Molecular Advances in Breeding for Durable Resistance against Pests and Diseases in Wheat: Opportunities and Challenges
Abstract
:1. Introduction
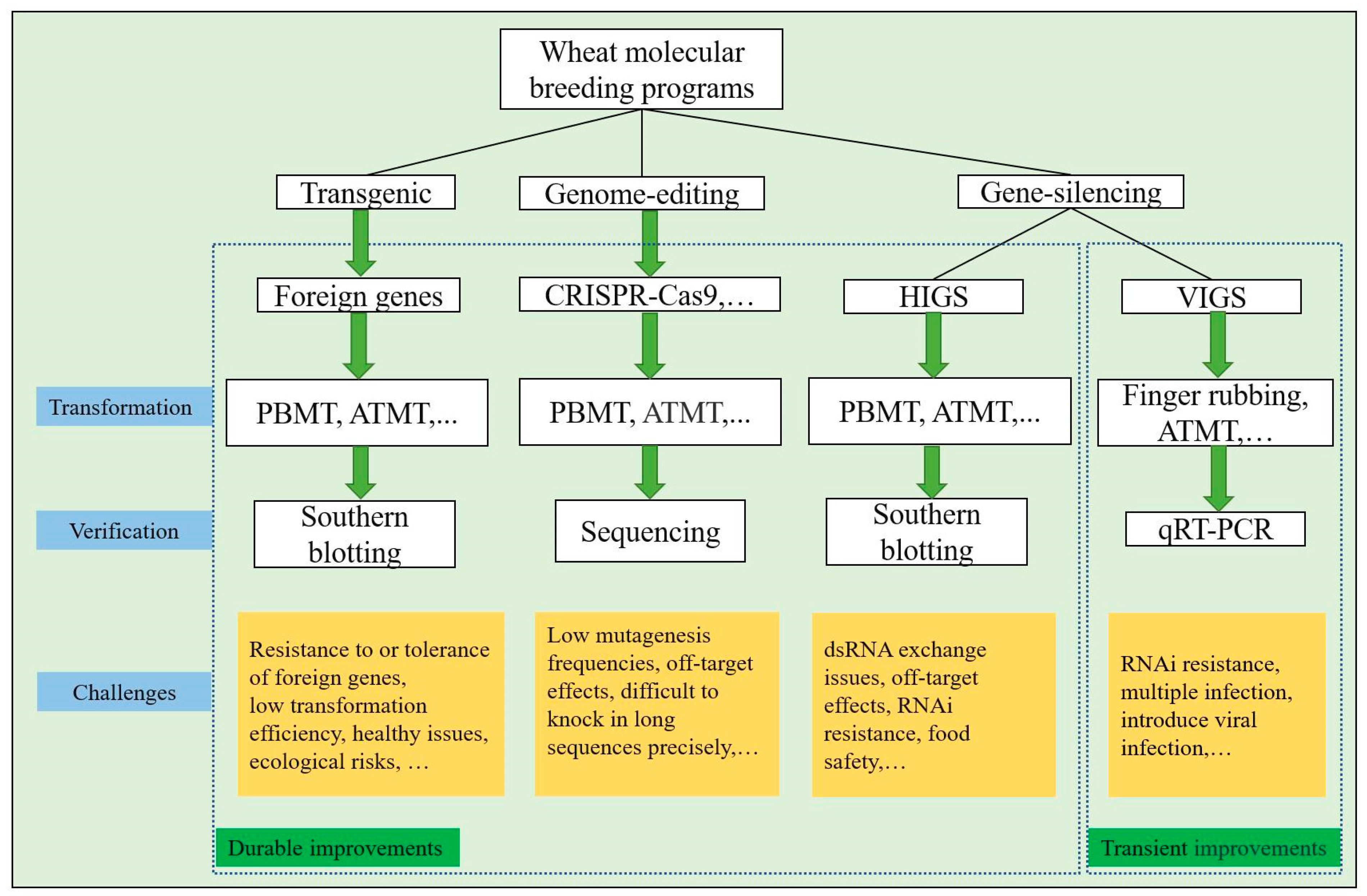
2. Challenges in Transgenic-Based Wheat Cultivar Improvements Forward to Biotic Stress Resistance
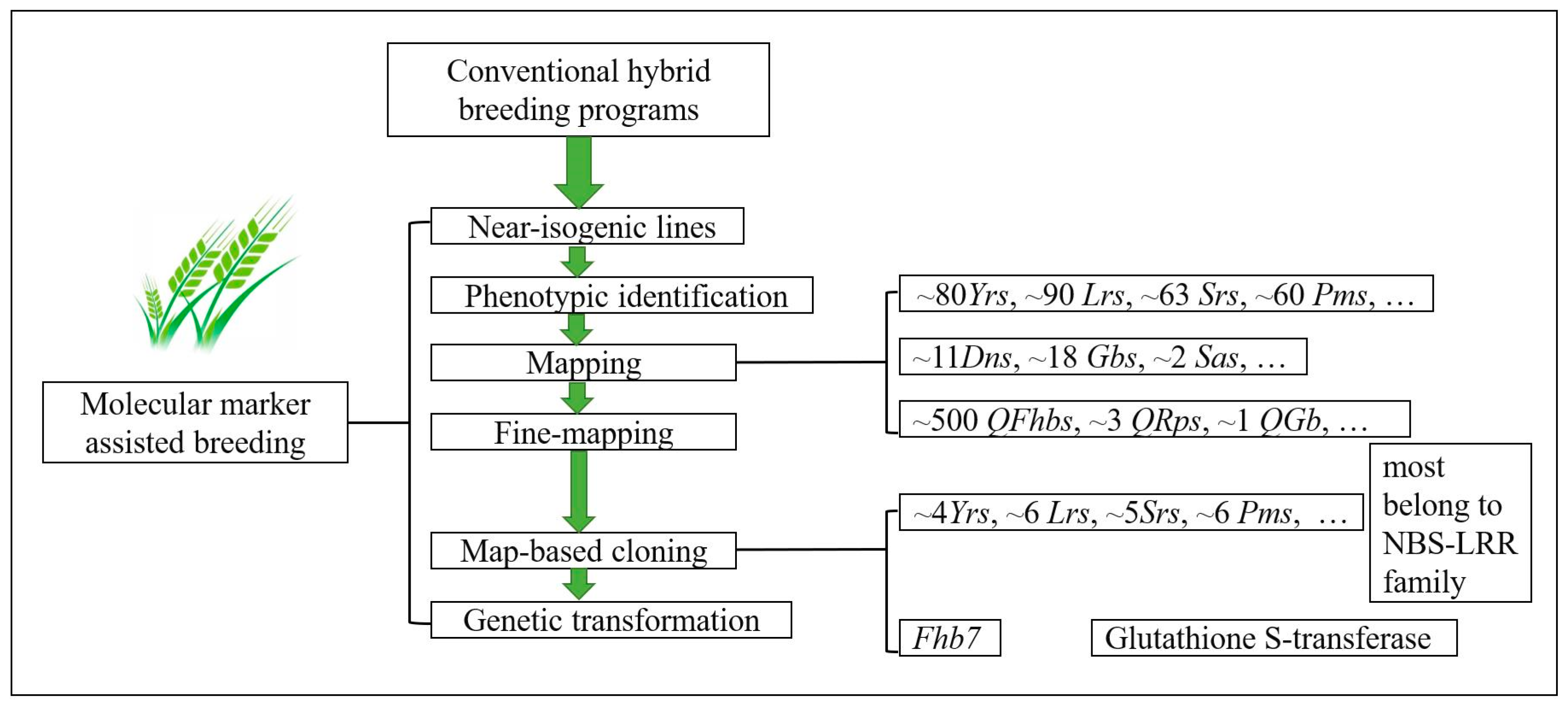
3. Molecular Markers Applied in Selecting Resistance Genes and Their Use in Cultivar Improvements
4. Map-Based Cloning of Resistance Genes and Its Application in Wheat Improvements
5. Gene Silencing-Based Cultivar Improvements against for Biotic Stresses
6. Genome Editing Involved in Crop Breeding Forward to Biotic Stress Resistance
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 2016, 242, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Abdul Fiyaz, R.; Ajay, B.C.; Ramya, K.T.; Aravind Kumar, J.; Sundaram, R.M.; Subba Rao, L.V. Speed Breeding: Methods and Applications. In Accelerated Plant Breeding, Volume 1: Cereal Crops; Gosal, S.S., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 31–49. [Google Scholar] [CrossRef]
- De Zutter, N.; Audenaert, K.; Ameye, M.; De Boevre, M.; De Saeger, S.; Haesaert, G.; Smagghe, G. The plant response induced in wheat ears by a combined attack of Sitobion avenae aphids and Fusarium graminearum boosts fungal infection and deoxynivalenol production. Mol. Plant Pathol. 2017, 18, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Drakulic, J.; Caulfield, J.; Woodcock, C.; Jones, S.P.T.; Linforth, R.; Bruce, T.J.A.; Ray, R.V. Sharing a host plant (wheat [Triticum aestivum]) increases the fitness of Fusarium graminearum and the severity of Fusarium head blight but reduces the fitness of grain aphids (Sitobion avenae). Appl. Environ. Microbiol. 2015, 81, 3492–3501. [Google Scholar] [CrossRef] [PubMed]
- Trail, F. For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 2009, 149, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.P.; Paul, V.L.; Slater, R.; Warren, A.; Denholm, I.; Field, L.M.; Williamson, M.S. A mutation (L1014F) in the voltage-gated sodium channel of the grain aphid, Sitobion avenae, is associated with resistance to pyrethroid insecticides. Pest. Manag. Sci. 2014, 70, 1249–1253. [Google Scholar] [CrossRef]
- Jones, H.D. Transformation and Transgene Expression. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Brian, T., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 248–254. [Google Scholar] [CrossRef]
- Dai, S.; Zheng, P.; Marmey, P.; Zhang, S.; Tian, W.; Chen, S.; Beachy, R.N.; Fauquet, C. Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol. Breed. 2001, 7, 25–33. [Google Scholar] [CrossRef]
- Hu, T.; Metz, S.; Chay, C.; Zhou, H.P.; Biest, N.; Chen, G.; Cheng, M.; Feng, X.; Radionenko, M.; Lu, F.; et al. Agrobacterium-mediated large-scale transformation of wheat (Triticum aestivum L.) using glyphosate selection. Plant Cell Rep. 2003, 21, 1010–1019. [Google Scholar] [CrossRef]
- Wu, H.; Sparks, C.; Amoah, B.; Jones, H.D. Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep. 2003, 21, 659–668. [Google Scholar] [CrossRef]
- Zhao, T.; Zhao, S.; Chen, H.; Zhao, Q.; Hu, Z.; Hou, B.; Xia, G. Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep. 2006, 25, 1199–1204. [Google Scholar] [CrossRef]
- Weir, B.; Gu, X.; Wang, M.; Upadhyaya, N.; Elliott, A.R.; Brettell, R.I.S. Agrobacterium tumefaciens-mediated transformation of wheat using suspension cells as a model system and green fluorescent protein as a visual marker. Funct. Plant Biol. 2001, 28, 807–818. [Google Scholar] [CrossRef]
- Chen, W.P.; Chen, P.D.; Liu, D.J.; Kynast, R.; Friebe, B.; Velazhahan, R.; Muthukrishnan, S.; Gill, B.S. Development of wheat scab symptoms is delayed in transgenic wheat plants that constitutively express a rice thaumatin-like protein gene. Theor. Appl. Genet 1999, 99, 755–760. [Google Scholar] [CrossRef]
- Shin, S.; Mackintosh, C.A.; Lewis, J.; Heinen, S.J.; Radmer, L.; Dill-Macky, R.; Baldridge, G.D.; Zeyen, R.J.; Muehlbauer, G.J. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 2008, 59, 2371–2378. [Google Scholar] [CrossRef]
- Bliffeld, M.; Mundy, J.; Potrykus, I.; Fütterer, J. Genetic engineering of wheat for increased resistance to powdery mildew disease. Theor. Appl. Genet. 1999, 98, 1079–1086. [Google Scholar] [CrossRef]
- Ward, E.R.; George, B.P.; Moyer, M.B.; Williams, S.C.; Dincher, S.S.; Kevin, C.S.; James, J.B.; Hope, T.T.; Ahl-Goy, P.; Meins, F.; et al. Differential regulation of β-1,3-glucanase messenger RNAs in response to pathogen infection. Plant Physiol. 1991, 96, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Aradottir, G.I.; Crespo-Herrera, L. Host plant resistance in wheat to barley yellow dwarf viruses and their aphid vectors: A review. Curr. Opin. Insect Sci. 2021, 45, 59–68. [Google Scholar] [CrossRef]
- Yu, Y.; Wei, Z. Increased oriental armyworm and aphid resistance in transgenic wheat stably expressing Bacillus thuringiensis (Bt) endotoxin and Pinellia ternate agglutinin (PTA). Plant Cell Tiss. Org. 2008, 94, 33–44. [Google Scholar] [CrossRef]
- Michiels, K.; Van Damme, E.J.; Smagghe, G. Plant-insect interactions: What can we learn from plant lectins? Arch. Insect Biochem. Physiol. 2010, 73, 193–212. [Google Scholar] [CrossRef]
- Christou, P.; Capell, T.; Kohli, A.; Gatehouse, J.A.; Gatehouse, A.M.R. Recent developments and future prospects in insect pest control in transgenic crops. Trends Plant Sci. 2006, 11, 302–308. [Google Scholar] [CrossRef]
- Nayak, S.N.; Singh, V.K.; Varshney, R.K. Marker-Assisted Selection. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Thomas, B., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 183–197. [Google Scholar] [CrossRef]
- Gupta, P.K.; Roy, J.K.; Prasad, M. Single nucleotide polymorphisms: A new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr. Sci. 2001, 80, 524–535. [Google Scholar]
- Kassa, M.T.; You, F.M.; Fetch, T.G.; Fobert, P.; Sharpe, A.; Pozniak, C.J.; Menzies, J.G.; Jordan, M.C.; Humphreys, G.; Zhu, T.; et al. Genetic mapping of SrCad and SNP marker development for marker-assisted selection of Ug99 stem rust resistance in wheat. Theor. Appl. Genet. 2016, 129, 1373–1382. [Google Scholar] [CrossRef]
- Wu, J.; Zeng, Q.; Wang, Q.; Liu, S.; Yu, S.; Mu, J.; Huang, S.; Sela, H.; Distelfeld, A.; Huang, L.; et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018, 131, 1481–1496. [Google Scholar] [CrossRef]
- Singla, J.; Lüthi, L.; Wicker, T.; Bansal, U.; Krattinger, S.G.; Keller, B. Characterization of Lr75: A partial, broad-spectrum leaf rust resistance gene in wheat. Theor. Appl. Genet. 2017, 130, 1–12. [Google Scholar] [CrossRef]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2018, 218, 298–309. [Google Scholar] [CrossRef]
- Liu, X.L.; Yang, X.F.; Wang, C.Y.; Wang, Y.J.; Zhang, H.; Ji, W.Q. Molecular mapping of resistance gene to English grain aphid (Sitobion avenae F.) in Triticum durum wheat line C273. Theor. Appl. Genet. 2012, 124, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Luo, K.; Wang, L.; Zhao, H.; Zhang, G. Molecular mapping of resistance gene to the English grain aphid, Sitobion avenae, in a Chinese wheat line XN98-10-35. Mol. Breed. 2015, 35, 203. [Google Scholar] [CrossRef]
- Luo, K.; Zhao, H.; Wang, X.; Kang, Z. Prevalent Pest Management Strategies for Grain Aphids: Opportunities and Challenges. Front. Plant Sci. 2022, 12, 3252. [Google Scholar] [CrossRef]
- Lu, H.; Rudd, J.C.; Burd, J.D.; Weng, Y. Molecular mapping of greenbug resistance genes Gb2 and Gb6 in T1AL.1RS wheat-rye translocations. Plant Breeding 2010, 129, 472–476. [Google Scholar] [CrossRef]
- Bernardo, R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Young, P.R.; Vivier, M.A. 10—Genetics and genomic approaches to improve grape quality for winemaking. In Managing Wine Quality; Reynolds, A.G., Ed.; Woodhead Publishing: Sawston, UK, 2010; pp. 316–364. [Google Scholar] [CrossRef]
- Crespo-Herrera, L.A.; Akhunov, E.; Garkava-Gustavsson, L.; Jordan, K.W.; Smith, C.M.; Singh, R.P.; Åhman, I. Mapping resistance to the bird cherry-oat aphid and the greenbug in wheat using sequence-based genotyping. Theor. Appl. Genet. 2014, 127, 1963–1973. [Google Scholar] [CrossRef]
- Miedaner, T.; Wilde, F.; Steiner, B.; Buerstmayr, H.; Korzun, V.; Ebmeyer, E. Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Theor. Appl. Genet. 2006, 112, 562–569. [Google Scholar] [CrossRef]
- Xue, S.; Li, G.; Jia, H.; Lin, F.; Cao, Y.; Xu, F.; Tang, M.; Wang, Y.; Wu, X.; Zhang, Z.; et al. Marker-assisted development and evaluation of near-isogenic lines for scab resistance QTLs of wheat. Mol. Breed. 2010, 25, 397–405. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Dong, Z.; Zhao, L.; Ren, Y.; Zhang, N.; Chen, F. The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 2020, 18, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Joukhadar, R.; El-Bouhssini, M.; Jighly, A.; Ogbonnaya, F.C. Genome-wide association mapping for five major pest resistances in wheat. Mol. Breed. 2013, 32, 943–960. [Google Scholar] [CrossRef]
- Eshed, Y.; Zamir, D. Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 1996, 143, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Marcel, T.C.; Aghnoum, R.; Durand, J.; Varshney, R.K.; Niks, R.E. Dissection of the Barley 2L1.0 Region Carrying the ‘Laevigatum’ Quantitative Resistance Gene to Leaf Rust Using Near-Isogenic Lines (NIL) and subNIL. Mol. Plant Microbe. Interact. 2007, 20, 1604–1615. [Google Scholar] [CrossRef]
- Wan, W.; Xiao, J.; Li, M.; Tang, X.; Wen, M.; Cheruiyot, A.K.; Li, Y.; Wang, H.; Wang, X. Fine mapping of wheat powdery mildew resistance gene Pm6 using 2B/2G homoeologous recombinants induced by the ph1b mutant. Theor. Appl. Genet. 2020, 133, 1265–1275. [Google Scholar] [CrossRef]
- Huang, L.; Brooks, S.A.; Li, W.; Fellers, J.P.; Trick, H.N.; Gill, B.S. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 2003, 164, 655–664. [Google Scholar] [CrossRef]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef]
- Ling, H.Q.; Zhu, Y.; Keller, B. High-resolution mapping of the leaf rust disease resistance gene Lr1 in wheat and characterization of BAC clones from the Lr1 locus. Theor. Appl. Genet 2003, 106, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, D.; Zeng, Q.; Duan, Y.; Yuan, F.; Shi, J.; Wang, Q.; Wu, J.; Huang, L.; Kang, Z.; et al. Fine mapping of wheat stripe rust resistance gene Yr26 based on collinearity of wheat with Brachypodium distachyon and rice. PLoS ONE 2013, 8, e57885. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, L.; Holdgate, S.; James, L.; Thomas, J.; Mackay, I.J.; Cockram, J. The evolving battle between yellow rust and wheat: Implications for global food security. Theor. Appl. Genet. 2022, 135, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, Y.; Li, Y.; Fahima, T.; Shen, Q.; Xie, C. Introgression of the powdery mildew resistance genes Pm60 and Pm60b from Triticum urartu to common wheat using Durum as a ‘Bridge’. Pathogens 2021, 11, 25. [Google Scholar] [CrossRef]
- Lukasik, E.; Takken, F.L. STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 2009, 12, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Xue, J.; Wang, Q.; Wang, B.; Chen, J. Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, D.A.; Jander, G. The role of protein effectors in plant-aphid interactions. Curr. Opin. Plant Biol. 2013, 16, 451–456. [Google Scholar] [CrossRef]
- Jaouannet, M.; Rodriguez, P.A.; Thorpe, P.; Lenoir, C.J.G.; MacLeod, R.; Escudero-Martinez, C.; Bos, J.I.B. Plant immunity in plant-aphid interactions. Front. Plant Sci. 2014, 5, 663. [Google Scholar] [CrossRef]
- Xu, H.; Qian, L.; Wang, X.; Shao, R.; Hong, Y.; Liu, S.; Wang, X. A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 490–495. [Google Scholar] [CrossRef]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef]
- Rossi, M.; Goggin, F.L.; Milligan, S.B.; Kaloshian, I.; Ullman, D.E.; Williamson, V.M. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 1998, 95, 9750–9754. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Chuang, W. Plant resistance to aphid feeding: Behavioral, physiological, genetic and molecular cues regulate aphid host selection and feeding. Pest. Manag. Sci. 2014, 70, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Zhang, G.; Wang, C.; Ouellet, T.; Wu, J.; Zhu, Q.; Zhao, H. Candidate genes expressed in tolerant common wheat with resistant to English grain aphid. J. Econ. Entomol. 2014, 107, 1977–1984. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Yao, X.; Luo, C.; Hu, X.; Wang, C.; Wang, Y.; Hu, Z.; Zhang, G.; Zhao, H. Biological and morphological features associated with English grain aphid and bird cherry-oat aphid tolerance in winter wheat line XN98-10-35. J. Plant Growth Regul. 2019, 38, 46–54. [Google Scholar] [CrossRef]
- Stein, N.; Feuillet, C.; Wicker, T.; Schlagenhauf, E.; Keller, B. Subgenome chromosome walking in wheat: A 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc. Natl. Acad. Sci. USA 2000, 97, 13436–13441. [Google Scholar] [CrossRef]
- Wang, H.Y.; Liu, D.Q.; Yang, W.X. A wheat disease resistance gene analog of the NBS-LRR class: Identification and analysis. J. Plant Dis. Protect. 2011, 118, 63–68. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Borisjuk, N.; Kishchenko, O.; Eliby, S.; Schramm, C.; Anderson, P.; Jatayev, S.; Kurishbayev, A.; Shavrukov, Y.; Kirti, P.B. Genetic modification for wheat improvement: From transgenesis to genome editing. Biomed. Res. Int. 2019, 2019, 6216304. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, R.; Matzke, M.; Matzke, A.J.M. Plants, RNAi, and the Nobel Prize. Science 2006, 314, 1242–1243. [Google Scholar] [CrossRef]
- Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-induced gene silencing: A powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci. 2019, 20, 206. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Geng, S.; Li, A.; Mao, Y.; Mao, L. RNAi technology for plant protection and its application in wheat. aBIOTECH 2021, 2, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ang, L.; Weilin, Z. Current understanding of the molecular players involved in resistance to rice planthoppers. Pest. Manag. Sci. 2019, 75, 2566–2574. [Google Scholar] [CrossRef]
- Chung, S.H.; Jing, X.; Luo, Y.; Douglas, A.E. Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem. Mol. Biol. 2018, 95, 55–63. [Google Scholar] [CrossRef]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.X.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef]
- Chen, W.; Kastner, C.; Nowara, D.; Oliveira-Garcia, E.; Rutten, T.; Zhao, Y.; Deising, H.B.; Kumlehn, J.; Schweizer, P. Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 2016, 67, 4979–4991. [Google Scholar] [CrossRef]
- Benedito, V.A.; Visser, P.B.; Angenent, G.C.; Krens, F.A. The potential of virus-induced gene silencing for speeding up functional characterization of plant genes. Genet. Mol. Res. 2004, 3, 323–341. [Google Scholar]
- Qi, T.; Zhu, X.; Tan, C.; Liu, P.; Guo, J.; Kang, Z.; Guo, J. Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 2018, 16, 797–807. [Google Scholar] [CrossRef]
- Koch, A.; Wassenegger, M. Host-induced gene silencing-mechanisms and applications. New Phytol. 2021, 231, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Panwar, V.; McCallum, B.; Bakkeren, G. Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 2013, 81, 595–608. [Google Scholar] [CrossRef]
- Yin, C.; Jurgenson, J.E.; Hulbert, S.H. Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2010, 24, 554–561. [Google Scholar] [CrossRef]
- Schaefer, L.K.; Parlange, F.; Buchmann, G.; Jung, E.; Wehrli, A.; Herren, G.; Müller, M.C.; Stehlin, J.; Schmid, R.; Wicker, T.; et al. Cross-kingdom RNAi of pathogen effectors leads to quantitative adult plant resistance in wheat. Front. Plant Sci. 2020, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Song, X.; Li, H.; Cao, L.; Sun, K.; Qiu, X.; Xu, Y.; Yang, P.; Huang, T.; Zhang, J.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Duan, X.; Lv, Y.; Zhang, X.; Nie, Z.; Xie, C.; Ni, Z.; Liang, R. Silencing of an aphid carboxylesterase gene by use of plant-mediated RNAi impairs Sitobion avenae tolerance of phoxim insecticides. Transgenic Res. 2014, 23, 389–396. [Google Scholar] [CrossRef]
- Abdellatef, E.; Will, T.; Koch, A.; Imani, J.; Vilcinskas, A.; Kogel, K. Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 2015, 13, 849–857. [Google Scholar] [CrossRef]
- Zhao, Y.; Sui, X.; Xu, L.; Liu, G.; Lu, L.; You, M.; Xie, C.; Li, B.; Ni, Z.; Liang, R. Plant-mediated RNAi of grain aphid CHS1 gene confers common wheat resistance against aphids. Pest. Manag. Sci. 2018, 74, 2754–2760. [Google Scholar] [CrossRef]
- Xu, L.; Hou, Q.; Zhao, Y.; Lu, L.; Li, B.; Ni, Z.; Liang, R. Silencing of a lipase maturation factor 2-like gene by wheat-mediated RNAi reduces the survivability and reproductive capacity of the grain aphid, Sitobion avenae. Arch. Insect Biochem. 2017, 95, e21392. [Google Scholar] [CrossRef]
- Sun, Y.; Sparks, C.; Jones, H.; Riley, M.; Francis, F.; Du, W.; Xia, L. Silencing an essential gene involved in infestation and digestion in grain aphid through plant-mediated RNA interference generates aphid-resistant wheat plants. Plant Biotechnol. J. 2019, 17, 852–854. [Google Scholar] [CrossRef]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genomics 2018, 18, 31–41. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Schaart, J.G.; van de Wiel, C.C.M.; Smulders, M.J.M. Genome editing of polyploid crops: Prospects, achievements and bottlenecks. Transgenic Res. 2021, 30, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rodriguez, D.; Ramirez-Solis, R.; Garza-Elizondo, M.A.; de Lourdes Garza-Rodriguez, M.D.L.; Barrera-Saldana, H.A. Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases (Review). Int. J. Mol. Med. 2019, 43, 1559–1574. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Schaefer, K.A.; Wu, W.; Colgan, D.F.; Tsang, S.H.; Bassuk, A.G.; Mahajan, V.B. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods 2017, 14, 547–548. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; He, G.; Ma, L.; Deng, X.W. CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J. Genet. Genom. 2020, 47, 263–272. [Google Scholar] [CrossRef]
- Song, G.; Jia, M.; Chen, K.; Kong, X.; Khattak, B.; Xie, C.; Li, A.; Mao, L. CRISPR/Cas9: A powerful tool for crop genome editing. Crop J. 2016, 4, 75–82. [Google Scholar] [CrossRef]
- Wang, N.; Fan, X.; He, M.; Hu, Z.; Tang, C.; Zhang, S.; Lin, D.; Gan, P.; Wang, J.; Huang, X.; et al. Transcriptional repression of TaNOX10 by TaWRKY19 compromises ROS generation and enhances wheat susceptibility to stripe rust. Plant Cell 2022, 34, 1784–1803. [Google Scholar] [CrossRef] [PubMed]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, R.; Gao, J.; Qi, Y.; Song, G.; Li, W.; Li, Y.; Li, G. Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J. 2021, 19, 427–429. [Google Scholar] [CrossRef]
- Cram, D.; Kulkarni, M.; Buchwaldt, M.; Rajagopalan, N.; Bhowmik, P.; Rozwadowski, K.; Parkin, I.A.P.; Sharpe, A.G.; Kagale, S. WheatCRISPR: A web-based guide RNA design tool for CRISPR/Cas9-mediated genome editing in wheat. BMC Plant Biol. 2019, 19, 474. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zhang, W.; Fernie, A.R.; Wen, W. Combining novel technologies with interdisciplinary basic research to enhance horticultural crops. Plant J. 2022, 109, 35–46. [Google Scholar] [CrossRef]
- Chen, J.; Hu, X.; Shi, T.; Yin, H.; Sun, D.; Hao, Y.; Xia, X.; Luo, J.; Fernie, A.R.; He, Z.; et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol. J. 2020, 18, 1722–1735. [Google Scholar] [CrossRef]
- Brauer, E.K.; Balcerzak, M.; Rocheleau, H.; Leung, W.; Schernthaner, J.; Subramaniam, R.; Ouellet, T. Genome editing of a deoxynivalenol-induced transcription factor confers resistance to Fusarium graminearum in wheat. Mol. Plant Microbe Interact. 2020, 33, 553–560. [Google Scholar] [CrossRef]
- Zhu, L.C.; Smith, C.M.; Fritz, A.; Boyko, E.; Voothuluru, P.; Gill, B.S. Inheritance and molecular mapping of new greenbug resistance genes in wheat germplasms derived from Aegilops tauschii. Theor. Appl. Genet. 2005, 111, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Tanguy, S.; Dedryver, C.A. Reduced BYDV-PAV transmission by the grain aphid in a Triticum monococcum line. Eur. J. Plant Pathol. 2009, 123, 281–289. [Google Scholar] [CrossRef]
- Ni, X.; Quisenberry, S.S. Diuraphis noxia and Rhopalosiphum padi (Hemiptera: Aphididae) interactions and their injury on resistant and susceptible cereal seedlings. J. Econ. Entomol. 2006, 99, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Ouellet, T.; Zhao, H.; Wang, X.; Kang, Z. Wheat–Fusarium graminearum Interactions Under Sitobion avenae Influence: From Nutrients and Hormone Signals. Front. Nutr. 2021, 8, 703293. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, K.; He, D.; Guo, J.; Li, G.; Li, B.; Chen, X. Molecular Advances in Breeding for Durable Resistance against Pests and Diseases in Wheat: Opportunities and Challenges. Agronomy 2023, 13, 628. https://doi.org/10.3390/agronomy13030628
Luo K, He D, Guo J, Li G, Li B, Chen X. Molecular Advances in Breeding for Durable Resistance against Pests and Diseases in Wheat: Opportunities and Challenges. Agronomy. 2023; 13(3):628. https://doi.org/10.3390/agronomy13030628
Chicago/Turabian StyleLuo, Kun, Dejia He, Jiao Guo, Guangwei Li, Boliao Li, and Xiulin Chen. 2023. "Molecular Advances in Breeding for Durable Resistance against Pests and Diseases in Wheat: Opportunities and Challenges" Agronomy 13, no. 3: 628. https://doi.org/10.3390/agronomy13030628
APA StyleLuo, K., He, D., Guo, J., Li, G., Li, B., & Chen, X. (2023). Molecular Advances in Breeding for Durable Resistance against Pests and Diseases in Wheat: Opportunities and Challenges. Agronomy, 13(3), 628. https://doi.org/10.3390/agronomy13030628








