Transcriptome and Proteome Analysis Identifies Salt Stress Response Genes in Bottle Gourd Rootstock-Grafted Watermelon Seedlings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Design
2.3. Measurement of Fresh and Dry Weight
2.4. RNA Extraction and Transcriptome Sequencing
2.5. RNA Sequence Data Analysis
2.6. Protein Extraction
2.7. Two-Dimensional Electrophoresis (2-DE) and Staining
2.8. Image Acquisition and Spot Identification
2.9. Statistical Analysis
3. Results
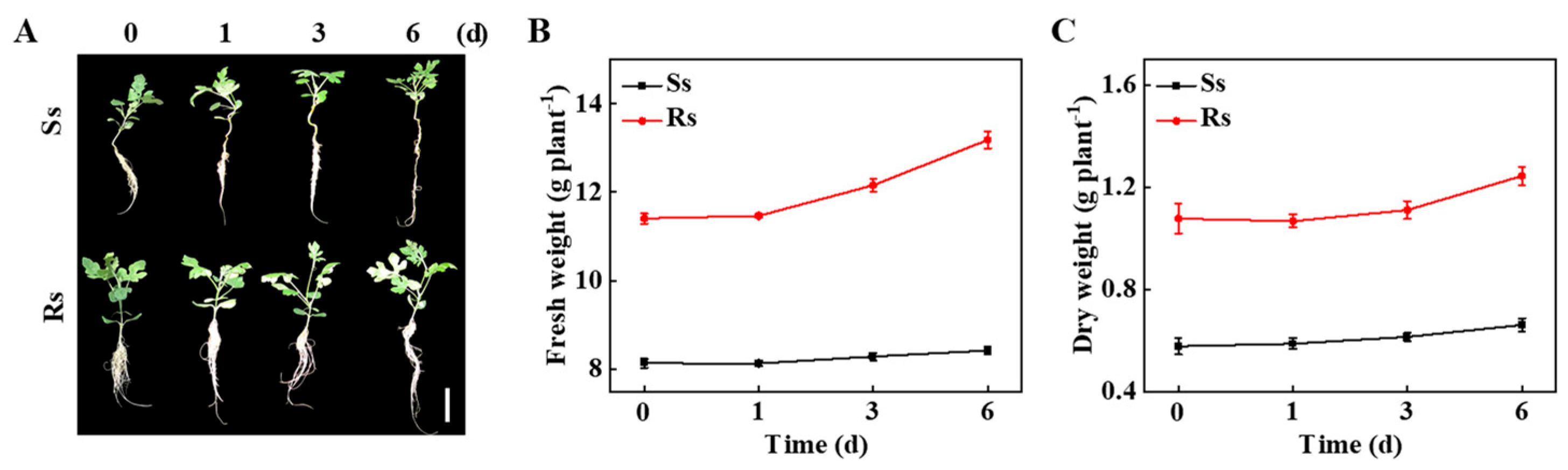
3.1. Bottle Gourd Rootstock Enhanced the Tolerance of Grafted Watermelon to Salt Stress
3.2. Transcriptome Sequencing Identification of DEGs in Salt Stressed Grafted Watermelon Plants
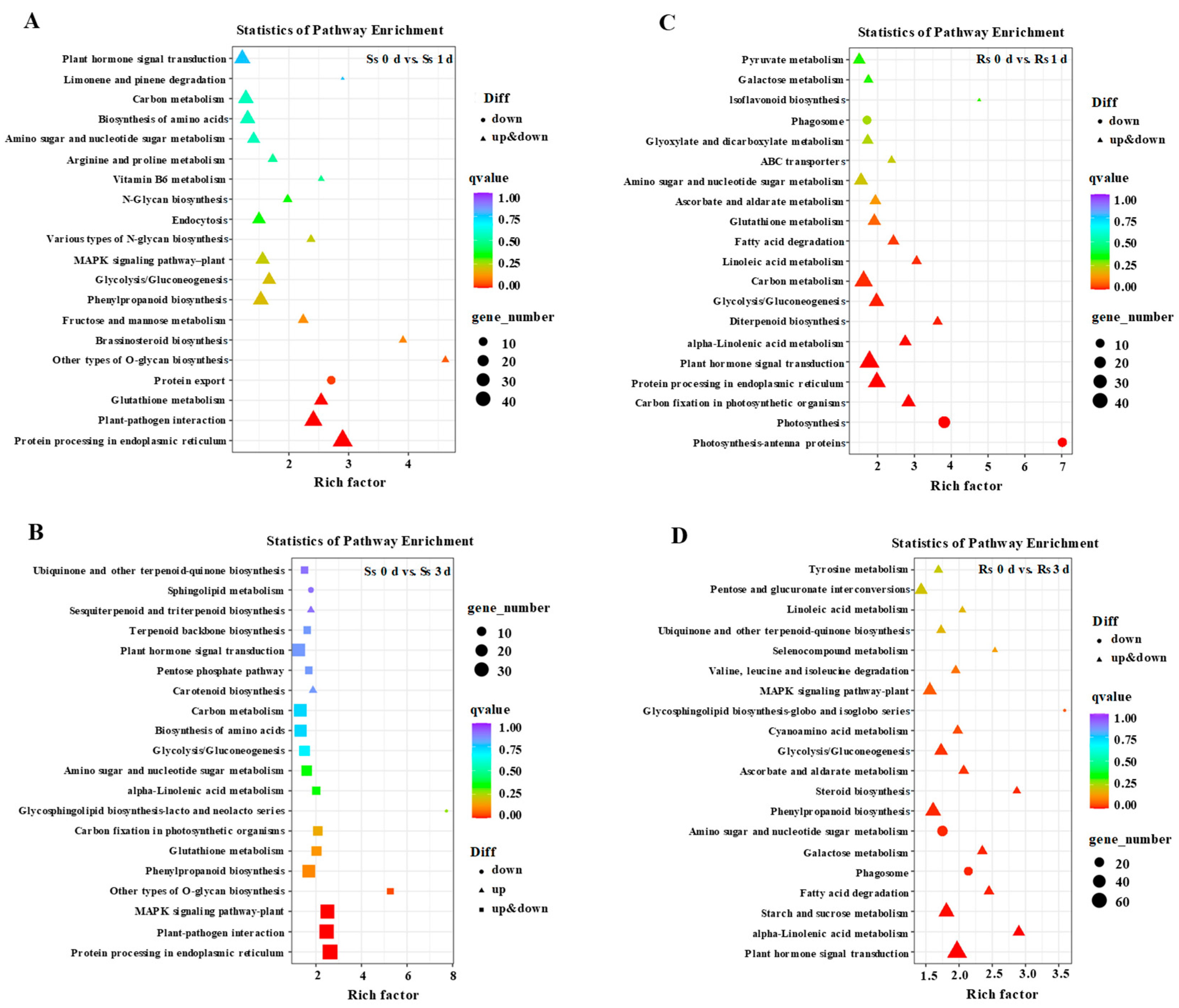
3.3. GO and KEGG Pathway Analyses of DEGs
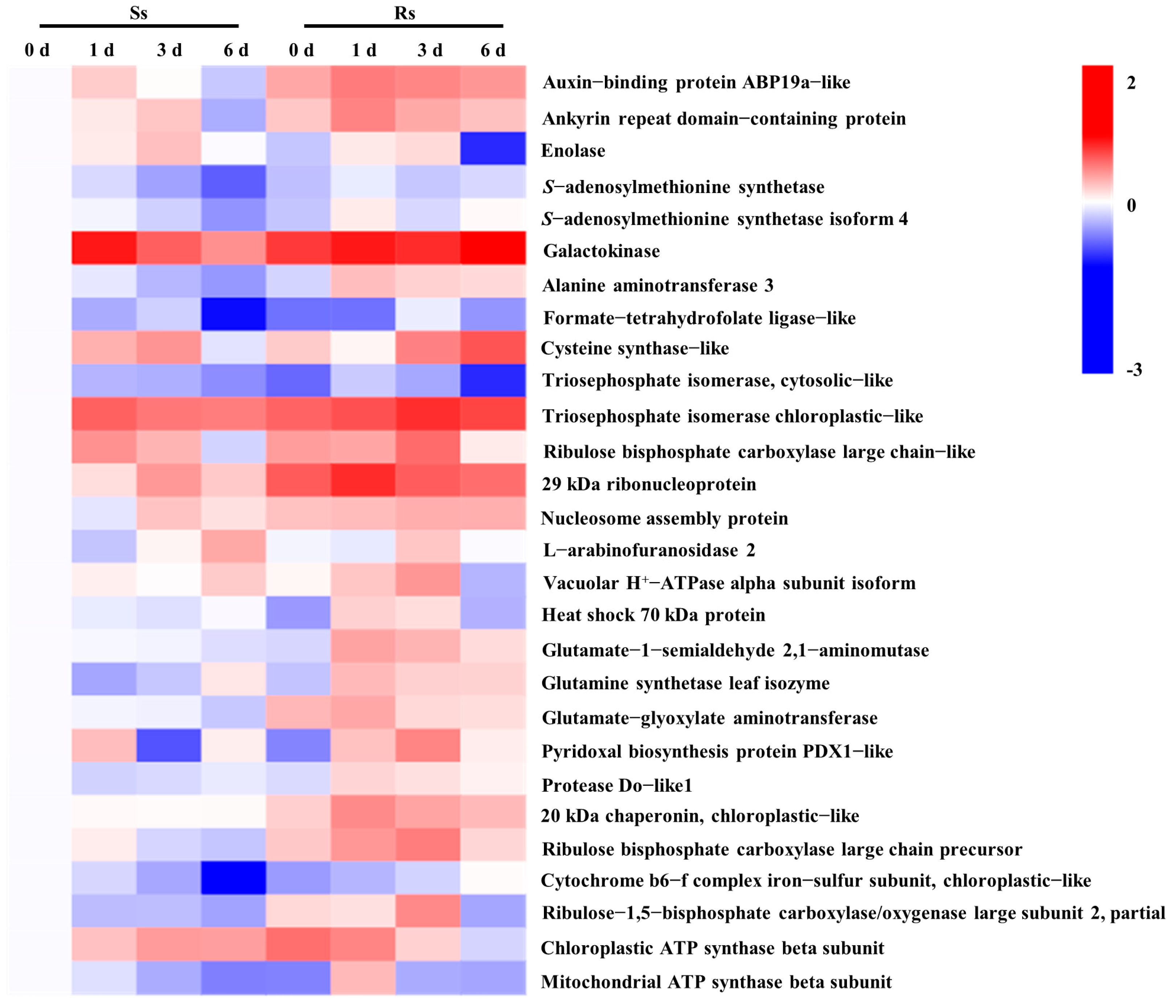
3.4. 2-DE Analysis of Differentially Expressed Proteins (DEPs)
3.5. Combined Analysis of DEGs and DEPs
4. Discussion
4.1. DEGs and DEPs Associated with Plant Hormones
4.2. DEGs and DEPs Associated with Photosynthesis
4.3. DEGs and DEPs Associated with Amino Acid Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Landi, S.; Hausman, J.F.; Guerriero, G.; Esposito, S. Poaceae vs. abiotic stress: Focus on drought and salt stress, recent insights and perspectives. Front. Plant Sci. 2017, 8, 1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Rizwan, M.; Qayyum, M.F.; Ok, Y.S.; Ibrahim, M.; Riaz, M.; Arif, M.S.; Hafeez, F.; Al-Wabel, M.I.; Shahzad, A.N. Biochar soil amendment on alleviation of drought and salt stress in plants: A critical review. Environ. Sci. Pollut. Res. 2017, 24, 12700–12712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.; Zhang, S.; Yin, Z.; Zhu, W.; Li, J.; Meng, L.; Zhong, H.; Xu, N.; Wu, Y.; et al. Rootstock alleviates salt stress in grafted mulberry seedlings: Physiological and PSII function responses. Front. Plant Sci. 2018, 9, 1806. [Google Scholar] [CrossRef] [PubMed]
- Koleska, I.; Hasanagic, D.; Todorovic, V.; Murtic, S.; Maksimovic, I. Grafting influence on the weight and quality of tomato fruit under salt stress. Ann. Appl. Biol. 2018, 172, 187–196. [Google Scholar] [CrossRef]
- Guo, Z.; Qin, Y.; Lv, J.; Wang, X.; Dong, H.; Dong, X.; Zhang, T.; Du, N.; Piao, F. Luffa rootstock enhances salt tolerance and improves yield and quality of grafted cucumber plants by reducing sodium transport to the shoot. Environ. Pollut. 2023, 316, 120521. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Cao, H.; Peng, Z.; Zhou, L.; Sohail, H.; Cui, L.; Yang, L.; Huang, Y.; Bie, Z. Transcriptomic and functional characterization reveals CsHAK5;3 as a key player in K plus homeostasis in grafted cucumbers under saline conditions. Plant Sci. 2023, 326, 111509. [Google Scholar] [CrossRef] [PubMed]
- Penella, C.; Nebauer, S.G.; Lopez-Galarza, S.; Quinones, A.; Bautista, A.S.; Calatayud, A. Grafting pepper onto tolerant rootstocks: An environmental-friendly technique overcome water and salt stress. Sci. Hortic. 2017, 226, 33–41. [Google Scholar] [CrossRef]
- Ulas, A.; Aydin, A.; Ulas, F.; Yetisir, H.; Miano, T.F. Cucurbita rootstocks improve salt tolerance of melon scions by inducing physiological, biochemical and nutritional responses. Horticulturae 2020, 6, 66. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, S.; Wei, M.; Gong, B.; Shi, Q. Effect of different rootstocks on the salt stress tolerance in watermelon seedlings. Hortic. Plant J. 2018, 4, 239–249. [Google Scholar] [CrossRef]
- Kacjan Marsic, N.; Stolfa, P.; Vodnik, D.; Kosmelj, K.; Mikulic-Petkovsek, M.; Kump, B.; Vidrih, R.; Kokalj, D.; Piskernik, S.; Ferjancic, B.; et al. Physiological and biochemical responses of ungrafted and grafted bell pepper plants (Capsicum annuum L. var. grossum (L.) Sendtn.) grown under moderate salt stress. Plants 2021, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, W.; Wang, T.; Zhang, J.; Li, X.; Zhang, W. Systemic long-distance signaling and communication between rootstock and scion in grafted vegetables. Front. Plant Sci. 2020, 11, 460. [Google Scholar] [CrossRef]
- Buesa, I.; Perez-Perez, J.G.; Visconti, F.; Strah, R.; Intrigliolo, D.S.; Bonet, L.; Gruden, K.; Pompe-Novak, M.; de Paz, J.M. Physiological and transcriptional responses to saline irrigation of young ’Tempranillo’ vines grafted onto different rootstocks. Front. Plant Sci. 2022, 13, 866053. [Google Scholar] [CrossRef]
- Zhu, H.; Zhao, S.; Lu, X.; He, N.; Gao, L.; Dou, J.; Bie, Z.; Liu, W. Genome duplication improves the resistance of watermelon root to salt stress. Plant Physiol. Biochem. 2018, 133, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, J.; Chen, H.; Wang, Z.; Gu, X.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Exogenous melatonin confers salt stress tolerance to watermelon by improving photosynthesis and redox homeostasis. Front. Plant Sci. 2017, 8, 295. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Shamimuzzaman, M.; Sun, H.; Salse, J.; Sui, X.L.; Wilder, A.; Wu, Z.J.; Levi, A.; Xu, Y.; Ling, K.S.; et al. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 2017, 92, 963–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mkhize, P.; Mashilo, J.; Shimelis, H. Progress on genetic improvement and analysis of bottle gourd [Lagenaria siceraria (Molina) Standl.] for agronomic traits, nutrient compositions, and stress tolerance: A review. Front. Sustain. Food Syst. 2021, 5, 683635. [Google Scholar] [CrossRef]
- Davis, A.R.; Perkins-Veazie, P.; Sakata, Y.; Lopez-Galarza, S.; Maroto, J.V.; Lee, S.G.; Huh, Y.C.; Sun, Z.; Miguel, A.; King, S.R.; et al. Cucurbit grafting. Crit. Rev. Plant Sci. 2008, 27, 50–74. [Google Scholar] [CrossRef]
- Yang, Y.J.; Lu, X.M.; Yan, B.; Li, B.; Sun, J.; Guo, S.; Tezuka, T. Bottle gourd rootstock-grafting affects nitrogen metabolism in NaCl-stressed watermelon leaves and enhances short-term salt tolerance. J. Plant Physiol. 2013, 170, 653–661. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Liu, W.; Ahammed, G.J.; Wen, W.; Guo, S.; Shu, S.; Sun, J. Auxin is involved in arbuscular mycorrhizal fungi-promoted tomato growth and NADP-malic enzymes expression in continuous cropping substrates. BMC Plant Biol. 2021, 21, 48. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotech. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurkman, W.J.; Tanaka, C.K. Solubilization of plant membrane-proteins for analysis by two-dimensional gel-electrophoresis. Plant Physiol. 1986, 81, 802–806. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lu, X.; Tian, J.; Yang, Y.; Li, B.; Li, J.; Guo, S. Proteomic analysis of the effects of exogenous calcium on hypoxic-responsive proteins in cucumber roots. Proteome Sci. 2012, 10, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological responses to drought, salinity, and heat stress in plants: A review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rathod, S.; Manohara, K.K.; Gireesh, C.; Anantha, M.S.; Sakhare, A.S.; Parmar, B.; Yadav, B.K.; Bandumula, N.; Raihan, F.; et al. Salt stress in plants and mitigation approaches. Plants 2022, 11, 717. [Google Scholar] [CrossRef] [PubMed]
- Behera, T.K.; Krishna, R.; Ansari, W.A.; Aamir, M.; Kumar, P.; Kashyap, S.P.; Pandey, S.; Kole, C. Approaches involved in the vegetable crops salt stress tolerance improvement: Present status and way ahead. Front. Plant Sci. 2022, 12, 787292. [Google Scholar] [CrossRef] [PubMed]
- Mashilo, J.; Shimelis, H.; Ngwepe, R.M. Genetic resources of bottle gourd (Lagenaria siceraria (Molina) Standl.] and citron watermelon (Citrullus lanatus var. citroides (L.H. Bailey) Mansf. ex Greb.)- implications for genetic improvement, product development and commercialization: A review. S. Afr. J. Bot. 2022, 145, 28–47. [Google Scholar] [CrossRef]
- Chen, X.L.; Guo, P.J.; Wang, Z.Y.; Liang, J.Y.; Li, G.H.; He, W.W.; Zhen, A. Grafting improves growth and nitrogen-use efficiency by enhancing NO3− uptake, photosynthesis, and gene expression of nitrate transporters and nitrogen metabolizing enzymes in watermelon under reduced nitrogen application. Plant Soil 2022, 480, 305–327. [Google Scholar] [CrossRef]
- Pal, S.; Rao, E.S.; Hebbar, S.S.; Sriram, S.; Pitchaimuthu, M.; Rao, V.K. Assessment of Fusarium wilt resistant Citrullus sp. rootstocks for yield and quality traits of grafted watermelon. Sci. Hortic. 2020, 272, 109497. [Google Scholar] [CrossRef]
- Ribba, T.; Garrido-Vargas, F.; Antonio O’Brien, J. Auxin-mediated responses under salt stress: From developmental regulation to biotechnological applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef]
- Verma, S.; Negi, N.P.; Pareek, S.; Mudgal, G.; Kumar, D. Auxin response factors in plant adaptation to drought and salinity stress. Physiol. Plant. 2022, 174, e13714. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongue, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, e107678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt stress affects the redox status of Arabidopsis root meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Shen, L.; Guo, J.; Jing, W.; Qu, Y.; Li, W.; Bi, R.; Xuan, W.; Zhang, Q.; Zhang, W. Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell 2019, 31, 250–271. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.W.; Deng, W.; Tang, N.; Wang, X.M.; Yan, F.; Lin, D.B.; Li, Z.G. Overexpression of ZmAFB2, the maize homologue of AFB2 gene, enhances salt tolerance in transgenic tobacco. Plant Cell Tiss. Organ. Cult. 2013, 112, 171–179. [Google Scholar] [CrossRef]
- Kirungu, J.N.; Magwanga, R.O.; Lu, P.; Cai, X.Y.; Zhou, Z.L.; Wang, X.X.; Peng, R.H.; Wang, K.B.; Liu, F. Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 2019, 20, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, L.; Yu, B.; Guo, J.; Zhao, Y.; Zhu, Y. Expression analysis of AUX/IAA family genes in apple under salt stress. Biochem. Genet. 2022, 60, 1205–1221. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, C.; Lian, W.; Zhu, X.; Yu, Q.; Jia, Y.; Jia, H.; Xie, L. NtIAA26 positively regulates salt tolerance in tobacco by modulating potassium uptake and antioxidant activity. Plant Growth Regul. 2022, 97, 559–569. [Google Scholar] [CrossRef]
- Zhang, A.Y.; Yang, X.; Lu, J.; Song, F.Y.; Sun, J.H.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B.C. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Amjad, M.; Akhtar, J.; Anwar-ul-Haq, M.; Yang, A.; Akhtar, S.S.; Jacobsen, S.E. Integrating role of ethylene and ABA in tomato plants adaptation to salt stress. Sci. Hortic. 2014, 172, 109–116. [Google Scholar] [CrossRef]
- Wang, G.L.; Ren, X.Q.; Liu, J.X.; Yang, F.; Wang, Y.P.; Xiong, A.S. Transcript profiling reveals an important role of cell wall remodeling and hormone signaling under salt stress in garlic. Plant Physiol. Biochem. 2019, 135, 87–98. [Google Scholar] [CrossRef]
- Feng, X.Y.; Chen, S.Y.; Yang, S.M.; An, X.; Liu, Y.Y.; Lu, H.L.; Yang, C.Q.; Qin, Y.G. Effects of exogenous abscisic acid on salt tolerance of watermelon seedlings under NaCl stress. Appl. Ecol. Environ. Res. 2022, 20, 4515–4524. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, Y.; Zhang, Y.; Shu, S.; Sun, J.; Guo, S. Overexpression of transglutaminase from cucumber in tobacco increases salt tolerance through regulation of photosynthesis. Int. J. Mol. Sci. 2019, 20, 894. [Google Scholar] [CrossRef] [Green Version]
- Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteom. 2005, 4, 1122–1133. [Google Scholar] [CrossRef] [Green Version]
- Rasoulnia, A.; Bihamta, M.R.; Peyghambari, S.A.; Alizadeh, H.; Rahnama, A. Proteomic response of barley leaves to salinity. Mol. Biol. Rep. 2011, 38, 5055–5063. [Google Scholar] [CrossRef] [PubMed]
- de Abreu, C.E.B.; Araújo, G.D.; Monteiro-Moreira, A.C.D.; Costa, J.H.; Leite, H.D.; Moreno, F.B.M.B.; Prisco, J.T.; Gomes-Filho, E. Proteomic analysis of salt stress and recovery in leaves of Vigna unguiculata cultivars differing in salt tolerance. Plant Cell Rep. 2014, 33, 1289–1306. [Google Scholar] [CrossRef] [PubMed]
- Zaffagninia, M.; Michelet, L.; Sciabolini, C.; Di Giacinto, N.; Morisse, S.; Marchand, C.H.; Trost, P.; Fermani, S.; Lemaire, S.D. High-resolution crystal structure and redox properties of chloroplastic triosephosphate isomerase from Chlamydomonas reinhardtii. Mol. Plant 2014, 7, 101–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro-Torres, E.; Jimenez-Sandoval, P.; Fernandez-de Gortari, E.; Lopez-Castillo, M.; Baruch-Torres, N.; Lopez-Hidalgo, M.; Peralta-Castro, A.; Diaz-Quezada, C.; Sotelo-Mundo, R.R.; Benitez-Cardoza, C.G.; et al. Structural basis for the limited response to oxidative and thiol-conjugating agents by triosephosphate isomerase from the photosynthetic bacteria synechocystis. Front. Mol. Biosci. 2018, 5, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Castillo, L.M.; Jimenez-Sandoval, P.; Baruch-Torres, N.; Trasvina-Arenas, C.H.; Diaz-Quezada, C.; Lara-Gonzalez, S.; Winkler, R.; Brieba, L.G. Structural basis for redox regulation of cytoplasmic and chloroplastic triosephosphate isomerases from Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro-Torres, E.; Jimenez-Sandoval, P.; Romero-Romero, S.; Fuentes-Pascacio, A.; Lopez-Castillo, L.M.; Diaz-Quezada, C.; Fernandez-Velasco, D.A.; Torres-Larios, A.; Brieba, L.G. Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications. Plant J. 2019, 99, 950–964. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Takahashi, S.; Makino, A.; Price, G.D.; Badger, M.R.; von Caemmerer, S. The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 2011, 155, 956–962. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Ballesta, M.C.; Alcaraz-Lopez, C.; Muries, B.; Mota-Cadenas, C.; Carvajal, M. Physiological aspects of rootstock-scion interactions. Sci. Hortic. 2010, 127, 112–118. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004, 166, 3–16. [Google Scholar] [CrossRef]
- Valderrama-Martin, J.M.; Ortigosa, F.; Avila, C.; Canovas, F.M.; Hirel, B.; Canton, F.R.; Canas, R.A. A revised view on the evolution of glutamine synthetase isoenzymes in plants. Plant J. 2022, 110, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Canas, R.A.; Betti, M. Is plastidic glutamine synthetase essential for C-3 plants? A tale of photorespiratory mutants, ammonium tolerance and conifers. New Phytol. 2022, 234, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Zavala, A.A.; Carrasco-Miranda, J.S.; Ramirez-Aguirre, C.D.; Lopez-Hidalgo, M.; Benitez-Cardoza, C.G.; Ochoa-Leyva, A.; Cardona-Felix, C.S.; Diaz-Quezada, C.; Rudino-Pinera, E.; Sotelo-Mundo, R.R.; et al. Structural insights from a novel invertebrate triosephosphate isomerase from Litopenaeus vannamei. BBA-Proteins Proteom. 2016, 1864, 1696–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roje, S. S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Muller, M. Foes or friends: ABA and ethylene interaction under abiotic stress. Plants 2021, 10, 448. [Google Scholar] [CrossRef]
- Wang, W.; Paschalidis, K.; Feng, J.C.; Song, J.; Liu, J.H. Polyamine catabolism in plants: A universal process with diverse functions. Front. Plant Sci. 2019, 10, 561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- He, M.W.; Wang, Y.; Wu, J.Q.; Shu, S.; Sun, J.; Guo, S.R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, L.; Guo, S.; Li, J.; Yang, Y.J.; Yan, B.; Sun, J.; Li, J. Proteomics reveal cucumber Spd-responses under normal condition and salt stress. Plant Physiol. Biochem. 2013, 67, 7–14. [Google Scholar] [CrossRef]



| GO Classification | GO Term | Total Number of DEGs | Number of DEGs in Ss 0 d vs. Ss 1 d | Number of DEGs in Ss 0 d vs. Ss 3 d | Number of DEGs in Rs 0 d vs. Rs 1 d | Number of DEGs in Rs 0 d vs. Rs 3 d |
|---|---|---|---|---|---|---|
| biological process | metabolic process | 6103 | 597 | 556 | 588 | 935 |
| biological process | cellular process | 5004 | 516 | 459 | 482 | 747 |
| biological process | single-organism process | 3033 | 312 | 304 | 338 | 506 |
| biological process | localization | 1251 | 149 | 121 | 124 | 187 |
| biological process | biological regulation | 1103 | 134 | 117 | 123 | 177 |
| biological process | response to stimulus | 954 | 114 | 96 | 103 | 166 |
| biological process | signaling | 318 | 43 | 32 | 25 | 49 |
| biological process | cellular component organization or biogenesis | 609 | 36 | 46 | 41 | 89 |
| biological process | multicellular organismal process | 262 | 26 | 21 | 17 | 40 |
| biological process | developmental process | 298 | 23 | 21 | 24 | 52 |
| biological process | reproduction | 146 | 9 | 8 | 7 | 22 |
| biological process | reproductive process | 140 | 8 | 8 | 7 | 20 |
| biological process | detoxification | 111 | 8 | 16 | 9 | 23 |
| biological process | multi-organism process | 69 | 7 | 8 | 7 | 8 |
| biological process | growth | 40 | 5 | 4 | 6 | 15 |
| biological process | cell killing | 4 | 2 | 1 | 2 | 2 |
| biological process | immune system process | 18 | 0 | 2 | 1 | 0 |
| biological process | biological adhesion | 3 | 0 | 0 | 0 | 1 |
| biological process | rhythmic process | 5 | 0 | 1 | 0 | 1 |
| cellular component | membrane | 4238 | 469 | 421 | 404 | 699 |
| cellular component | membrane part | 3751 | 423 | 372 | 366 | 622 |
| cellular component | cell | 3531 | 303 | 257 | 311 | 432 |
| cellular component | cell part | 3531 | 303 | 257 | 311 | 432 |
| cellular component | organelle | 2425 | 214 | 191 | 213 | 306 |
| cellular component | organelle part | 774 | 55 | 68 | 68 | 73 |
| cellular component | macromolecular complex | 725 | 32 | 27 | 51 | 30 |
| cellular component | cell junction | 65 | 9 | 5 | 8 | 14 |
| cellular component | extracellular region | 82 | 6 | 7 | 10 | 20 |
| cellular component | membrane-enclosed lumen | 105 | 6 | 3 | 5 | 5 |
| cellular component | extracellular region part | 12 | 1 | 2 | 0 | 1 |
| cellular component | virion | 26 | 0 | 1 | 0 | 1 |
| cellular component | virion part | 26 | 0 | 1 | 0 | 1 |
| cellular component | supramolecular complex | 3 | 0 | 0 | 1 | 1 |
| molecular function | catalytic activity | 5639 | 489 | 554 | 552 | 975 |
| molecular function | binding | 5829 | 487 | 444 | 521 | 840 |
| molecular function | transporter activity | 496 | 55 | 48 | 53 | 76 |
| molecular function | nucleic acid binding transcription factor activity | 338 | 49 | 45 | 47 | 62 |
| molecular function | structural molecule activity | 276 | 11 | 6 | 21 | 19 |
| molecular function | antioxidant activity | 106 | 8 | 16 | 9 | 23 |
| molecular function | molecular function regulator | 141 | 8 | 7 | 16 | 21 |
| molecular function | electron carrier activity | 20 | 1 | 2 | 1 | 0 |
| molecular function | molecular transducer activity | 25 | 1 | 2 | 2 | 2 |
| Spot ID 1 | Accession No. 2 | Protein Name 3 | Group 4 | Mr (kDa)/ pI 5 | Peptide Count | Score 6 | Protein Score C.I.% 7 |
|---|---|---|---|---|---|---|---|
| A2 | Cla97C09G162960 | Auxin-binding protein ABP19a-like | Hormone metabolism | 21.50/6.38 | 3 | 196 | 100 |
| B1 | Cla97C02G036490 | Ankyrin repeat domain-containing protein | Protein interaction | 38.87/4.52 | 11 | 150 | 100 |
| B2 | Cla97C04G076580 | Enolase | Carbon metabolism | 48.15/5.71 | 12 | 126 | 100 |
| B3 | Cla97C10G194620 | S-adenosylmethionine synthetase | Amino acid synthesis | 43.70/5.59 | 18 | 482 | 100 |
| B4 | Cla97C09G167000 | S-adenosylmethionine synthetase isoform 4 | Amino acid synthesis | 43.65/5.35 | 16 | 515 | 100 |
| B5 | Cla97C05G088280 | Galactokinase | Carbon metabolism | 55.10/6 | 6 | 185 | 100 |
| B6 | Cla97C05G107900 | Alanine aminotransferase 3 | Amino acid synthesis | 53.79/5.52 | 8 | 97 | 99.98 |
| B7 | Cla97C03G064150 | Formate-tetrahydrofolate ligase-like | Stress defence | 68.58/7.21 | 9 | 123 | 100 |
| B8 | Cla97C02G042210 | Cysteine synthase-like | Amino acid synthesis | 34.57/5.92 | 10 | 267 | 100 |
| B9 | Cla97C04G071440 | Triosephosphate isomerase, cytosolic-like | Carbon metabolism | 27.50/5.61 | 8 | 75 | 96.70 |
| B10 | Cla97C02G037100 | Triosephosphate isomerase chloroplastic-like | Photosynthesis | 33.00/7.01 | 12 | 209 | 100 |
| B11 | Cla97C07G134850 | Ribulose bisphosphate carboxylase large chain-like | Photosynthesis | 16.81/4.78 | 5 | 267 | 100 |
| B12 | Cla97C10G203440 | 29 kDa ribonucleoprotein | Protein synthesis | 30.48/5.84 | 5 | 112 | 100 |
| C1 | Cla97C11G208080 | Nucleosome assembly protein | Chromosome assembly | 43.03/4.31 | 7 | 240 | 100 |
| C2 | Cla97C09G177130 | L-arabinofuranosidase 2 | Carbon metabolism | 85.45/6.99 | 13 | 168 | 100 |
| C3 | Cla97C09G163740 | Vacuolar H+-ATPase alpha subunit isoform | Energy metabolism | 68.81/5.2 | 13 | 198 | 100 |
| C4 | Cla97C10G192810 | Heat shock 70 kDa protein | Stress defence | 73.25/5.69 | 15 | 140 | 100 |
| C5 | Cla97C09G178080 | Glutamate-1-semialdehyde 2,1-aminomutase | Amino acid synthesis | 49.84/5.67 | 11 | 161 | 100 |
| C6 | Cla97C05G086890 | Glutamine synthetase leaf isozyme | Amino acid synthesis | 48.03/7.62 | 16 | 179 | 100 |
| C7 | Cla97C05G107870 | Glutamate-glyoxylate aminotransferase | Amino acid synthesis | 53.83/6.5 | 11 | 117 | 100 |
| C8 | Cla97C10G205390 | Pyridoxal biosynthesis protein PDX1-like | Amino acid synthesis | 33.23/5.85 | 11 | 79 | 98.69 |
| C9 | Cla97C09G180430 | Protease Do-like1 | Protein degradation | 46.92/7.13 | 18 | 460 | 100 |
| C10 | Cla97C03G065070 | 20 kDa chaperonin, chloroplastic-like | Stress defence | 26.88/7.85 | 8 | 165 | 100 |
| C11 | Cla97C03G060940 | Ribulose bisphosphate carboxylase large chain precursor | Photosynthesis | 56.55/9.04 | 15 | 345 | 100 |
| C12 | Cla97C02G027850 | Cytochrome b6-f complex iron-sulfur subunit, chloroplastic-like | Photosynthesis | 24.21/8.53 | 3 | 123 | 100 |
| C13 | Cla97C03G051890 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit 2, partial | Photosynthesis | 19.70/6.18 | 5 | 112 | 100 |
| C14 | Cla97C01G006310 | Chloroplastic ATP synthase beta subunit | Energy metabolism | 51.92/5.07 | 11 | 100 | 99.99 |
| C15 | Cla97C05G104100 | Mitochondrial ATP synthase beta subunit | Energy metabolism | 59.89/5.90 | 10 | 73 | 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhou, J.; Wen, W.; Sun, J.; Shu, S.; Guo, S. Transcriptome and Proteome Analysis Identifies Salt Stress Response Genes in Bottle Gourd Rootstock-Grafted Watermelon Seedlings. Agronomy 2023, 13, 618. https://doi.org/10.3390/agronomy13030618
Wang Y, Zhou J, Wen W, Sun J, Shu S, Guo S. Transcriptome and Proteome Analysis Identifies Salt Stress Response Genes in Bottle Gourd Rootstock-Grafted Watermelon Seedlings. Agronomy. 2023; 13(3):618. https://doi.org/10.3390/agronomy13030618
Chicago/Turabian StyleWang, Yu, Junqian Zhou, Wenxu Wen, Jin Sun, Sheng Shu, and Shirong Guo. 2023. "Transcriptome and Proteome Analysis Identifies Salt Stress Response Genes in Bottle Gourd Rootstock-Grafted Watermelon Seedlings" Agronomy 13, no. 3: 618. https://doi.org/10.3390/agronomy13030618





