Abstract
In plants, the essential roles played by the regulator of chromosome condensation 1 (RCC1) in diverse biological processes, including UV-B (ultraviolet-B radiation) response, hormonal signal transduction, cold tolerance and phenotypic plasticity, have been identified. No comprehensive study on the evolution and function of RCC1 gene family in rice has been carried out. A genome-wide analysis of this gene family is thus required. In this study, a total of 26 OsRCC1s unevenly distributed across 10 chromosomes were identified in rice. Based on their phylogenetic relationship and sequence composition, the OsRCC1 family could be classified into six groups. Members within the same group share a similar gene structure and protein motif/domain composition. Gene duplication analysis revealed that segmental duplication might be the main contributor to the expansion of the RCC1 gene family in rice. Several cis-regulatory elements (CREs) relevant to light, abscisic acid (ABA) and methyl jasmonate (MeJA) are abundant in the promoters of OsRCC1s. A large number of microRNA (miRNA) target sites were present in OsRCC1 mRNAs. Additionally, we used data from gene microarray and qRT-PCR to analyze the expression of OsRCC1 genes during various developmental stages and under abiotic stress conditions. OsRCC1s were found to be highly expressed in panicles and seeds, and most OsRCC1s were differentially expressed under abiotic stresses. Taken together, our study provides a systematic characterization of OsRCC1s and preliminarily explores their diversity as well as their biological functions. Evidence demonstrates that OsRCC1s may play vital roles in both development and abiotic stress response. The results presented here lay a foundation for further investigating the functions of OsRCC1s.
1. Introduction
RCC1 is known as a chromosomal protein that functions as a guanine nucleotide exchange factor (GEF) for the small GTPase protein Ran (a Ras-like nuclear protein) in the nucleus. RCC1 can convert Ran-GDP to Ran-GTP and establish a Ran-GTP gradient between the nucleus and cytoplasm [1,2], thereby regulating a variety of cellular physiological activities such as spindle formation, nuclear membrane assembly, nucleocytoplasmic transport and the G1/S phase transition in the cell cycle [3]. Furthermore, RCC1 is associated with DNA damage repair reaction [2]. RCC1 can be recruited to DNA damage repair sites and participate in the homologous recombination (HR) repair pathway. Overexpression of RCC1 can accelerate cell proliferation and DNA damage repair, as well as inhibit cell senescence caused by DNA damage [4].
Previous studies on RCC1s have primarily focused on mammals and yeast. Although few studies on RCC1s have been conducted in plants, recent studies in Arabidopsis thaliana have revealed that RCC1s are essential in abiotic stress signaling and responses. The first to be functionally characterized in A. thaliana was UVR8, a specific photoreceptor for UV-B. UV-B absorption induces an instantaneous conversion of UVR8 from dimer to monomer. Interaction with COP1, the central regulator of light signaling, also regulates plant responses to UV-B [5,6]. RUG3 is another RCC1 family protein that is located in the mitochondria. It can interact with ATM to synergistically regulate alternative splicing of the nad2 precursor mRNA, mitochondrial reactive oxygen species (ROS) accumulation, DNA damage response and cell cycle progression, so as to maintain mitochondrial function and redox homeostasis [7,8]. This function was confirmed by DEK47 (a functional homolog of AtRUG3 in maize). Furthermore, DEK47 was found to be closely related to seed development [9]. TCF1 could be rapidly induced and activate the expression of BCB during cold acclimation, thereby modulating lignin biosynthesis and subsequent cell wall remodeling. It regulates cold acclimation and freezing resistance through a chromatin-based regulatory mechanism (a CBF-independent pathway) [10]. SAB1 was discovered to be a key component in ABA signal transduction, negatively regulating ABI5 via a multi-dimensional mechanism [11]. RCC1 also regulates root development as well as phenotypic plasticity. For example, RLD regulates polar auxin transport by interacting with LZY. Gravity signals first appear in gravity-sensing cells in the form of LZY3 polarity distribution. It then regulates the flow of auxin via RLD1 polarity distribution and PIN3 repositioning to influence gravitropic set point angles of roots [12]. CmRCC1 from pumpkin, which is highly homologous to AtRLD1 and AtRLD4, also participates in root architecture control and PIN expression regulation. Moreover, overexpression of CmRCC1 can enhance the cold tolerance of transgenic tobacco [13]. PROTON1 was identified to be involved in controlling rosette size plasticity in response to nitrogen during initial vegetative growth of A. thaliana [14]. Six previously reported RCC1s in A. thaliana were evaluated, and it was found that there was no GEF activity in UVR8, TCF1 and RUG3, suggesting that RCC1s may have distinct functions in plants and animals [8,10,15]. In plants, RCC1s could mediate multiple responses to environmental stimuli, participating in UV-B photomorphogenic response, ROS accumulation, cold signal transduction and hormone signaling pathways, etc., thereby, affecting plant morphogenesis and adaptation to the environment by influencing a variety of biological processes such as downstream gene transcription and epigenetic modifications.
Rice is an important crop that feeds more than half of the population in the world and serves as a model plant of monocot. However, the function of only one RCC1 (OsRLR4) has been clarified in rice so far. It directly binds to the promoter of OsAUX1 and interacts with OsTrx1 to promote the accumulation of H3K4me3 on the OsAUX1 promoter and positively modulate the OsAUX1 expression, thus, regulating auxin accumulation in primary root (PR) tip and root apical meristem activity to control PR development [16]. Despite some progress that has been made in understanding the role of RCC1 family in A. thaliana, information about the rice RCC1 gene family is relatively rare. It is necessary to investigate the relationship between RCC1s and growth and development, as well as environmental stresses in rice.
In this paper, a total of 26 rice RCC1 family members were identified and characterized. Their phylogeny, physical-chemical properties, gene structures, motif patterns, domain compositions, CREs in promoter, chromosome distribution, gene duplication events, miRNAs targets and gene ontology (GO) enrichments were comprehensively determined. Finally, we examined the expression patterns of OsRCC1s in various organs/tissues and under different abiotic stresses by analyzing gene microarray and qRT-PCR data. Our study offers a theoretical basis for understanding the function of RCC1s in rice evolution, growth, development and survival, as well as a reference for the subsequent functional validation.
2. Materials and Methods
2.1. Identification of RCC1s in Rice
The Arabidopsis RCC1 protein sequences were downloaded from the TAIR database (https://www.arabidopsis.org/) (accessed on 30 April 2022) and used as queries for a BLASTP search in the rice genome of Ensembl Plants database (http://plants.ensembl.org/Multi/Tools/Blast) (accessed on 30 April 2022) to screen potential OsRCC1s with E-value ≦ 1 × 10−5 and identity ≧ 50% as standards [17,18,19]. The Domain Search function of Rice Genome Annotation Project database (MSU-RGAP, http://rice.uga.edu/analyses_search_domain.shtml) (accessed on 30 April 2022) was used to search the proteins containing RCC1 domains (accession numbers: PF00415 and PF13540) in rice [20]. Verification was performed by the SMART website (http://smart.embl-heidelberg.de/) (accessed on 1 May 2022) as well as the Pfam database (http://pfam.xfam.org/) (accessed on 1 May 2022) to remove proteins without RCC1 domains Supplementary Table S1 [21,22,23].
2.2. Phylogenetic Analysis
To explore the evolutionary relationships of RCC1s among rice, Arabidopsis and maize (Zea mays), multiple sequence alignment of RCC1s from the three species was carried out using the ClustalW program in MEGA7 software [24]. Then, a phylogenetic tree of RCC1s was constructed by the Neighbor-joining (NJ) method using Poisson model. The bootstrap value was set to 1000, and the other parameters were all default values. Finally, Evolview (http://www.evolgenius.info/evolview/#/) (accessed on 5 May 2022) was used to display the evolutionary tree.
2.3. Physicochemical Properties of OsRCC1s
The computational physical and chemical properties of OsRCC1s were evaluated using the ProtParam website (https://web.expasy.org/protparam/) (accessed on 1 May 2022), including the number of amino acids, molecular weight (MW), theoretical isoelectric point (pI) and grand average of hydrophilicity (GRAVY) [25]. The subcellular localization was predicted using the Plant-mPLoc website (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (accessed on 1 May 2022) [26]. The detailed information of all OsRCC1s were listed in Table 1.

Table 1.
Detailed information of the identified rice RCC1s.
2.4. Structural Analysis
Gene structures and conserved motifs of OsRCC1s were identified using the TBtools software [27]. The GFF3 file (Generic Feature Format version3 Data) of rice and protein sequences of OsRCC1s were used as input, respectively. Except for the number of motifs, which was set to 12, all other parameters were default values. Annotations of identified motifs were obtained from the InterPro database (https://www.ebi.ac.uk/interpro/search/sequence/) (accessed on 3 May 2022). Furthermore, conserved domains in OsRCC1s were identified using the HMMER database (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) (accessed on 5 May 2022) by submitting protein sequences of OsRCC1s. Gene structures, conserved motifs and domains of OsRCC1s were visualized using the TBtools software.
2.5. CREs Prediction in Promoter Regions of OsRCC1s
For each OsRCC1, 2000 bp sequence upstream of the start codon (ATG) was downloaded from RAP-DB (https://rapdb.dna.affrc.go.jp/download/irgsp1.html) (accessed on 10 May 2022) and submitted to the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 10 May 2022) to extract CREs [28]. The obtained prediction information was categorized and summarized. Then, the CREs were displayed using TBtools software and Excel 2019 software.
2.6. Chromosome Distribution, Gene Duplication and Selective Pressure Analysis
Genome data for rice, Arabidopsis, tomato (Solanum lycopersicum), barley (Hordeum vulgare) and wheat (Triticum aestivum) were downloaded from the Ensembl Plants database (http://plants.ensembl.org/info/data/ftp/index.html) (accessed on 13 May 2022) and the Phytozome v13 database (https://phytozome-next.jgi.doe.gov/) (accessed on 13 May 2022) [29]. The TBtools software was used to analyze and visualize the chromosomal distribution, gene duplication events within the rice genome, as well as the gene level collinearity among plant genomes. The Simple Ka/Ks Calculator (NG) tool in TBtools software was used to calculate the ratio of non-synonymous to synonymous substitution (Ka/Ks) of duplicated gene pairs.
2.7. miRNA Target Predictions in OsRCC1s
Potential OsRCC1-targeting miRNAs were predicted using the psRNATarget database (https://www.zhaolab.org/psRNATarget/) (accessed on 10 May 2022) [30]. The coding sequences of OsRCC1 genes of rice were used as input in psRNATarget against the rice reference miRNAs. All the parameters were left as default values [30]. The Cytoscape v3.9.1 software was used to construct an interaction network between the identified miRNA and the corresponding targeted OsRCC1s mRNA [31].
2.8. Expression Profile and GO Annotation Analysis
For microarray analysis of OsRCC1s, Affymetrix GeneChip rice genome arrays (http://www.ncbi.nlm.nih.gov/geo/; Gene Expression Omnibus platform accession nos. GSE6893 and GSE6901) (accessed on 15 May 2022) were used. The Affymetrix values in 15 various organs/tissues of rice, including seedling roots, mature leaves, young leaves, stem apical meristems (SAM), panicles and seeds, as well as conditions of three different abiotic stresses (salt, drought and low temperature), can be queried through the Rice eFP Browser (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi) (accessed on 15 May 2022). Heat maps that were sorted based on hierarchical clustering were created with the TBtools software.
GO enrichment analysis for OsRCC1s was performed by agriGO v2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/index.php) (accessed on 15 May 2022) [32]. The histogram was generated using GraphPad Prism v8.3 software (GraphPad Software Inc., La Jolla, CA, USA).
2.9. Plant Material, Stress Treatment, RNA Extraction and Quantitative RT-PCR Analysis
Oryza sativa L. japonica cv. Nipponbare seeds were sterilized (2.5% NaClO) and germinated at 30 °C. Seedlings were cultivated in a greenhouse at 27 ± 2 °C with a 14 h light/10 h dark photoperiod. For stress treatments, the two-week-old seedlings grown in Yoshida medium (YM) were treated with YM supplemented with 150 mM NaCl, 20% (m/V) PEG6000, cold (4 °C), heat (40 °C) and 50 μM ABA, respectively. In each experiment, there were 3 replicates with 96 rice plants. The durations of all the stress treatments were 2, 4, 8 and 12 h, and untreated seedlings were taken as the experimental control. Rice leaf samples were collected randomly from more than 3 rice plants and quickly frozen in liquid nitrogen. Then, they were stored at −80 °C.
Total RNA was extracted by an OminiPlant RNA Kit (CWBIO, Beijing, China), and cDNA synthesis was performed using Hifair® II 1st Strand cDNA Synthesis SuperMix for qPCR (YEASEN, Shanghai, China) according to the manufacturer’s instructions. The Hieff® qPCR SYBR Green Master Mix (YEASEN, Shanghai, China) was used to conduct qRT-PCR amplification in the CFX96 Touch TM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). All primers were designed using the Primer Premier 5 software. Their specificity was then confirmed by using the Ensembl Plants database (http://plants.ensembl.org/Multi/Tools/Blast) (accessed on 20 May 2022) for BLASTN search in rice genome. OsActin1 (LOC_Os03g50885) and OsUBQ5 (LOC_Os01g22490) were used as the internal reference genes. The sequence information of all primers was listed in Supplementary Table S2. Three biological replicates were performed with three technical replicates for each sample. The relative expression level was calculated by the 2−ΔΔCt method [33]. Gene expressions for selected OsRCC1s were visualized with heatmaps using the TBtools software.
3. Results
3.1. Identification of RCC1s in Rice
After removing redundant and incomplete sequences and verifying domains, a total of 26 RCC1s were identified in the whole rice genome (designated as OsRCC1-1~OsRCC1-26 according to their chromosomal location). The physicochemical properties of OsRCC1s differ greatly. The average protein length of OsRCC1s is 699.5 aa (range from 395 to 1086 aa). The average MW is 75,527.4 Da (range from 41,731.5 to 117,823.8 Da). The theoretical pI ranges from 5.05 (OsRCC1-2) to 9.18 (OsRCC1-20), with an average pI value of 7.2. All of the OsRCC1s possess negative GRAVY values (range from −0.654 to −0.055), with an average value of −0.305, indicating the hydrophilic nature of OsRCC1s. Subcellular localization prediction showed that OsRCC1-4 may be located in both cytoplasm and nucleus, OsRCC1-26 in both cell wall and nucleus, and other OsRCC1s in nucleus. All the characteristics of OsRCC1s are shown in Table 1.
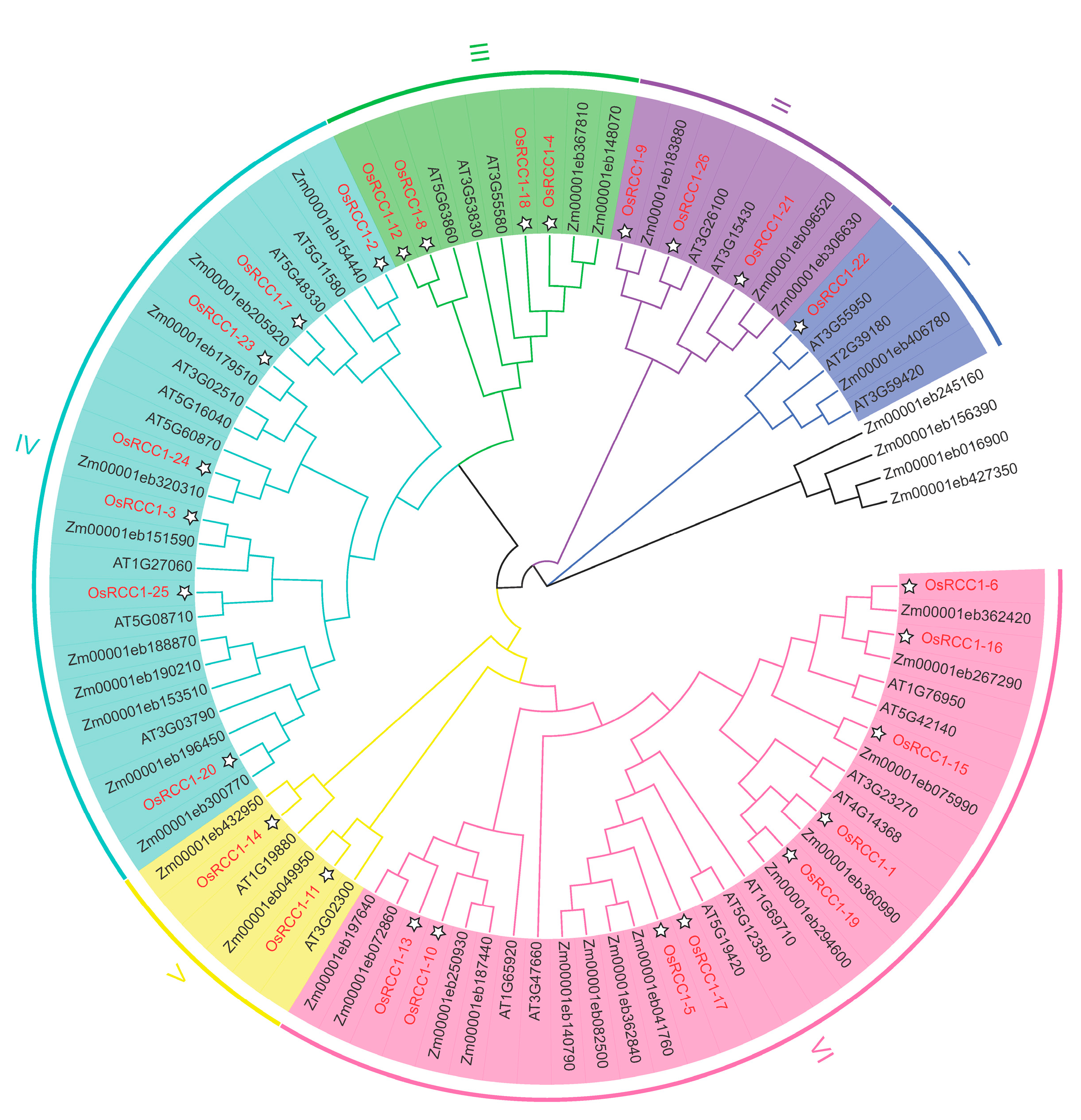
Phylogenetic relationships are crucial for understanding the structure and evolution of gene families. By comparing RCC1 proteins among several plant species (rice, Arabidopsis and maize), a phylogenetic tree was constructed using the NJ method. As shown in Figure 1, RCC1s of rice and maize are more clustered together within the same branch than Arabidopsis, showing a closer genetic relationship. Based on phylogenetic analysis and motif composition, OsRCC1s could be divided into six groups (Group I~VI) of different sizes. Group VI is the largest, containing nine OsRCC1s, while Group I is the smallest, containing only one OsRCC1 (OsRCC1-22).
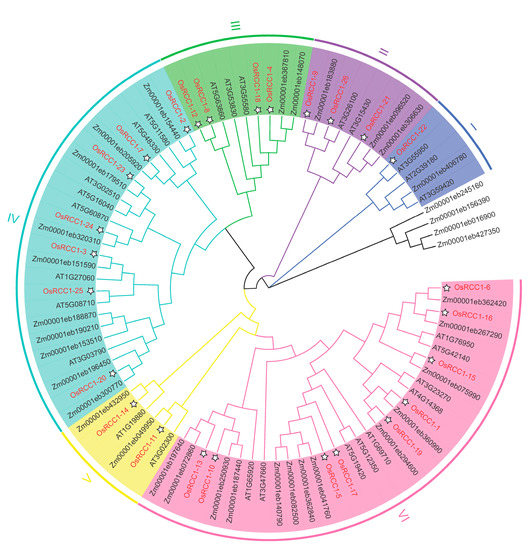
Figure 1.
Phylogenetic analysis of OsRCC1s and their homologs from various organisms. AT represents A. thaliana, Zm represents Z. mays, and Os represents O. sativa L. The phylogenetic tree was generated using the NJ method by MEGA7. OsRCC1s are classified into 6 groups (Group I–VI) and distinguished by different colors.
3.2. Sequence Structure Analysis of OsRCC1s
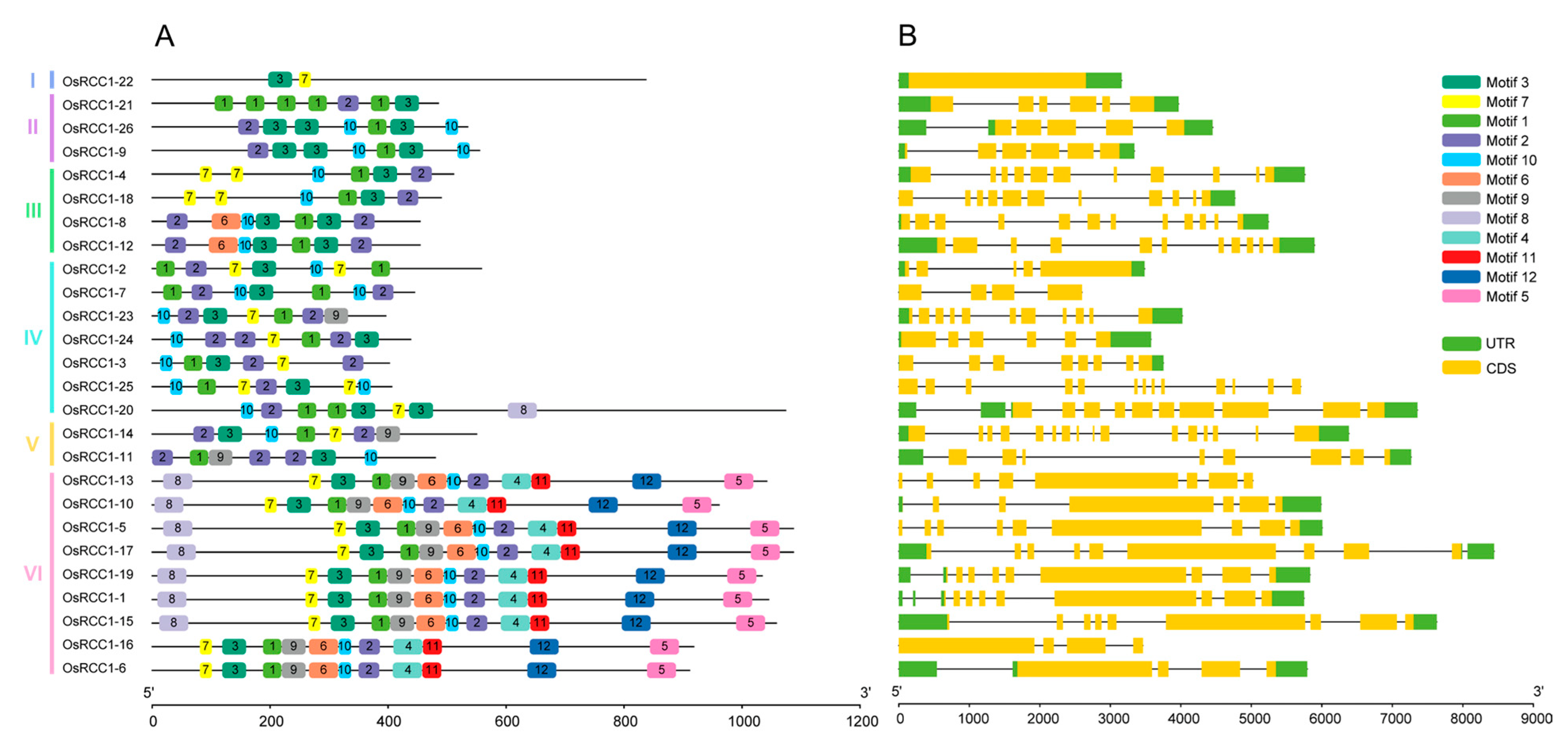
To investigate the structural characteristics of OsRCC1s, we analyzed their exon–intron structures (Figure 2B). The results revealed that there is no non-coding region in OsRCC1-22, which is the only member of Group I. Three members in Group II contain 5~6 exons and 5 introns. A total of four members in Group III contain 11~12 exons and 10~11 introns. Five members in Group VI contain nine exons. The last four exons of all nine members in Group VI are similar in length. In general, OsRCC1s possess multiple exons and introns with the exception of OsRCC1-22, and members within the same group tend to share more similar gene structures and exon lengths.
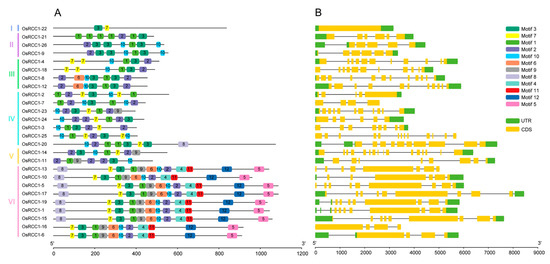
Figure 2.
Conserved motifs and gene structures of OsRCC1s. (A) Motif patterns of OsRCC1s. Boxes with different colors represent various conserved motifs. (B) Exon/intron architectures of OsRCC1s. Introns, exons and untranslated regions are represented by black lines, yellow boxes and green boxes, respectively.
The conserved domains and motifs of OsRCC1s were also examined. The entire RCC1 family is characterized by the presence of at least one conserved RCC1 domains. Some OsRCC1s also have other domains (Supplementary Figure S1). For instance, only OsRCC1-22 contains the protein kinase domain (PF00069) as well as the protein tyrosine and serine/threonine kinase domain (PF07714). Only OsRCC1-20 contains ankyrin-repeat (Ank) domain (PF00023, PF12796, PF13637), and only OsRCC1-15 contains the pleckstrin homology (PH) domain (PF16457). Moreover, nine members of Group VI all contain the FYVE zinc finger domain (PF01363) and BREVIS RADIX (BRX) domain (PF08381, PF13713, PF16627), demonstrating a close evolutionary relationship and potential functional similarity.
We identified 12 conserved motifs in OsRCC1s (Figure 2A), and motif annotations were predicted (Supplementary Table S3). Motifs 1, 2, 3, 6, 7, 9 and 10 belong to RCC1 domains. Motif 4 belongs to FYVE zinc finger domain, motif 5 belongs to the BRX domain, and motif 12 belongs to N-terminal BRX domain. Motifs 8 and 11 could not be annotated. Therefore, their functions remain unknown. Almost all OsRCC1s contain motifs 1, 2, 3, 7 and 10. Motifs 4, 5, 11 and 12 are only present in all members of Group VI; motif 6 is only present in 2 members of Group III and all members of Group VI; motif 8 is only in one member of Group IV and seven members of Group VI; and motif 9 is only present in one member of Group IV and all members of Group V and Group VI. These above results suggest that there are many group-specific motifs in OsRCC1s, which may be related to specific functions.
3.3. CREs Analysis in Promoter of OsRCC1s
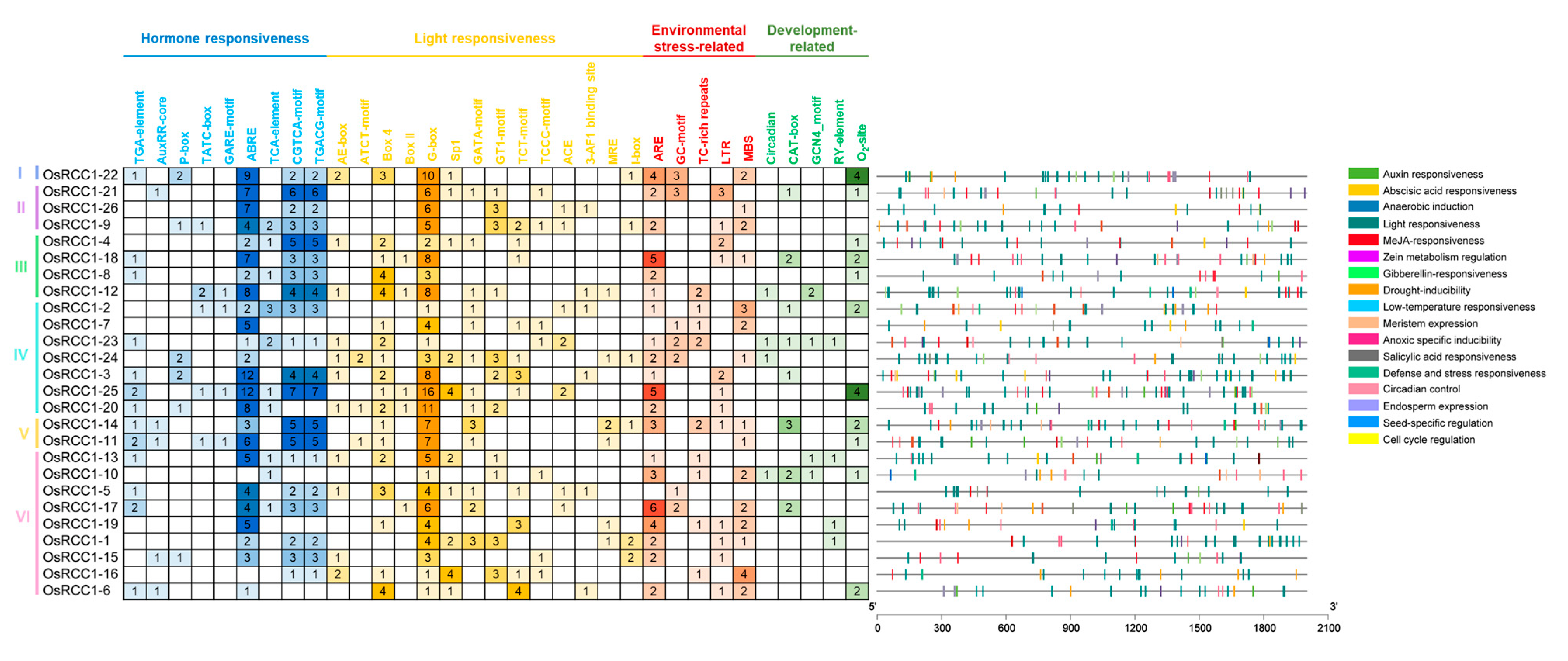
As a crop that is sensitive to various biotic and abiotic stresses, rice needs to rapidly adapt to the constantly changing environment [34]. Altering gene transcription via CREs in the promoter region remains a crucial mechanism of gene expression regulation. CREs within promoters (2000 bp upstream of initiation codon “ATG”) of OsRCC1s were predicted, so as to speculate on the possible factors affecting OsRCC1 gene expression and the regulatory pathways in which OsRCC1s may be involved. As shown in Figure 3, in addition to the basic CREs (such as CAAT-box and TATA-box), the other functional CREs found in the promoters of OsRCC1s could be grouped into four categories: (1) hormone-responsive elements, such as ABA-responsive element (ABRE), auxin-responsive elements (AuxRR-core, TGA-element), salicylic acid-responsive element (TCA-element), MeJA-responsive element (CGTCA motif/TGACG motif), etc.; (2) light-responsive elements, such as Box 4, G-box, GT1-motif, GATA-motif, etc.; (3) stress-responsive elements, such as drought-responsive element (MBS), low temperature-responsive element (LTR), anaerobic-responsive element (ARE), etc.; (4) growth and development regulation elements, such as zein metabolism regulation-related element (O2-site), meristem expression-related element (CAT-box), etc. OsRCC1s have the most light-responsive elements distributed within promoter regions (309), and they all contain G-box elements (135). Furthermore, 121 ABRE, 65 CGTCA-motif/TGACG-motif, 51 ARE, 27 MBS and 21 O2-sites were found in the promoter regions of OsRCC1s. CREs that response to phytohormone, light and environmental stresses were detected in all of the 26 OsRCC1s, whereas only 19 OsRCC1s possess growth and development-related CREs. The above results imply that transcriptional profiles of OsRCC1s may vary depending on environmental, hormonal and developmental factors.
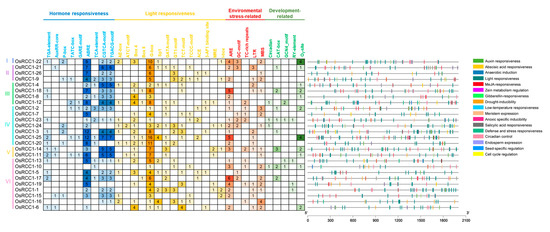
Figure 3.
CREs distribution in the promoter regions of OsRCC1s. CREs in the promoter of each OsRCC1 gene are classified based upon the putative functions, including hormone-responsive CREs, light-responsive CREs, environmental stress-responsive CREs and growth and development-responsive CREs. The positional distribution of various CREs on promoters is shown as vertical bars with different colors.
3.4. Chromosome Mapping and Gene Duplication Analysis of OsRCC1s
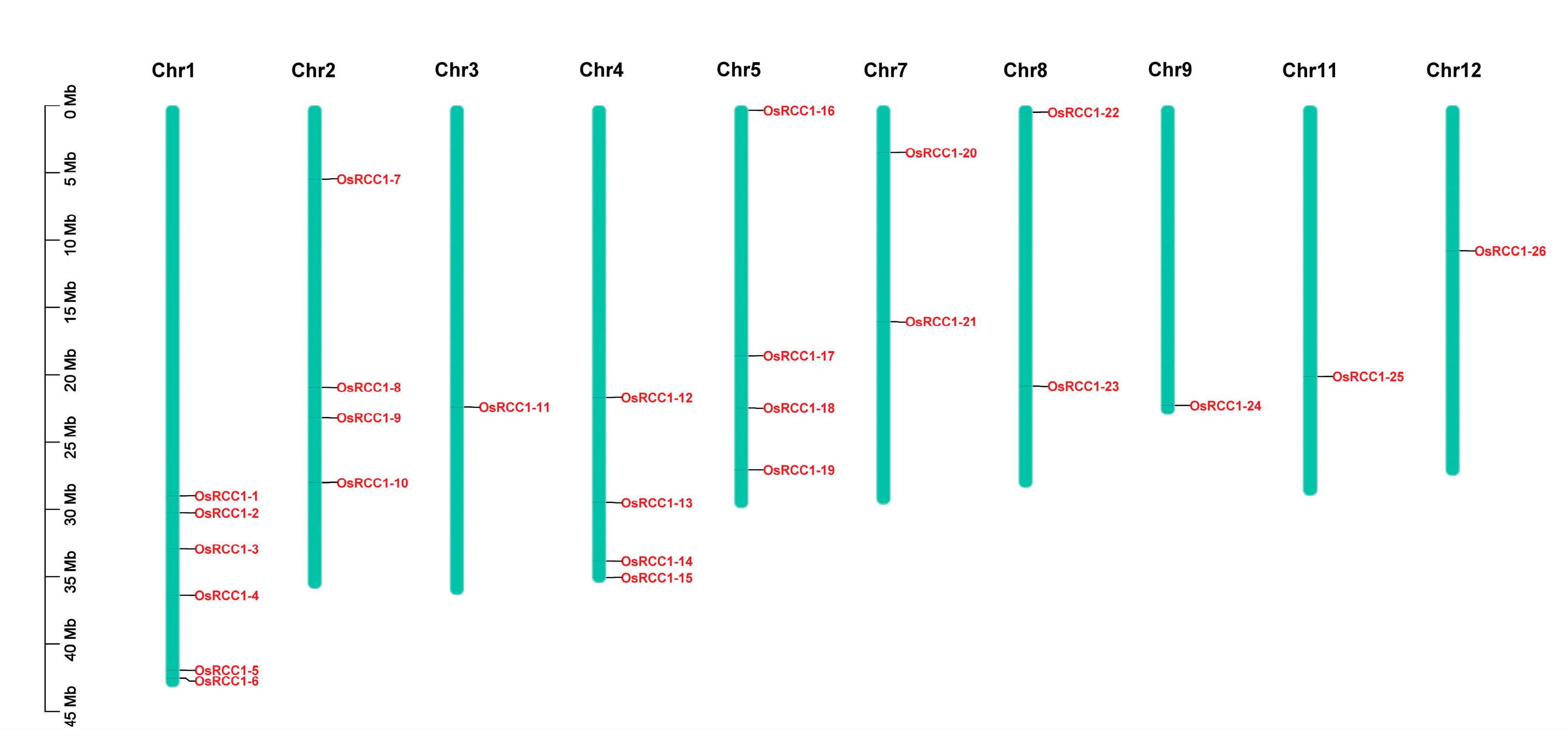
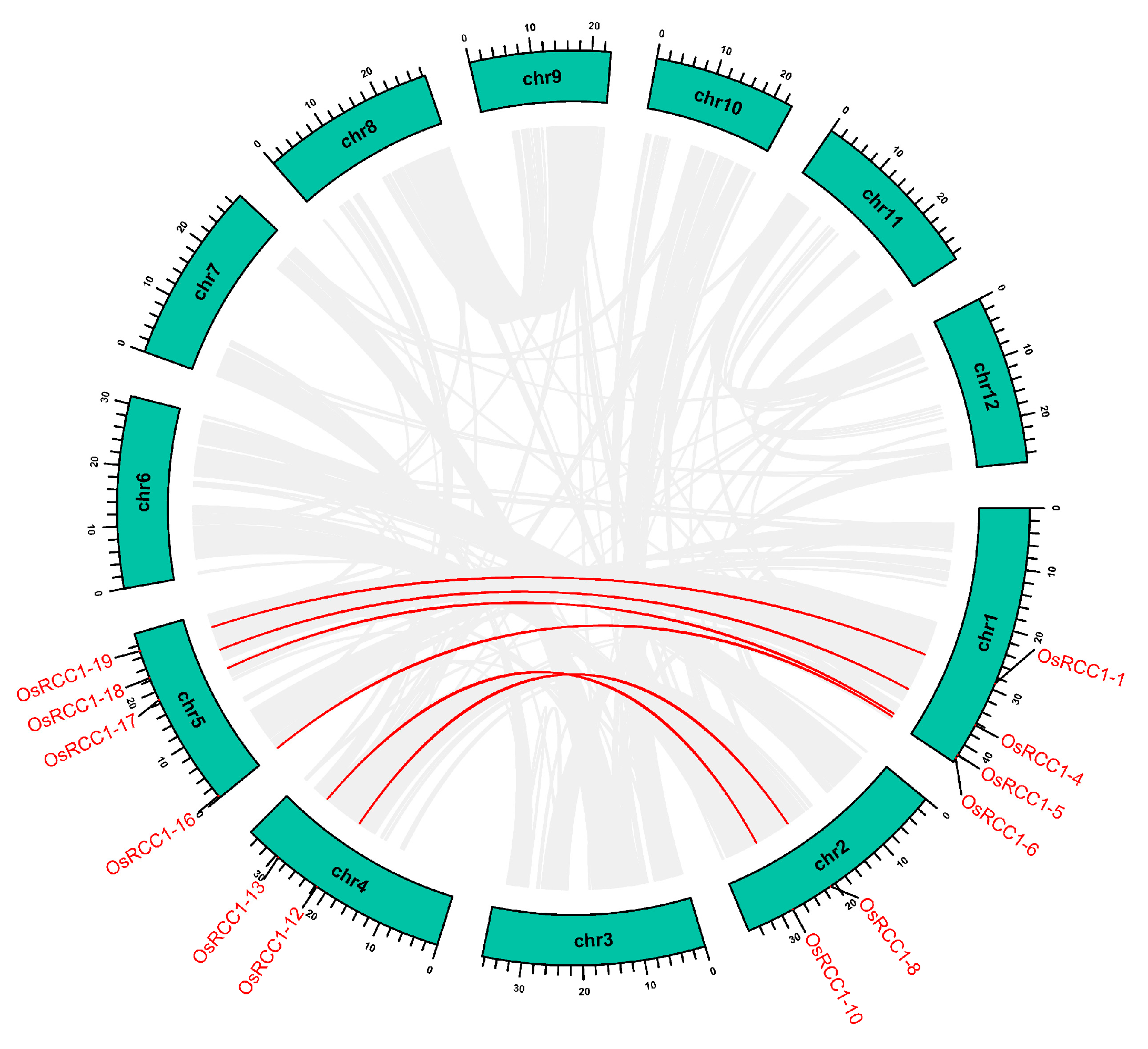
OsRCC1s are unevenly distributed on 10 of the 12 rice chromosomes (Figure 4). The number of OsRCC1s distributed on chromosome 1 is the largest, with a total of six. Chromosomes 2, 4 and 5 both have four OsRCC1s. Chromosomes 7 and 8 both have two OsRCC1s. The number of OsRCC1s distributed on chromosomes 3, 9, 11 and 12 is the least, with only one each. OsRCC1s are not found on chromosome 6 or 10.
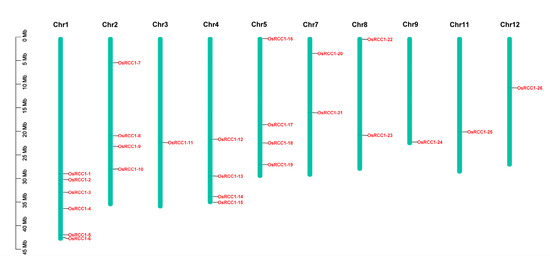
Figure 4.
Chromosome distribution of OsRCC1s. Chromosome numbers are shown at the top of each chromosome, and the name of each OsRCC1 is labeled at the right side of each chromosome. Scale bar on the left side indicates the chromosome lengths (Mb).
Gene duplication events, such as tandem and segmental duplication, provide major forces that drive the expansion of gene families and the evolution of the entire genome [35]. We used the TBtools software to examine the duplication events of OsRCC1s within the rice genome. Gene family members generated by tandem duplication are typically closely aligned on the same chromosome, forming a cluster of genes with similar sequences and functions. According to Holub’s description, a tandem duplication event is defined as three or more gene family members clustering within a 200-kb chromosome region, which means that duplicated genes are contiguous to each other [36]. However, members of OsRCC1 gene family are distant from each other on chromosomes, and tandem duplication events did not occur. Segmental duplication can be defined as the copies of whole blocks of genes from one chromosome to another. This event would result in gene duplication to unlinked sites, even when a segment is translocated to the same chromosome [37]. There are six pairs of segmental duplication genes in the OsRCC1 gene family (Figure 5). These genes are distributed on four chromosomes (chromosome 1, 2, 4 and 5), belonging to Group III (OsRCC1-4 & OsRCC1-18, OsRCC1-8 & OsRCC1-12) and Group VI (OsRCC1-1 & OsRCC1-19, OsRCC1-5 & OsRCC1-17, OsRCC1-6 & OsRCC1-16, OsRCC1-10 & OsRCC1-13). Hence, segmental duplication events may be a vital driving force for the expansion of OsRCC1 gene family. The Ka/Ks ratio is an important indicator reflecting the type and strength of selection pressure in evolution, with Ka/Ks <1 indicating negative selection pressure. The Ka/Ks ratios for segmental duplicated OsRCC1 gene pairs within the rice genome vary from 0.099 to 0.277, with an average value of 0.159 < 1. It is presumed that the OsRCC1 gene family had mainly been subjected to strong purifying selection during evolution, which was crucial for the functional conservation of OsRCC1 gene family (Supplementary Table S4).
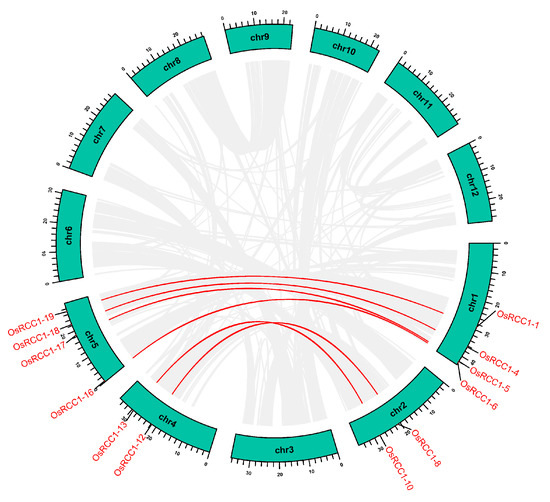
Figure 5.
Segmental duplication events of OsRCC1s. Segmental duplicated OsRCC1 gene pairs are linked by the red lines between chromosomes. Segmental duplicated gene pairs within the rice genome are linked by the gray lines. The chromosome numbers are shown at the center of each chromosome. The scale bar marked on the chromosome indicates chromosome lengths (Mb).
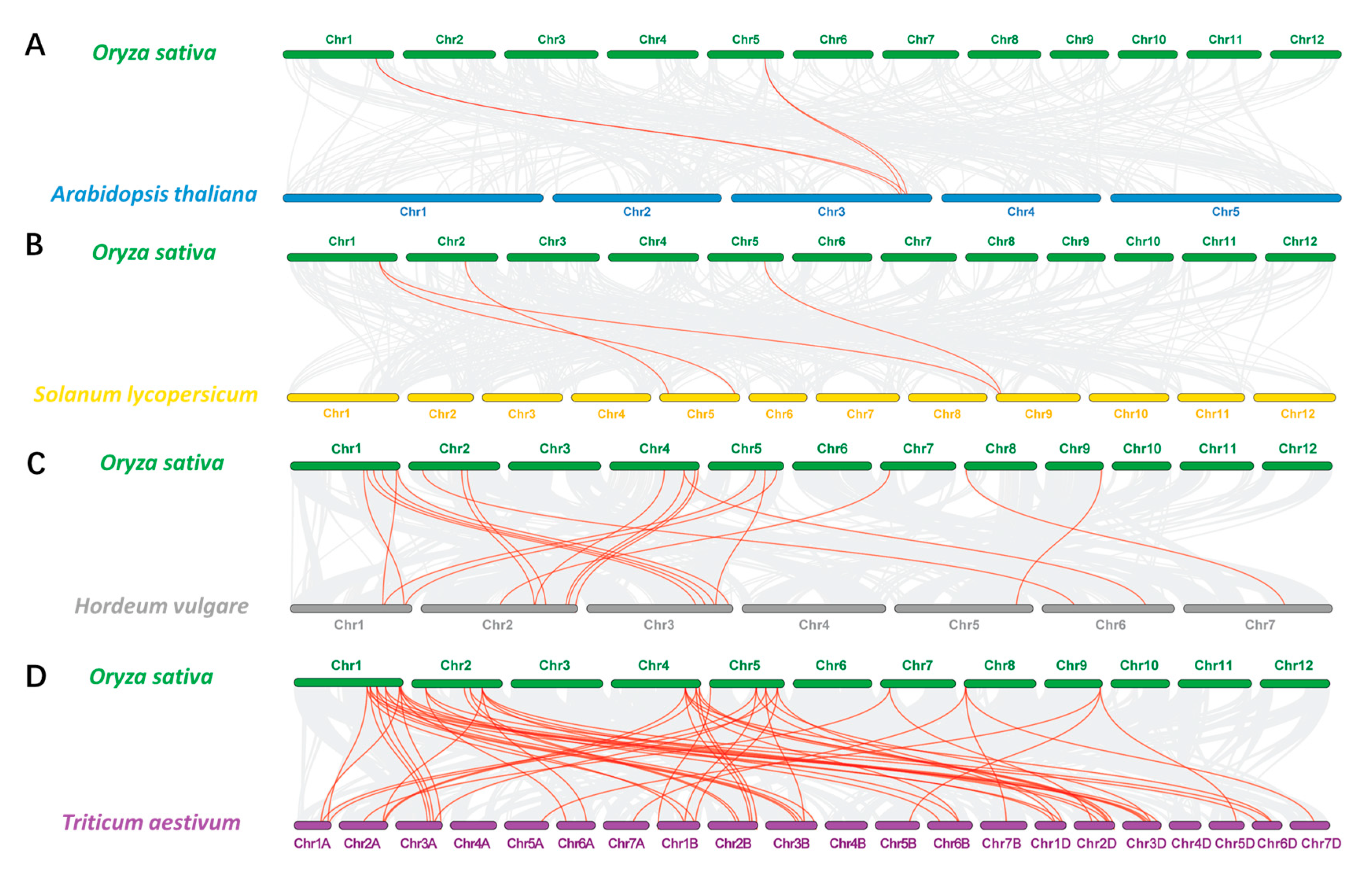
Syntenic relationships of RCC1s between rice and three other representative species, including two dicots (A. thaliana and tomato) and two monocots (wheat and barley), were also investigated (Figure 6). There are 57 collinear gene pairs between rice and wheat, 20 pairs between rice and barley, and 4 pairs between rice and both Arabidopsis and tomato (Supplementary Table S5). There is greater collinearity between OsRCC1s and TaRCC1s. Compared with dicots, OsRCC1s display a high level of identity with orthologs in other monocots with fewer evolutionary separation events. OsRCC1-4 and OsRCC1-18 exhibit multi-collinearity among species, suggesting that they are more conservative in evolution.
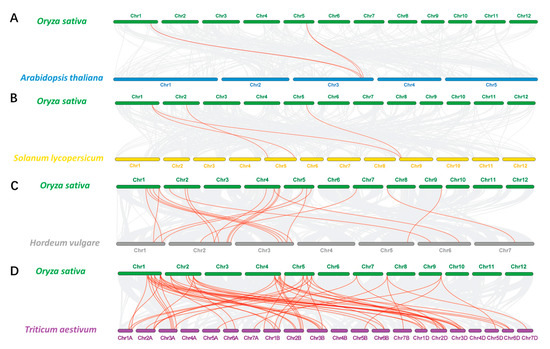
Figure 6.
Collinearity relationship of the RCC1 genes in rice and other four plant species, including Arabidopsis (A), tomato (B), barley (C) and wheat (D). Gray lines in the background indicate the syntenic blocks between rice and other plant genomes, while the red lines highlight the syntenic RCC1 gene pairs.
3.5. Putative miRNA Targeting OsRCC1s Prediction
miRNAs are a class of small non-coding single-stranded RNA that recognize target mRNAs via complementary base pairing. They then cause mRNA cleavage and/or translational inhibition to regulate target gene expression at the post-transcriptional level, thereby affecting plant growth and development, crop yield, seed quality, stress response, etc. [38]. The CDS of OsRCC1s was employed to distinguish target miRNAs by using the psRNATarget database. The results showed that a total of 336 mature miRNAs (19-24 nt) were identified (Supplementary Table S6). All of the OsRCC1s contain target miRNAs, but no major target miRNAs (Supplementary Figure S2). The number of target miRNAs ranges from a minimum of 3 (OsRCC1-8, OsRCC1-21) to a maximum of 44 (OsRCC1-1). A number of 274 miRNAs regulate the expression of OsRCC1s through the cleavage effect, while only 62 miRNAs through translational inhibition. It could be speculated that the cleavage effect, as the main function of miRNAs, plays a vital role in regulating the expression of OsRCC1s.
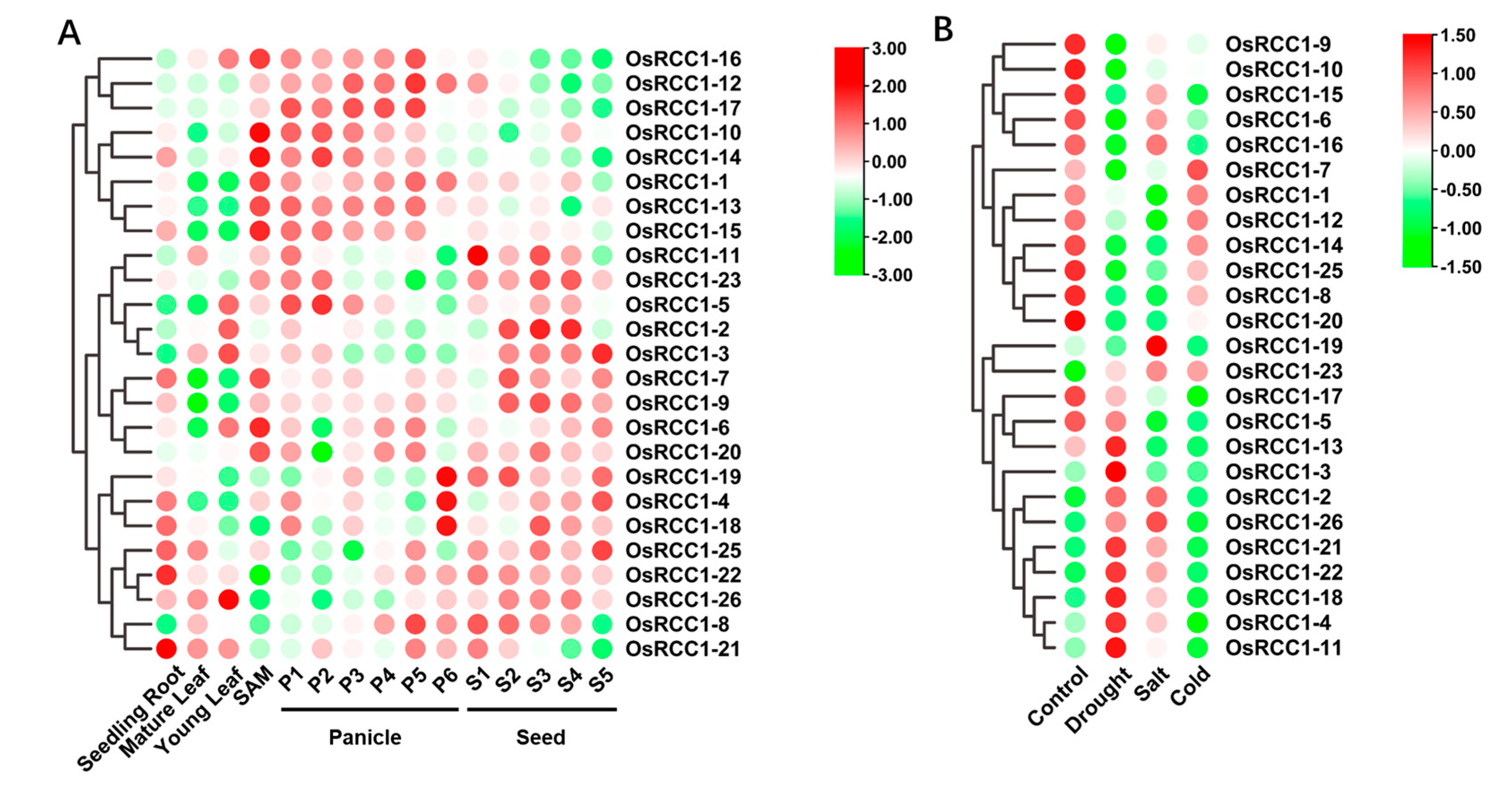
3.6. Expression Profiles of OsRCC1s in Various Tissues/Organs
The analysis of tissue-/stage-specific expression profiles could help to further unveil the potential functions of OsRCC1s in rice. The expression data of 25 OsRCC1s (OsRCC1-24 was not found) in different organs/tissues of indica rice variety IR64 were collected from the Rice eFP Browser database to generate a clustering heat map (Figure 7A). The results revealed that OsRCC1s mainly display two expression patterns among various tissues/organs. For instance, OsRCC1-1, OsRCC1-10 and OsRCC1-12~OsRCC1-17 exhibit similar expression patterns, which are highly expressed in SAM and panicles, but lower levels in other tissues/organs. Due to their obvious tissue-specific characteristics, these genes may be necessary for panicle morphogenesis. Other OsRCC1s (OsRCC1-2~OsRCC1-9, OsRCC1-11, OsRCC1-18~OsRCC1-23, OsRCC1-25~OsRCC1-26) were found to be predominantly expressed in seeds and at least one other tissue/organ. The above results suggest that OsRCC1s are differentially expressed in various tissues/organs and may participate in panicle and seed development.
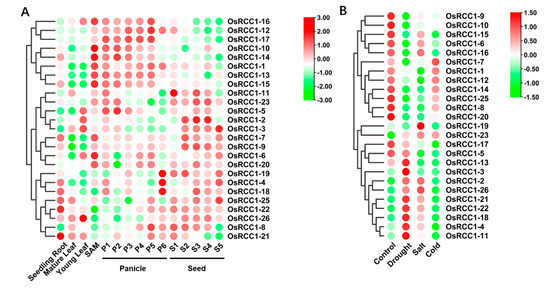
Figure 7.
Expression patterns of OsRCC1s from microarray data. (A) Expression patterns of OsRCC1s in diverse tissues and across various developmental stages. The sample sources are as follows: roots of 7-day-old seedling, mature leaf (collected before pollination), young leaf, shoot apical meristem (SAM), P1 (0–3 cm panicle), P2 (3–5 cm panicle), P3 (5–10 cm panicle), P4 (10–15 cm panicle), P5 (15–22 cm panicle), P6 (22–30 cm panicle), S1 (0–2 DAP), S2 (3–4 DAP), S3 (5–10 DAP), S4 (11–20 DAP) and S5 (21–29 DAP). Different stages of panicle and seed development have been categorized according to panicle length and days after pollination, respectively. (B) Expression patterns of OsRCC1s under three different abiotic stress conditions. Seven-day-old rice seedlings were subjected to salinity (200 mM NaCl), dehydration (dried between folds of tissue paper) and cold (4 °C) treatments for 3 h. The seedlings kept in water for 3 h at 28 ± 1 °C served as control. The color scale represents log2 of average signal values. The red color shows the higher and the green color shows the lower expression levels in expression bar.
3.7. Expression Profiles of OsRCC1s under Abiotic Stresses
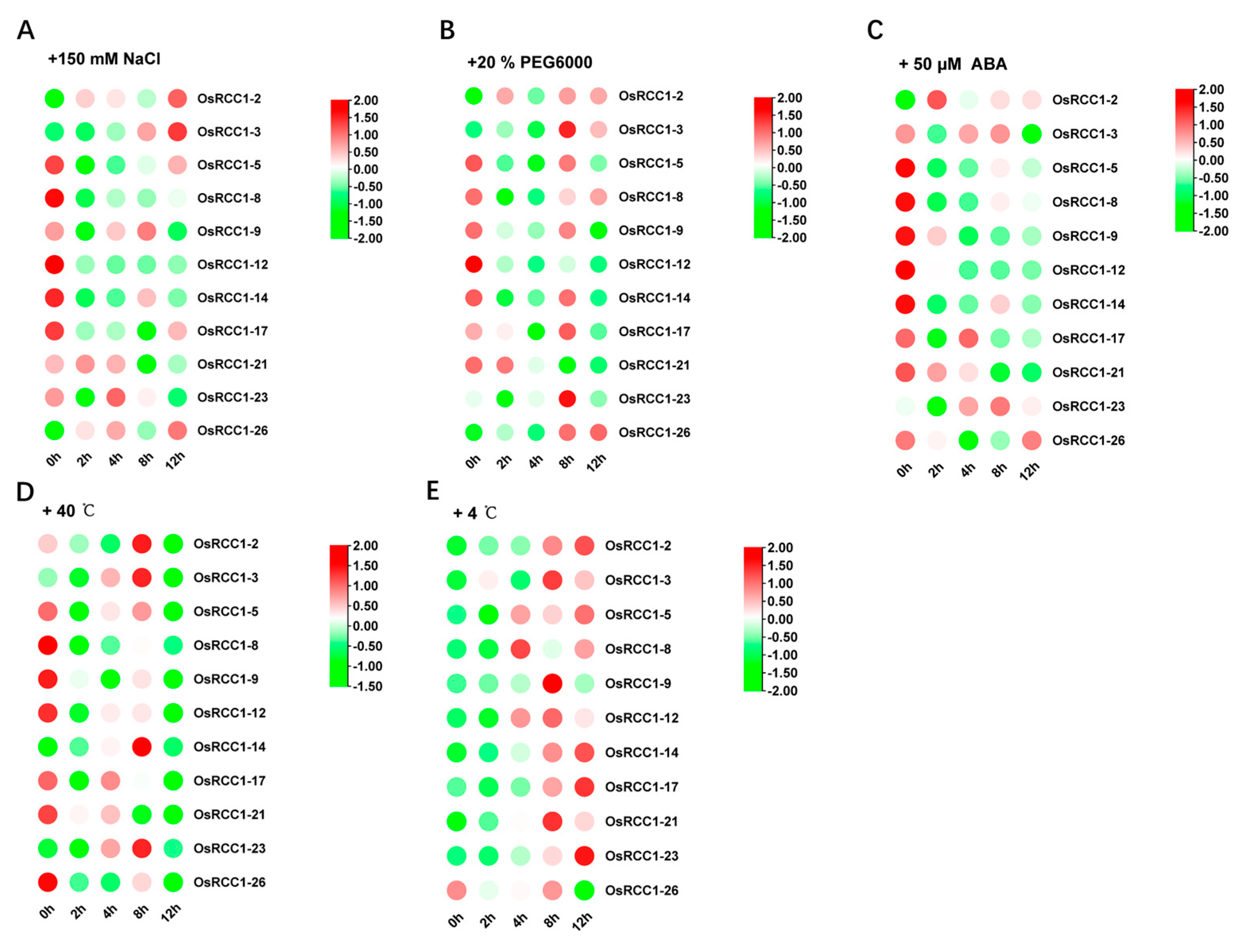
To investigate the potential role of OsRCC1s in response to abiotic stresses, microarray data of 7-day-old rice seedlings (indica variety IR64) under three abiotic stresses (salt, drought and cold) and control conditions for 3 h were obtained from the public database, Rice eFP Browser, and analyzed (OsRCC1-24 was not found). As shown in Figure 7B, all OsRCC1s, except for OsRCC1-24, were differentially expressed under different abiotic stress conditions, indicating that OsRCC1s play a role in the plant response to abiotic stress. The response patterns of RCC1s were different under varying types of stress conditions. For salt and drought stresses, 16 OsRCC1s (OsRCC1-1, OsRCC1-5~OsRCC1-10, OsRCC1-12~OsRCC1-17, OsRCC1-19~OsRCC1-20, OsRCC1-25) were down-regulated under at least one stress factor. Notably, the expression of most OsRCC1s were suppressed by chilling stress. Meanwhile, 2-week-old rice seedlings (Japonica variety Nipponbare) were treated with salt (150 mM NaCl), drought (20% (m/V) PEG6000), heat (40 °C), cold (4 °C) and ABA (50 μM) for 0, 2, 4, 8 and 12 h, respectively. A total of 11 OsRCC1s with higher expression levels in leaves at seedling stage were chosen from each group (according to the results shown in Figure 7A). Expression profiles of these genes in leaves under these five stresses were detected by qRT-PCR. The results showed that all selected OsRCC1s were differentially expressed over time, but with distinct response patterns under different stresses (Figure 8). After salt, drought, ABA and heat treatments, the expression levels of most OsRCC1s were down-regulated compared with the control at most time points. However, the expression of most OsRCC1s was up-regulated by chilling stress, reaching the peak value at 8 or 12 h. It was noted that OsRCC1-2 was significantly induced by all five stresses. All of the above findings pointed toward a potential involvement of OsRCC1s in the response to environmental stresses. Interestingly, the trends in expression of OsRCC1s before and after stresses derived by qRT-PCR were not consistent with the microarray data, which could be attributed to the differences in genetic background, seedling age and stress conditions.
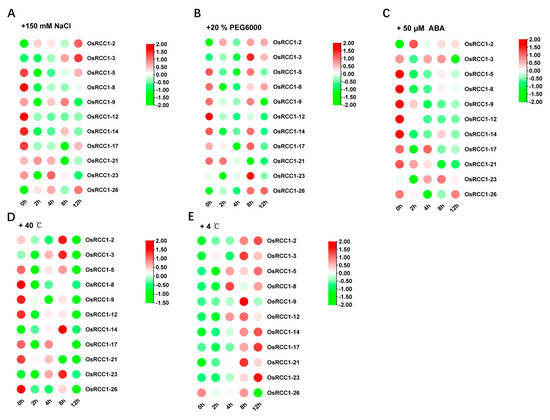
Figure 8.
qRT-PCR based expression analysis of 11 OsRCC1s under abiotic stresses, viz salt (A), drought (B), ABA (C), heat (D) and cold (E) stresses. The red color shows the higher and the green color shows the lower expression levels in expression bar.
4. Discussion
To date, the research on RCC1 gene family in plants is limited to the functional research on a particular RCC1 family member in A. thaliana [10,39], with sporadic reports in other plants like rice [16], wheat [40], maize [9], alfalfa (Medicago truncatula) [41], poplar (Populus euphratica) [6] and Spartina alterniflora [42]. Therefore, it is critical to identify and characterize the OsRCC1 family at a genome-wide level, which would help to elucidate their functions and evolutionary relationships in important crops. In this study, by using bioinformatics and experimental methods, we conducted a comprehensive analysis of OsRCC1s and investigated their potential functions in development and abiotic stress response.
The size of gene families is variable across species, which may be a functional result of adaptation or speciation [43,44]. Herein, the number of RCC1s in rice (26) is more than those identified in cacao trees (21) and grape (21), comparable to those in Arabidopsis (27), but lower than those in maize (35), soybean (55) and upland cotton (56) [45,46]. Hence, it could be inferred that RCC1 genes are more numerous in species with larger genome sizes.
Based on the phylogenetic analysis and structural characteristics, OsRCC1s can be divided into six groups (Group I~VI). The largest group is Group VI, which contains nine members. Most OsRCC1s within the same group are less variable in exon–intron structures, motif and domain distributions. Except for OsRCC1-22 in Group I (no intron), other OsRCC1s are composed of several exons and introns, which is consistent with RCC1s in upland cotton [46]. All the OsRCC1s have more than two motifs associated with the RCC1 domain (motifs 1, 2, 3, 6, 7, 9 and 10). Motif 4 (associated with FYVE zinc finger domain), 5 (associated with BRX domain), 11 (function is unknown) and 12 (associated with BRX N-terminal domain) only exist in Group VI. Motif 8 (function is unknown) exists in one member of Group IV and seven members of Group VI. In addition to RCC1 domains, Group VI also contains FYVE domains, BRX domains and PH domains, which are peculiar to the PRAF subfamily [47,48]. In terms of structural characteristics, Group VI differs significantly from other groups.
In the promoter regions of OsRCC1s, we found numerous CREs related to hormone response, light response, stress regulation and development. The number of light-responsive elements is the largest (309), followed by MeJA-responsive elements (CGTCA-motif/TGACG-motif, 65) and ABA-responsive elements (ABRE, 121). Several studies have shown that UVR8 is involved in the regulation of plant photomorphogenesis [5,49,50,51]. It can be speculated that there may be other photoreceptors in OsRCC1s. ABA and MeJA are crucial phytohormones that have been reported to regulate the stress resistance in rice [52,53]. Thus, OsRCC1s may participate in regulation of rice against environmental stress.
The gene duplication events may help to illuminate the mechanism about the expansion of gene family. Furthermore, gene duplication in plants can facilitate the generation of new genes, which contributes to the diversity of gene functions; thus, the ability of plants to adapt to a variety of adverse survival environments, such as drought, disease and extreme temperature is improved, and their reproductive development is ensured [54,55]. Our results showed that 46.15% (12/26) of OsRCC1s have segmental duplication, but there are no tandem duplication events, indicating that segmental duplication may be the predominant driving force for the expansion of the OsRCC1 gene family. Ka/Ks ratios of segmental duplicated OsRCC1 gene pairs are less than one, so we could infer that the OsRCC1 gene family has undergone strong purification selection during the evolutionary processes. The results of collinearity analysis among different species revealed that OsRCC1s show higher homology with counterparts from monocots, but less homology with dicots, reflecting the functional divergence with dicots. OsRCC1-4 and OsRCC1-18, which belong to Group III, formed the most colinear gene pairs, providing potential clues to the origin and evolution of OsRCC1s.
Numerous studies have previously demonstrated the role of miRNAs in regulating development and stress response in rice [56,57,58]. Mul-miR482a-5p may reduce the resistance of mulberry to biotic stress by inhibiting the expression of RCC1 [59]. In the present study, we identified 252 miRNAs that target OsRCC1s. Many OsRCC1s are targeted by multiple miRNAs, with up to 44 miRNAs targeting OsRCC1-1, suggesting that OsRCC1s may affect transcription regulation by miRNA target, with cleavage as a major function in miRNA–OsRCC1s interaction.
In order to better comprehend the possible functions of OsRCC1s, we examined the transcription patterns of OsRCC1s from various organs/tissues. According to gene microarray data, OsRCC1-1, OsRCC1-10 and OsRCC1-12~OsRCC1-17 are preferentially expressed in SAM and panicles, whereas other OsRCC1s are preferentially expressed in seeds and at least one other organ/tissue. These findings suggest that OsRCC1s may play regulatory roles in rice growth and development, especially in the reproductive developmental stage. Notably, several RCC1s, including MtZR1 [41], AtRUG3 [8], AtRLD [12], AtUVR8 [60] and OsRLR4 (OsRCC1-15) [16], are reported to be closely related to root architectural traits.
Previous studies have shown that RCC1s can function as regulators to mediate stress resistance in plants. For instance, AtTCF1 affects the cold tolerance by regulating lignin homeostasis in the cytoplasm [10]. Ectopic expression of GmTCF1a (homologous to AtTCF1 in soybean) in Arabidopsis confers enhanced chilling tolerance [45]. Overexpression of SaRCC1 in Arabidopsis resulted in a salt-sensitive phenotype, indicating that it negatively modulates salt tolerance [42]. The T-DNA insertion mutant of AtUVR8, as well as the VIGS lines of Gh_A05G3028 and Gh_D10G2310 (homologous to AtTCF1 and AtUVR8 in upland cotton), both exhibited salt-sensitive phenotypes [46]. The expression profiles of OsRCC1s under abiotic stresses were obtained from microarray data, and we also detected the transcript abundance of 11 OsRCC1s selected from each Group (except Group I) in leaves under abiotic stresses by qRT-PCR. qRT-PCR analysis revealed that the transcriptional expression of all 11 OsRCC1s was regulated by salt, drought, ABA, heat and chilling stresses. Most OsRCC1s were inhibited by salt, drought, heat and ABA, but induced by chilling stress. OsRCC1-2 was strongly induced by all five stresses, and OsRCC1-3 was induced by the other four stresses except for the ABA treatment. Furthermore, many stress-related elements (such as MBS, LTR and TC-rich repeats) were observed in the promoter regions of OsRCC1s. The above results reveal that OsRCC1s may be involved in rice response and resistance to abiotic stress.
We also conducted GO enrichment analysis of OsRCC1s using agriGO (Supplementary Figure S3). According to the annotation, there are no significant GO terms in the categories of “biological processes” and “cellular components”. In terms of molecular function, 10 OsRCC1s (OsRCC1-1, OsRCC1-5, OsRCC1-6, OsRCC1-10, OsRCC1-13, OsRCC1-15, OsRCC1-16, OsRCC1-17, OsRCC1-19, OsRCC1-22) are significantly enriched in GO terms related to binding (GO:0005488). Nine genes (other than OsRCC1-22) are involved in metal ion binding (GO:0046872), ion binding (GO:0043167), cation binding (GO:0043169), transition metal ion binding (GO:0046914) and zinc ion binding (GO:0008270) (Supplementary Table S7), which are not uncovered in previous studies. Taken together, our study systematically analyzed the characteristics and expression patterns of OsRCC1s, but the regulatory networks between OsRCC1s and development as well as stress response in rice remain unclear and require further exploration.
5. Conclusions
In this study, 26 OsRCC1s were identified from the rice genome. According to phylogenetic and structural analysis, OsRCC1s can be divided into six groups (Group I–VI). Members within the same group share similar structural characteristics. Gene duplication detection showed that segmental duplication and purifying selection contribute to the amplification and evolution of OsRCC1 gene family. In addition, the synteny analysis among rice, A. thaliana, barley, wheat and tomato showed different degrees of correlation. The expression of OsRCC1s may be influenced by various factors, including hormones, light, stress and miRNAs. OsRCC1s are highly expressed during panicle and seed development, implying that they may be involved in the regulation of reproductive growth in rice. Furthermore, several OsRCC1s exhibited differential expression patterns under different abiotic stresses, suggesting that OsRCC1s may be associated with the response to abiotic stresses. Overall, genome-wide identification and molecular characterization of OsRCC1s could provide novel insights into their evolutionary history, as well as their functional roles.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13030703/s1, Figure S1: Domain distribution of OsRCC1s; Figure S2: Network illustration of predicted miRNA-targeted OsRCC1s; Figure S3: GO analysis of OsRCC1s. (A) GO analysis of OsRCC1s based on molecular functions. (B) Bar chart of enriched GO terms in OsRCC1s based on molecular functions; Table S1: Validation of the RCC1 domains using the HMMER Search tool; Table S2: Primers used in this study; Table S3: Motif sequences of RCC1s in rice; Table S4: Ka/Ks ratios of segmental duplicated OsRCC1 gene pairs; Table S5: List of orthologous proteins of OsRCC1s in Arabidopsis, tomato, barley and wheat; Table S6: Predicted miRNA targeting OsRCC1 mRNAs; Table S7: The GO annotation and enrichment results of OsRCC1s.
Author Contributions
Y.F. and D.X. conceived and designed the research; Q.C., L.K., D.Z., X.Z. (Xian Zhang), Q.T., X.Z. (Xiaoqin Zhang), W.M. and C.D. performed the experiments and data analyses. Q.C., Y.F. and D.X. wrote and revised the manuscript. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Natural Science Foundation of Zhejiang province in China (LY21C130007; LY20C140003; LY19C130001), Hangzhou Scientific and Technological Major Project (202203A01) and Hangzhou Scientific and Technological Project (20201203B107), Inter-government science and technology innovation collaboration project, China national key R&D program (2022YFE0125600) and Hainan Yazhou Bay Seed Laboratory (B21HJ0220).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bischoff, F.R.; Ponstingl, H. Catalysis of guanine nucleotide exchange of Ran by RCC1 and stimulation of hydrolysis of Ran-bound GTP by Ran-GAP1. Methods Enzymol. 1995, 257, 135–144. [Google Scholar] [PubMed]
- Ren, X.; Jiang, K.; Zhang, F. The Multifaceted Roles of RCC1 in Tumorigenesis. Front. Mol. Biosci. 2020, 7, 225. [Google Scholar] [CrossRef] [PubMed]
- Hetzer, M.; Gruss, O.J.; Mattaj, I.W. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 2002, 4, E177–E184. [Google Scholar] [CrossRef] [PubMed]
- Cekan, P.; Hasegawa, K.; Pan, Y.; Tubman, E.; Odde, D.; Chen, J.Q.; Herrmann, M.A.; Kumar, S.; Kalab, P. RCC1-dependent activation of Ran accelerates cell cycle and DNA repair, inhibiting DNA damage-induced cell senescence. Mol. Biol. Cell 2016, 27, 1346–1357. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 Protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef]
- Mao, K.; Wang, L.; Li, Y.Y.; Wu, R. Molecular Cloning and Functional Analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus Euphratica. PLoS ONE 2015, 10, e0132390. [Google Scholar] [CrossRef]
- Kühn, K.; Carrie, C.; Giraud, E.; Wang, Y.; Meyer, E.H.; Narsai, R.; des Francs-Small, C.C.; Zhang, B.; Murcha, M.W.; Whelan, J. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J. 2011, 67, 1067–1080. [Google Scholar] [CrossRef]
- Su, C.; Zhao, H.; Zhao, Y.; Ji, H.; Wang, Y.; Zhi, L.; Li, X. UG3 and ATM synergistically regulate the alternative splicing of mitochondrial nad2 and the DNA damage response in Arabidopsis thaliana. Sci. Rep. 2017, 7, 43897. [Google Scholar] [CrossRef]
- Cao, S.K.; Liu, R.; Sayyed, A.; Sun, F.; Song, R.; Wang, X.; Xiu, Z.; Li, X.; Tan, B.C. Regulator of Chromosome Condensation 1-Domain Protein DEK47 Functions on the Intron Splicing of Mitochondrial Nad2 and Seed Development in Maize. Front. Plant Sci. 2021, 12, 695249. [Google Scholar] [CrossRef]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef]
- Ji, H.; Wang, S.; Cheng, C.; Li, R.; Wang, Z.; Jenkins, G.I.; Kong, F.; Li, X. The RCC1 family protein SAB1 negatively regulates ABI5 through multidimensional mechanisms during postgermination in Arabidopsis. New Phytol. 2019, 222, 907–922. [Google Scholar] [CrossRef]
- Furutani, M.; Hirano, Y.; Nishimura, T.; Nakamura, M.; Taniguchi, M.; Suzuki, K.; Oshida, R.; Kondo, C.; Sun, S.; Kato, K.; et al. Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat. Commun. 2020, 11, 76. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, S.; Lu, J.; Xu, A.; Huang, Y.; Bie, Z.; Cheng, F. CmRCC1 Gene From Pumpkin Confers Cold Tolerance in Tobacco by Modulating Root Architecture and Photosynthetic Activity. Front. Plant Sci. 2021, 12, 765302. [Google Scholar] [CrossRef] [PubMed]
- Duarte, G.T.; Pandey, P.K.; Vaid, N.; Alseekh, S.; Fernie, A.R.; Nikoloski, Z.; Laitinen, R.A.E. Plasticity of rosette size in response to nitrogen availability is controlled by an RCC1-family protein. Plant Cell Environ. 2021, 44, 3398–3411. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Cloix, C.; Jiang, G.H.; Kaiserli, E.; Herzyk, P.; Kliebenstein, D.J.; Jenkins, G.I. A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 2005, 102, 18225–18230. [Google Scholar] [CrossRef]
- Sun, C.; Li, D.; Gao, Z.; Gao, L.; Shang, L.; Wang, M.; Qiao, J.; Ding, S.; Li, C.; Geisler, M.; et al. OsRLR4 binds to the OsAUX1 promoter to negatively regulate primary root development in rice. J. Integr. Plant Biol. 2022, 64, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015, 15, 261. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pereira, A. Plant Abiotic Stress Challenges from the Changing Environment. Front. Plant Sci. 2016, 7, 1123. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Mandal, K.; Boro, P.; Chattopadhyay, S. Micro-RNA Based Gene Regulation: A Potential Way for Crop Improvements. Plant Gene 2021, 27, 100312. [Google Scholar] [CrossRef]
- Su, C.; Yuan, J.; Zhao, H.; Zhao, Y.; Ji, H.; Wang, Y.; Li, X. RUG3 is a negative regulator of plant responses to ABA in Arabidopsis thaliana. Plant Signal. Behav. 2017, 12, e1333217. [Google Scholar] [CrossRef]
- Duan, X.L.; Chen, H.Z.; Han, R. The Effects of Enhanced UV-B Radiation on the RCC1 in Wheat Somatic Cells. Russ. J. Plant Physiol. 2015, 62, 695–699. [Google Scholar] [CrossRef]
- Hopkins, J.; Pierre, O.; Kazmierczak, T.; Gruber, V.; Frugier, F.; Clement, M.; Frendo, P.; Herouart, D.; Boncompagni, E. MtZR1, a PRAF protein, is involved in the development of roots and symbiotic root nodules in Medicago truncatula. Plant Cell Environ. 2014, 37, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wen, J.; Song, Y.; Yuan, H.; Sun, B.; Wang, R.; Xu, S. SaRCC1, a Regulator of Chromosome Condensation 1 (RCC1) Family Protein Gene from Spartina alterniflora, Negatively Regulates Salinity Stress Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 8172. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Rooney, A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005, 39, 121–152. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Wang, H.; Li, X.; Ji, H. Enhancement of plant cold tolerance by soybean RCC1 family gene GmTCF1a. BMC Plant Biol. 2021, 21, 369. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, X.; Sun, C.; Rong, J. Identification and Expression Profiling of the Regulator of Chromosome Condensation 1 (RCC1) Gene Family in Gossypium Hirsutum L. under Abiotic Stress and Hormone Treatments. Int. J. Mol. Sci. 2019, 20, 1727. [Google Scholar] [CrossRef]
- Wywial, E.; Singh, S.M. Identification and structural characterization of FYVE domain-containing proteins of Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 157. [Google Scholar] [CrossRef]
- Xiao, S.; Shao, M.; Wang, D.; Li, W.; Liu, F. Identification and Evolution of FYVE Domain-Containing Proteins and Their Expression Patterns in Response to Abiotic Stresses in Rice. Plant Mol. Biol. Rep. 2016, 34, 1064–1082. [Google Scholar] [CrossRef]
- Carranco, R.; Prieto-Dapena, P.; Almoguera, C.; Jordano, J. A seed-specific transcription factor, HSFA9, anticipates UV-B light responses by mimicking the activation of the UV-B receptor in tobacco. Plant J. 2022, 111, 1439–1452. [Google Scholar] [CrossRef]
- Podolec, R.; Lau, K.; Wagnon, T.B.; Hothorn, M.; Ulm, R. A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2017284118. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Guan, Z.; Chang, H.; Ma, L.; Shen, C.; Qiu, L.; Yan, J.; Zhang, D.; Li, J.; et al. Structural insight into UV-B-activated UVR8 bound to COP1. Sci. Adv. 2022, 8, eabn3337. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Xu, H.; Su, C.; Wang, X.; Wang, L. Rice CIRCADIAN CLOCK ASSOCIATED 1 transcriptionally regulates ABA signaling to confer multiple abiotic stress tolerance. Plant Physiol. 2022, 190, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Mukherjee, P.; Dutta, S.; Meena, K.; Sarkar, S.K.; Mandal, A.B.; Dasgupta, T.; Mitra, J. Genomic insights into HSFs as candidate genes for high-temperature stress adaptation and gene editing with minimal off-target effects in flax. Sci. Rep. 2019, 9, 5581. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Sun, W.; Xu, X.H.; Li, Y.; Xie, L.; He, Y.; Li, W.; Lu, X.; Sun, H.; Xie, X. OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice. New Phytol. 2020, 226, 823–837. [Google Scholar] [CrossRef]
- Sun, M.; Shen, Y.; Chen, Y.; Wang, Y.; Cai, X.; Yang, J.; Jia, B.; Dong, W.; Chen, X.; Sun, X. Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 2022, 189, 2500–2516. [Google Scholar] [CrossRef]
- Yang, Z.; Hui, S.; Lv, Y.; Zhang, M.; Chen, D.; Tian, J.; Zhang, H.; Liu, H.; Cao, J.; Xie, W.; et al. miR395-regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice. Mol. Plant 2022, 15, 671–688. [Google Scholar] [CrossRef]
- Gai, Y.P.; Zhao, H.N.; Zhao, Y.N.; Zhu, B.S.; Yuan, S.S.; Li, S.; Guo, F.Y.; Ji, X.L. MiRNA-seq-based profiles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2018, 8, 812. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Chen, P.; Liang, T.; Li, X.; Liu, H. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J. 2020, 39, e101928. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).