA-Repair-related BrDMC1 in B. rapa
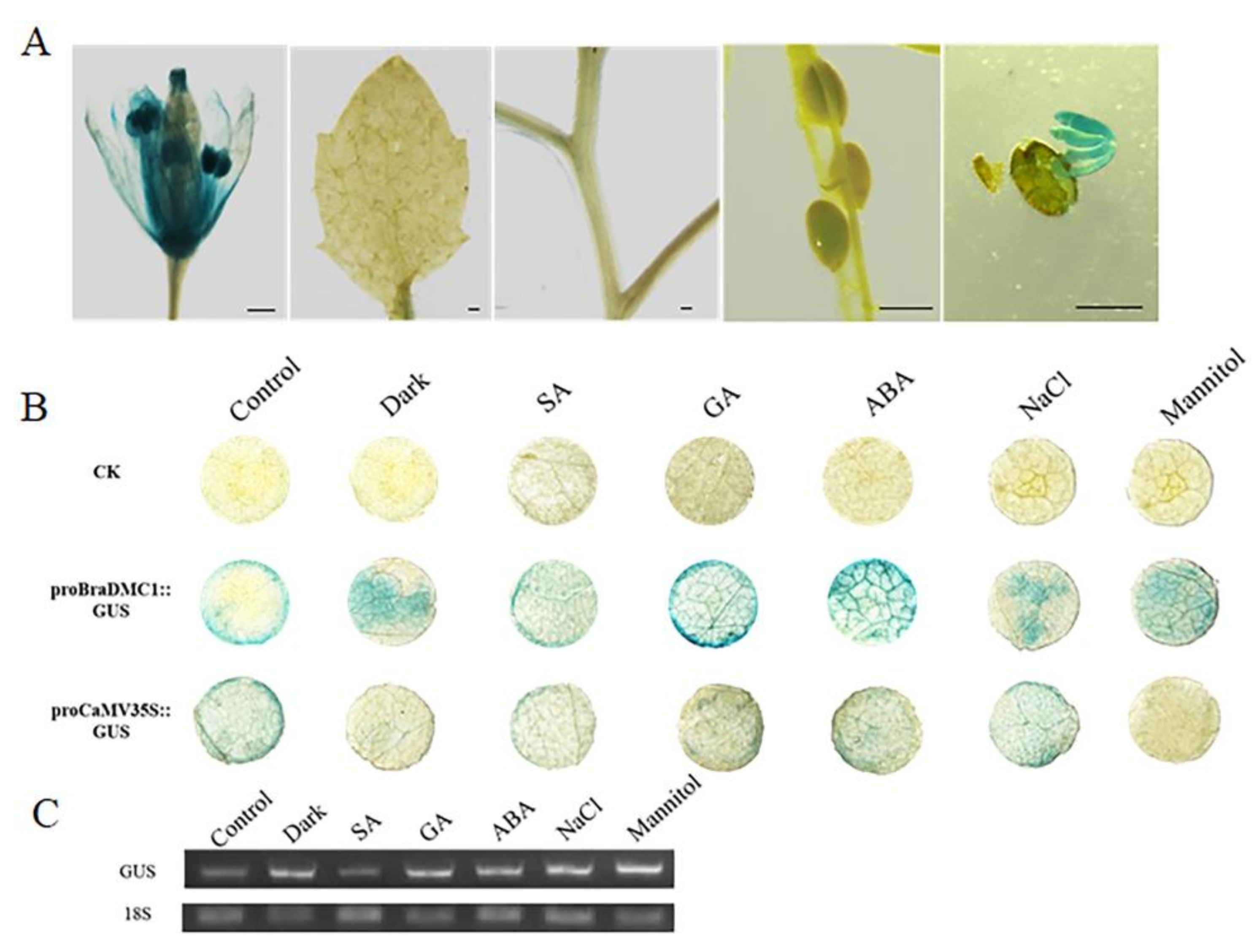
3.3. GUS Staining Analysis of Putative Promoter Regions of BrDMC1
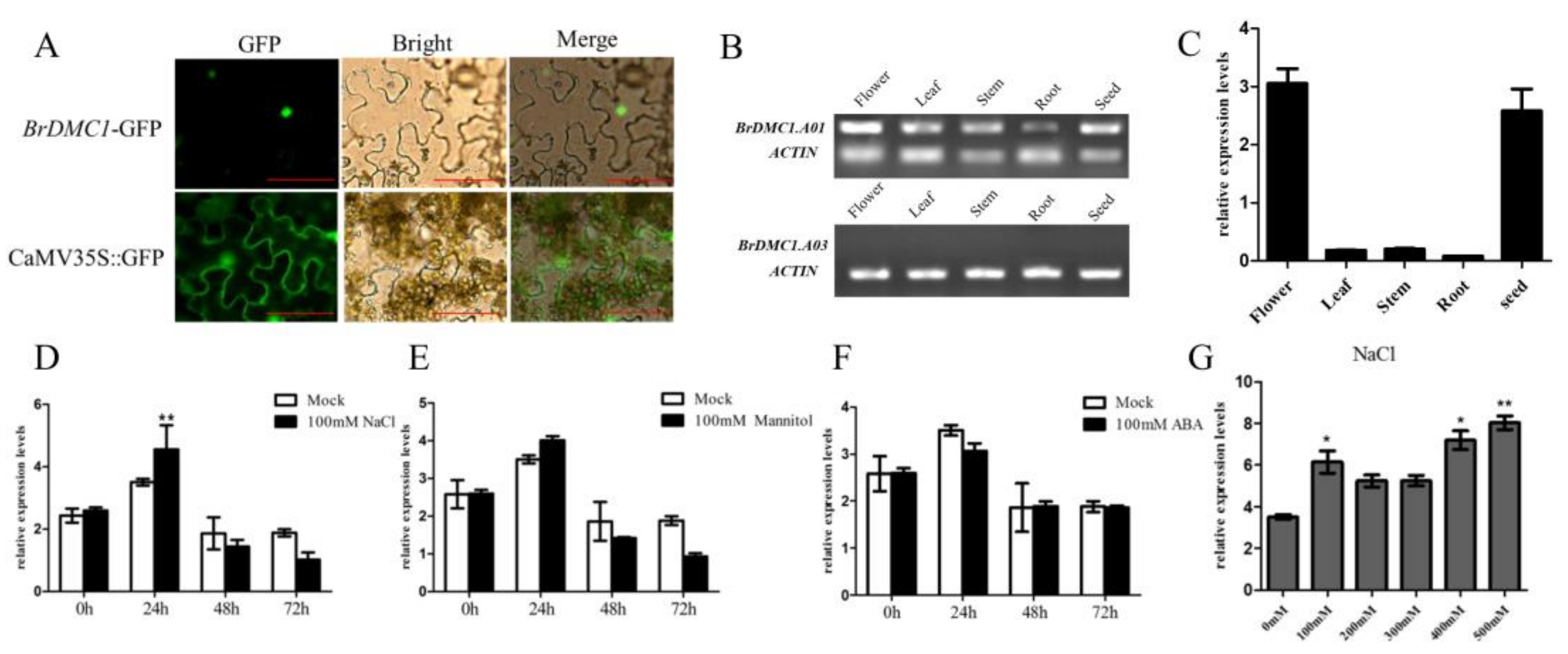
3.4. Expression Pattern, Salt Stress and Subcellular Localization of BrDMC1
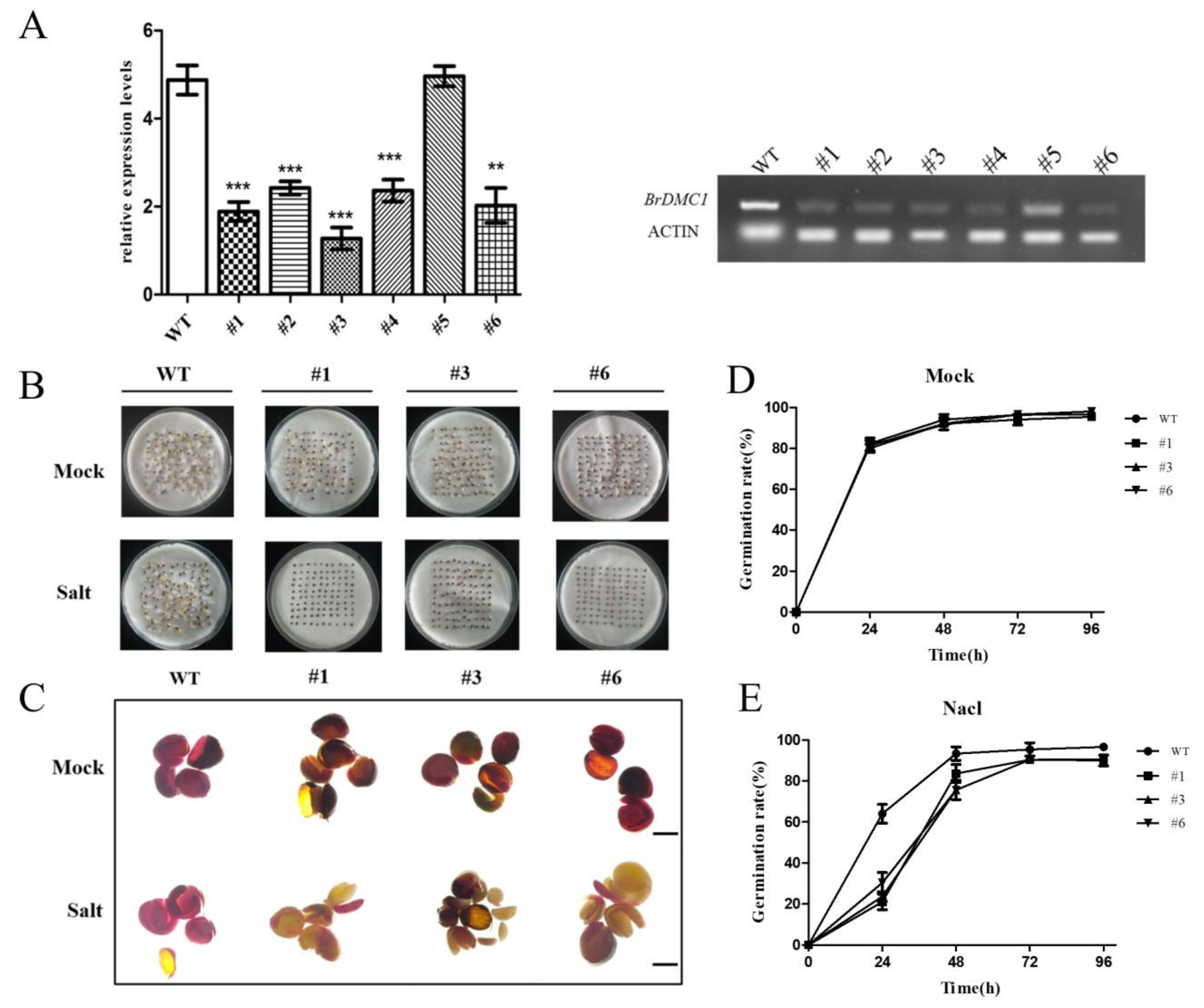
3.5. BrDMC1-RNAi Reduced Salt Tolerance in B. rapa Seed Germination
3.6. BrDMC1-RNAi Decreased Antioxidant Enzyme Activity under Salt Stress
3.7. BrDMC1 Differently Regulates the Expression of Stress-Related Genes and Damage Repair Genes under Salt Stress
4. Discussion
4.1. BrDMC1 Evolved to Retain a Single Copy Function in B.rapa
4.2. BrDMC1 May Play Essential Roles in Response to Stress
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| qBrDMC1.A01-F | GGAGGTGGGATTGAGACTGC |
| qBrDMC1.A01-R | GGACAATCCTATCAGGGCGG |
| qBrDMC1.A03-F | TCACTACTGGTTGCCAAGCTC |
| qBrDMC1.A03-R | TAAGCCACTTTTCCGTTCCCT |
| β-antin-F | ATCAACTACCAGCCTCCAAC |
| β-antin-R | CTGCTGTGTTGTTGCTGATC |
| ProBrDMC1.A01-F | AAGCAAATGTGTGCTGTGAAC |
| ProBrDMC1.A01-R | GAAGCGAGCAGAGATCGAAG |
| ProBrDMC1.A03-F | ATATGCACACCAAGTGAGACAG |
| ProBrDMC1.A03-R | CATCAAGAGCTTGGCAACCAG |
| GUS-F | TACCGACGAAAACGGCAAGA |
| GUS-R | ATGCCATGTTCATCTGCCCA |
| BrDMC1.A01-F | ATGCTTTCTGCTCTCAAATC |
| BrDMC1.A03-R | TTAAAAGGATATAGCTTCGC |
| BrDMC1(RNAi)-BP-F | TGTAATGGTCTCATGATGCATACCAAAAAGAACCTCACTGGAATC |
| BrDMC1(RNAi)-BP-R | CAGTGTCAATGTAAGCCACTTTCCCATTCCCACCTTTCATGCTTG |
| PDK(RNAi)-F | GACGAAGAAGATAAAAGTTGAGAGT |
| PDK(RNAi)-R | ACCTTGTTTATTCATGTTCGACTAA |
| P5C-F | GAACCGTACCTTCTCAAACT |
| P5C-R | CTCCATCGGTATCTGATGTT |
| TOP6B-F | CTCGTGGAAAGTTTGGTTTA |
| TOP6B-R | CTGTATTTCGGCTCCATGCC |
| NCED3-F | TCACCACTTCTTCGACGGAG |
| NCED3-R | GGTCGACTAAACCGGCTGCG |
| RAD51-F | GCAGCTTCAAAGCTGGTTCC |
| RAD51-R | CAGCTCCATTTAGTCCAAAC |
| XRCC3-F | GCTCTCGCGATCGTTTCATC |
| XRCC3-R | CACAGTTAGCCCAAGCCAATC |
| BRCA2-F | TCGCAATGATATAGCTCATG |
| BRCA2-R | CGTAGCTCTTGGACGCATTC |
Appendix B
References
- Wang, L.; Ma, R.; Yin, Y.; Jiao, Z. Role of Carbon Ion Beams Irradiation in Mitigating Cold Stress in Arabidopsis Thaliana. Ecotoxicol. Environ. Saf. 2018, 162, 341–347. [Google Scholar] [CrossRef]
- Jafari, A.; Kamarehie, B.; Ghaderpoori, M.; Khoshnamvand, N.; Birjandi, M. The Concentration Data of Heavy Metals in Iranian Grown and Imported Rice and Human Health Hazard Assessment. Data Brief 2018, 16, 453–459. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant Abiotic Stress Response and Nutrient Use Efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt Stress under the Scalpel—Dissecting the Genetics of Salt Tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity Tolerance Mechanisms in Glycophytes: An Overview with the Central Focus on Rice Plants. Rice 2012, 5, 11. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a Putative Calcium Sensor, Interacts with the Protein Kinase SOS2 to Protect Arabidopsis Shoots from Salt Stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Bray, C.M.; West, C.E. The Importance of Safeguarding Genome Integrity in Germination and Seed Longevity. J. Exp. Bot. 2015, 66, 3549–3558. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen Species, Abiotic Stress and Stress Combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Low-Temperature Stress: Is Phytohormones Application a Remedy? Environ. Sci. Pollut. Res. 2017, 24, 21574–21590. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Footitt, S.; Bray, C.M.; Finch-Savage, W.E.; West, C.E. DNA Damage Checkpoint Kinase ATM Regulates Germination and Maintains Genome Stability in Seeds. Proc. Natl. Acad. Sci. USA 2016, 113, 9647–9652. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Masnavi, G.; Bhardwaj, R.M.; Jiang, Q.; Bray, C.M.; West, C.E. A Plant DNA Ligase Is an Important Determinant of Seed Longevity. Plant J. 2010, 63, 848–860. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Drury, G.E.; Bray, C.M.; West, C.E. Repairing Breaks in the Plant Genome: The Importance of Keeping It Together. New Phytol. 2011, 192, 805–822. [Google Scholar] [CrossRef]
- Powell, A.; Matthews, S. Association of Official Seed Analysts Society of Commercial Seed Technologists (SCST) Seed Aging/Repair Hypothesis Leads to New Testing Methods. Seed Technol. 2012, 3, 2. [Google Scholar]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed Germination and Vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef]
- Dandoy, E.; Schnys, R.; Deltour, R.; Verly, W.G. Appearance and Repair of Apurinic/Apyrimidinic Sites in DNA during Early Germination of Zea Mays. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1987, 181, 57–60. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Latham, R.; Wang, D.; Alsharif, M.; West, C.E. Seed DNA Damage Responses Promote Germination and Growth in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2022, 119, 1–8. [Google Scholar] [CrossRef]
- Navashin, M.; Shkvarnikov, P. Process of Mutation in Resting Seeds Accelerated by Increased Temperature. Nature 1933, 132, 482–483. [Google Scholar] [CrossRef]
- Sequencing, G.; Consortium, P. The Genome of the Mesopolyploid Crop Species Brassica Rapa. Nat. Genet. 2011, 43, 1035–1040. [Google Scholar] [CrossRef]
- Lyu, T.; Liu, W.; Hu, Z.; Xiang, X.; Liu, T.; Xiong, X.; Cao, J. Molecular Characterization and Expression Analysis Reveal the Roles of Cys2/His2 Zinc-Finger Transcription Factors during Flower Development of Brassica Rapa Subsp. Chinensis. Plant Mol. Biol. 2020, 102, 123–141. [Google Scholar] [CrossRef]
- Devisetty, U.K.; Mayes, K.; Mayes, S. The RAD51 and DMC1 Homoeologous Genes of Bread Wheat: Cloning, Molecular Characterization and Expression Analysis. BMC Res. Notes 2010, 3, 245. [Google Scholar] [CrossRef]
- Prentiss, M.; Prévost, C.; Danilowicz, C.; Prentiss, M.; Prévost, C.; Structure, C.D.; Danilowicz, C.; Prentiss, M.; Pre, C. Structure/Function Relationships in RecA Protein- Mediated Homology Recognition and Strand Exchange Recognition and Strand Exchange. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 453–476. [Google Scholar] [CrossRef]
- Steinfeld, J.B.; Belán, O.; Kwon, Y.; Terakawa, T.; Al-zain, A.; Smith, M.J.; Crickard, J.B.; Qi, Z.; Zhao, W.; Rothstein, R.; et al. Defining the Influence of Rad51 and Dmc1 Lineage-Specific Amino Acids on Genetic Recombination. Genes Dev. 2019, 33, 1191–1207. [Google Scholar] [CrossRef]
- Lan, W.; Lin, S.; Kao, C.; Chang, W.; Yeh, H.; Chang, H. Rad51 Facilitates Filament Assembly of Meiosis-Specific. Proc. Natl. Acad. Sci. USA 2020, 117, 11257–11264. [Google Scholar] [CrossRef]
- Li, T.; Shi, Q.; Davies, B.; Donnelly, P.; Hinch, A.G.; Becker, P.W.; Li, T.; Moralli, D.; Zhang, G.; Bycroft, C.; et al. Article The Configuration of RPA, RAD51, and DMC1 Binding in Meiosis Reveals the Nature of Critical Recombination Intermediates Ll The Configuration of RPA, RAD51, and DMC1 Binding in Meiosis Reveals the Nature of Critical Recombination Intermediates. Mol. Cell 2020, 79, 689–701.e10. [Google Scholar] [CrossRef]
- Szurman-Zubrzycka, M.; Baran, B.; Stolarek-Januszkiewicz, M.; Kwaśniewska, J.; Szarejko, I.; Gruszka, D. The Dmc1 Mutant Allows an Insight into the DNA Double-Strand Break Repair during Meiosis in Barley (Hordeum Vulgare L.). Front. Plant Sci. 2019, 10, 761. [Google Scholar] [CrossRef]
- Couteau, F.; Belzile, F.; Horlow, C.; Grandjean, O.; Vezon, D.; Doutriaux, M.P. Random Chromosome Segregation without Meiotic Arrest in Both Male and Female Meiocytes of a Dmc1 Mutant of Arabidopsis. Plant Cell 1999, 11, 1623–1634. [Google Scholar] [CrossRef]
- Akada, I.S. Characterization of a DMC1 Homologue, RiLIM15, in Meiotic Panicles, Mitotic Cultured Cells and Mature Leaves of Rice (Oryza Sativa L.). Theor. Appl. Genet. 2001, 102, 1159–1163. [Google Scholar]
- Draeger, T.; Martin, A.C.; Kader, A.; Ali, A.; María, P.; Rey, D.; Shaw, P.; Moore, G. Dmc1 Is a Candidate for Temperature Tolerance during Wheat Meiosis. Theor. Appl. Genet. 2020, 133, 809–828. [Google Scholar] [CrossRef]
- Jeon, H.; Jin, Y.; Choi, M.H.; Lee, H.; Kim, M. Chloroplast-Targeted Bacterial RecA Proteins Confer Tolerance to Chloroplast DNA Damage by Methyl Viologen or UV-C Radiation in Tobacco (Nicotiana Tabacum) Plants. Physiol. Plant. 2013, 147, 218–233. [Google Scholar] [CrossRef]
- Muris, D.F.R.; Vreeken, K.; Carr, A.M.; Broughton, B.C.; Lehmann, A.R.; Lohman, P.H.M.; Pastink, A. Cloning the RAD51 Homologue of Schizosaccharomyces Pombe. Nucleic Acids Res. 1993, 21, 4586–4591. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.; Zhou, L.; Ali, A.; Jiang, H.; Zhu, S.; Li, X. DNA Repair Gene ZmRAD51A Improves Rice and Arabidopsis Resistance to Disease. Int. J. Mol. Sci. 2019, 20, 807. [Google Scholar] [CrossRef]
- Cao, G.; Jiang, W.; Shi, G.; Tian, Z.; Shang, J.; Xie, Z.; Chen, W.; Tian, B.; Wei, X.; Wei, F.; et al. Br PARP1, a Poly (ADP-Ribose) Polymerase Gene, Is Involved in Root Development in Brassica Rapa under Drought Stress. Horticulturae 2022, 8, 78. [Google Scholar] [CrossRef]
- Cao, G.; Gu, H.; Jiang, W.; Tian, Z.; Shi, G.; Chen, W.; Tian, B.; Wei, X.; Zhang, L.; Wei, F.; et al. BrDHC1, a Novel Putative DEAD-Box Helicase Gene, Confers Drought Tolerance in Transgenic Brassica Rapa. Horticulturae 2022, 8, 707. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, Transient Expression of Fluorescent Fusion Proteins in Tobacco Plants and Generation of Stably Transformed Plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Wang, C.Q.; Guthrie, C.; Sarmast, M.K.; Dehesh, K. BBX19 Interacts with CONSTANS to Repress FLOWERING LOCUS T Transcription, Defining a Flowering Time Checkpoint in Arabidopsis. Plant Cell 2014, 26, 3589–3602. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-Mediated Transformation of Arabidopsis Thaliana Using the Floral Dip Method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Leclercq, J.; Martin, F.; Sanier, C.; Clément-Vidal, A.; Fabre, D.; Oliver, G.; Lardet, L.; Ayar, A.; Peyramard, M.; Montoro, P. Over-Expression of a Cytosolic Isoform of the HbCuZnSOD Gene in Hevea Brasiliensis Changes Its Response to a Water Deficit. Plant Mol. Biol. 2012, 80, 255–272. [Google Scholar] [CrossRef]
- Kong, W.; Liu, F.; Zhang, C.; Zhang, J.; Feng, H. Non-Destructive Determination of Malondialdehyde (MDA) Distribution in Oilseed Rape Leaves by Laboratory Scale NIR Hyperspectral Imaging. Sci. Rep. 2016, 6, 35393. [Google Scholar] [CrossRef]
- Ahmad, H.; Hayat, S.; Ali, M.; Liu, T.; Cheng, Z. The Combination of Arbuscular Mycorrhizal Fungi Inoculation (Glomus Versiforme) and 28-Homobrassinolide Spraying Intervals Improves Growth by Enhancing Photosynthesis, Nutrient Absorption, and Antioxidant System in Cucumber (Cucumis Sativus L.) under Sali. Ecol. Evol. 2018, 8, 5724–5740. [Google Scholar] [CrossRef]
- Michaelidou, K.; Tzovaras, A.; Missitzis, I.; Ardavanis, A.; Scorilas, A. The Expression of the CEACAM19 Gene, a Novel Member of the CEA Family, Is Associated with Breast Cancer Progression. Int. J. Oncol. 2013, 42, 1770–1777. [Google Scholar] [CrossRef]
- Freeling, M. Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, J.; Wang, X. Genome Triplication Drove the Diversification of Brassica Plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef]
- Wu, P.; Wang, W.; Duan, W.; Li, Y.; Hou, X. Comprehensive Analysis of the CDPK-SnRK Superfamily Genes in Chinese Cabbage and Its Evolutionary Implications in Plants. Front. Plant Sci. 2017, 8, 162. [Google Scholar] [CrossRef]
- McGrew, D.A.; Knight, K.L. Molecular Design and Functional Organization of the RecA Protein. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 385–432. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional Divergence of Duplicated Genes Formed by Polyploidy during Arabidopsis Evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Rensing, S.A. Gene Duplication as a Driver of Plant Morphogenetic Evolution. Curr. Opin. Plant Biol. 2014, 17, 43–48. [Google Scholar] [CrossRef]
- Wang, H.; Hu, Q.; Tang, D.; Liu, X.; Du, G.; Shen, Y.; Li, Y.; Cheng, Z. OsDMC1 Is Not Required for Homologous Pairing in Rice Meiosis. Plant Physiol. 2016, 171, 230–241. [Google Scholar] [CrossRef]
- Kurzbauer, M.T.; Uanschou, C.; Chen, D.; Schlögelhofer, P. The Recombinases DMC1 and RAD51 Are Functionally and Spatially Separated during Meiosis in Arabidopsis. Plant Cell 2012, 24, 2058–2070. [Google Scholar] [CrossRef]
- Kobayashi, W.; Liu, E.; Ishii, H.; Matsunaga, S.; Schlögelhofer, P.; Kurumizaka, H. Homologous Pairing Activities of Arabidopsis Thaliana RAD51 and DMC1. J. Biochem. 2019, 165, 289–295. [Google Scholar] [CrossRef]
- Aranda, J.; Bardina, C.; Beceiro, A.; Rumbo, S.; Cabral, M.P.; Barbé, J.; Bou, G. Acinetobacter Baumannii Reca Protein in Repair of DNA Damage, Antimicrobial Resistance, General Stress Response, and Virulence. J. Bacteriol. 2011, 193, 3740–3747. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xie, Z.; Tian, Z.; Wang, S.; Shi, G.; Chen, W.; Cao, G.; Tian, B.; Wei, X.; Zhang, L.; et al. BrDMC1, a Recombinase Gene, Is Involved in Seed Germination in Brassica rapa under Salt Stress. Agronomy 2023, 13, 595. https://doi.org/10.3390/agronomy13020595
Wang X, Xie Z, Tian Z, Wang S, Shi G, Chen W, Cao G, Tian B, Wei X, Zhang L, et al. BrDMC1, a Recombinase Gene, Is Involved in Seed Germination in Brassica rapa under Salt Stress. Agronomy. 2023; 13(2):595. https://doi.org/10.3390/agronomy13020595
Chicago/Turabian StyleWang, Xulin, Zhengqing Xie, Zhaoran Tian, Shuaipeng Wang, Gongyao Shi, Weiwei Chen, Gangqiang Cao, Baoming Tian, Xiaochun Wei, Luyue Zhang, and et al. 2023. "BrDMC1, a Recombinase Gene, Is Involved in Seed Germination in Brassica rapa under Salt Stress" Agronomy 13, no. 2: 595. https://doi.org/10.3390/agronomy13020595
APA StyleWang, X., Xie, Z., Tian, Z., Wang, S., Shi, G., Chen, W., Cao, G., Tian, B., Wei, X., Zhang, L., & Wei, F. (2023). BrDMC1, a Recombinase Gene, Is Involved in Seed Germination in Brassica rapa under Salt Stress. Agronomy, 13(2), 595. https://doi.org/10.3390/agronomy13020595






