Comparative Transcriptome Analysis Identified Potential Genes and Transcription Factors for Flower Coloration in Kenaf (Hibiscus cannabinus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Biochemical Analysis
2.2. RNA Extraction and cDNA Library Construction
2.3. De Novo Assembly and Annotation
2.4. Differentially Expressed Genes (DEG) and Functional Enrichment Analysis
2.5. Quantitative Real-Time PCR Analysis
3. Results
3.1. Composition of the Phenolic Compounds in Three Kenaf Flowers
3.2. Transcriptome Analysis
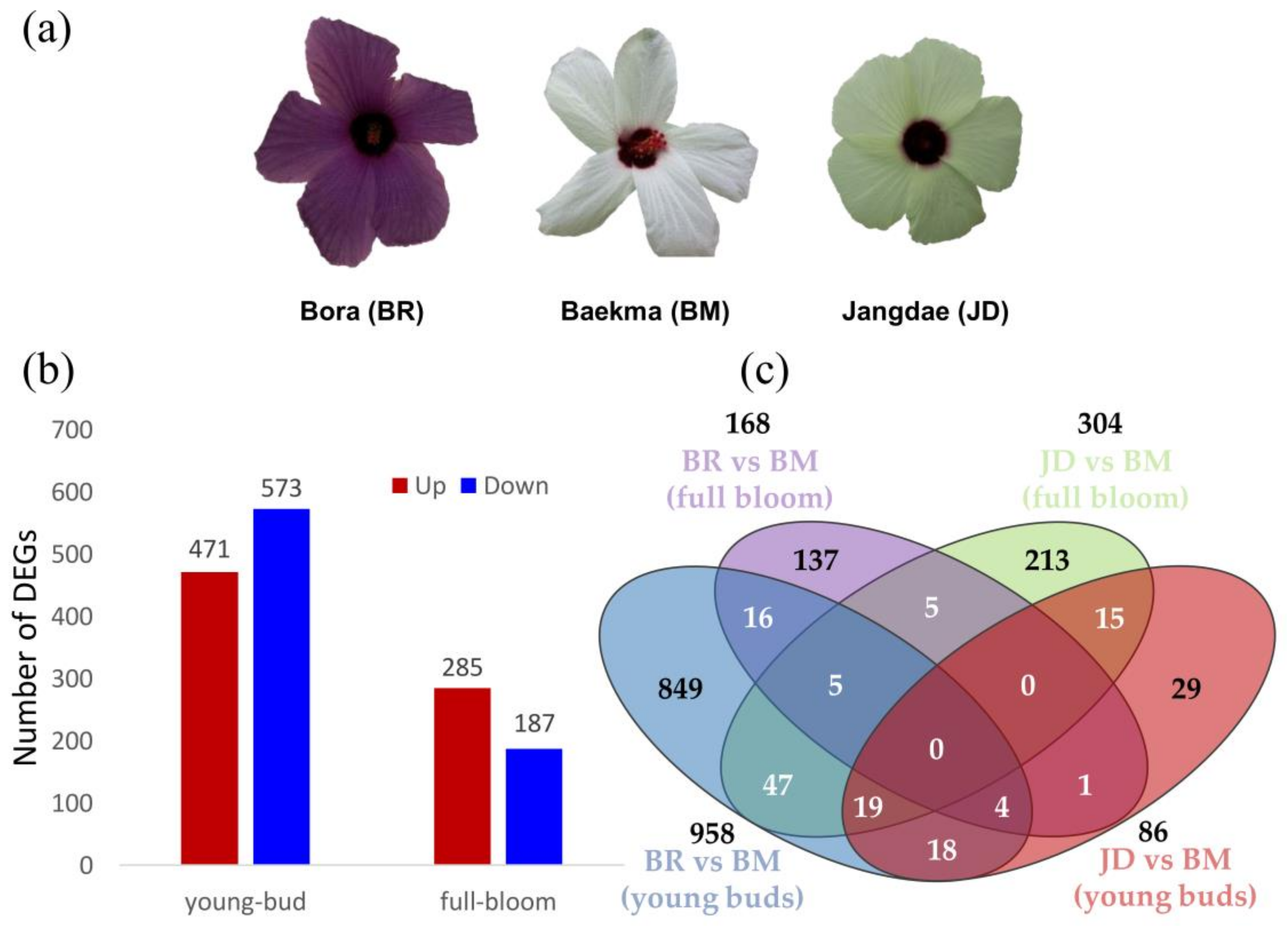
3.3. Comparison of DEGs among the Three Different Kenaf Flowers
3.4. Identification of Putative Genes Involved in the Phenylpropanoid Pathway
3.5. Validation of RNA-seq Data Using qRT-PCR
4. Discussion
4.1. Levels of Phenolic Compounds in Kenaf Flowers
4.2. Transcriptome Analysis of Different Kenaf Flowers
4.3. Putative Candidate Genes Involved in the Phenylpropanoid Pathway
4.4. Key Regulators of Differentially Expressed Genes during Flower Coloration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Alexopoulou, E.; Papatheohari, Y.; Christou, M.; Monti, A. Origin, Description, Importance, and Cultivation Area of Kenaf. In Kenaf: A Multi-Purpose Crop for Several Industrial Applications; New insights from the Biokenaf Project; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–15. [Google Scholar] [CrossRef]
- Agbor, G.A.; Oben, J.E.; Nkegoum, B.; Takala, J.P.; Ngogang, J.Y. Hepatoprotective activity of Hibiscus cannabinus (Linn.) against carbon tetrachloride and paracetamol induced liver damage in rats. Pak. J. Biol. Sci. 2005, 8, 1397–1401. [Google Scholar]
- Yazan, L.S.; Foo, J.B.; Chan, K.W.; Tahir, P.M.; Ismail, M. Kenaf seed oil from supercritical carbon dioxide fluid extraction shows cytotoxic effects towards various cancer cell lines. Afr. J. Biotechnol. 2011, 10, 5381–5388. [Google Scholar]
- Yusri, N.M.; Chan, K.W.; Iqbal, S.; Ismail, M. Phenolic content and antioxidant activity of Hibiscus cannabinus L. seed extracts after sequential solvent extraction. Molecules 2012, 17, 12612–12621. [Google Scholar] [CrossRef] [Green Version]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, M.E.K.; Choi, B.; Cho, B.-K.; Kim, J.B.; Park, S.U.; Natarajan, S.; Lim, H.-S.; Bae, H. Regulation of 4CL, encoding 4-coumarate: Coenzyme A ligase, expression in kenaf under diverse stress conditions. Plant Omics 2013, 6, 254–262. [Google Scholar]
- Jeong, M.-J.; Choi, B.S.; Bae, D.W.; Shin, S.C.; Park, S.U.; Lim, H.-S.; Kim, J.; Kim, J.B.; Cho, B.-K.; Bae, H. Differential expression of kenaf phenylalanine ammonia-lyase (PAL) ortholog during developmental stages and in response to abiotic stresses. Plant Omics 2012, 5, 392–399. [Google Scholar]
- Kim, J.; Choi, B.; Natarajan, S.; Bae, H. Expression analysis of kenaf cinnamate 4-hydroxylase (C4H) ortholog during developmental and stress responses. Plant Omics 2013, 6, 65–72. [Google Scholar]
- Zhao, S.; Li, X.; Cho, D.H.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.U. Accumulation of Kaempferitrin and Expression of Phenyl-Propanoid Biosynthetic Genes in Kenaf (Hibiscus cannabinus). Molecules 2014, 19, 16987–16997. [Google Scholar] [CrossRef] [Green Version]
- Lyu, J.I.; Choi, H.-I.; Ryu, J.; Kwon, S.-J.; Jo, Y.D.; Hong, M.J.; Kim, J.-B.; Ahn, J.-W.; Kang, S.-Y. Transcriptome Analysis and Identification of Genes Related to Biosynthesis of Anthocyanins and Kaempferitrin in Kenaf (Hibiscus cannabinus L.). J. Plant Biol. 2020, 63, 51–62. [Google Scholar] [CrossRef]
- Lyu, J.I.; Ramekar, R.; Kim, D.-G.; Kim, J.M.; Lee, M.-K.; Hung, N.N.; Kim, J.-B.; Ahn, J.-W.; Kang, S.-Y.; Choi, I.-Y. Characterization of gene isoforms related to cellulose and lignin biosynthesis in Kenaf (Hibiscus cannabinus L.) mutant. Plants 2020, 9, 631. [Google Scholar] [CrossRef]
- Li, H.; Li, D.; Chen, A.; Tang, H.; Li, J.; Huang, S. RNA-seq for comparative transcript profiling of kenaf under salinity stress. J. Plant Res. 2017, 130, 365–372. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Deng, Y.; Zhang, G.; Li, J.; Xiao, A.; Zhao, L.; Chen, A.; Tang, H.; Chang, L.; Pan, G.; et al. Comparative Transcriptome and Proteome Analysis Provides New Insights Into the Mechanism of Protein Synthesis in Kenaf (Hibiscus cannabinus L.) Leaves. Front. Plant Sci. 2022, 13, 879874. [Google Scholar] [CrossRef]
- Chen, P.; Chen, T.; Li, Z.; Jia, R.; Luo, D.; Tang, M.; Lu, H.; Hu, Y.; Yue, J.; Huang, Z. Transcriptome analysis revealed key genes and pathways related to cadmium-stress tolerance in Kenaf (Hibiscus cannabinus L.). Ind. Crops Prod. 2020, 158, 112970. [Google Scholar] [CrossRef]
- Chen, P.; Li, Z.; Luo, D.; Jia, R.; Lu, H.; Tang, M.; Hu, Y.; Yue, J.; Huang, Z. Comparative transcriptomic analysis reveals key genes and pathways in two different cadmium tolerance kenaf (Hibiscus cannabinus L.) cultivars. Chemosphere 2021, 263, 128211. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 1. [Google Scholar] [CrossRef] [Green Version]
- Lucas, A. Another Multidimensional Analysis Package. 2006. Available online: http://rdrr.io/cran/amap/ (accessed on 23 February 2023).
- Niu, X.; Chen, M.; Huang, X.; Chen, H.; Tao, A.; Xu, J.; Qi, J. Reference gene selection for qRT-PCR normalization analysis in kenaf (Hibiscus cannabinus L.) under abiotic stress and hormonal stimuli. Front. Plant Sci. 2017, 8, 771. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Badal, S.; Delgoda, R. Pharmacognosy; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Ryu, J.; Kwon, S.-J.; Ahn, J.-W.; Jo, Y.D.; Kim, S.H.; Jeong, S.W.; Lee, M.K.; Kim, J.-B.; Kang, S.-Y. Phytochemicals and antioxidant activity in the kenaf plant (Hibiscus cannabinus L.). J. Plant Biotechnol. 2017, 44, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Park, C.H.; Kwon, S.-J.; Kim, N.S.; Baek, S.-A.; Yeo, H.J.; Park, Y.E.; Chung, Y.S.; Kim, J.K.; Park, S.U. Metabolic analysis of carotenoids and phenolic compounds found in green and purple kenaf. Nat. Prod. Commun. 2020, 15, 1934578X20971138. [Google Scholar] [CrossRef]
- Hansawasdi, C.; Kawabata, J.; Kasai, T. α-Amylase Inhibitors from Roselle (Hibiscus sabdariffa Linn.) Tea. Biosci. Biotechnol. Biochem. 2000, 64, 1041–1043. [Google Scholar] [CrossRef]
- Zheoat, A.M.; Gray, A.I.; Igoli, J.O.; Ferro, V.A.; Drummond, R.M. Hibiscus acid from Hibiscus sabdariffa (Malvaceae) has a vasorelaxant effect on the rat aorta. Fitoterapia 2019, 134, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Portillo-Torres, L.A.; Bernardino-Nicanor, A.; Gómez-Aldapa, C.A.; González-Montiel, S.; Rangel-Vargas, E.; Villagómez-Ibarra, J.R.; González-Cruz, L.; Cortés-López, H.; Castro-Rosas, J. Hibiscus Acid and Chromatographic Fractions from Hibiscus Sabdariffa Calyces: Antimicrobial Activity against Multidrug-Resistant Pathogenic Bacteria. Antibiotics 2019, 8, 218. [Google Scholar]
- Jeong, S.W.; Kwon, S.-J.; Ryu, J.; Kim, J.-B.; Ahn, J.-W.; Kim, S.H.; Jo, Y.D.; Choi, H.-I.; Im, S.B.; Kang, S.-Y. Development of EST-SSR markers through de novo RNA sequencing and application for biomass productivity in kenaf (Hibiscus cannabinus L.). Genes Genom. 2017, 39, 1139–1156. [Google Scholar] [CrossRef]
- Kim, J.M.; Lyu, J.I.; Lee, M.-K.; Kim, D.-G.; Kim, J.-B.; Ha, B.-K.; Ahn, J.-W.; Kwon, S.-J. Cross-species transferability of EST-SSR markers derived from the transcriptome of kenaf (Hibiscus cannabinus L.) and their application to genus Hibiscus. Genet. Resour. Crop Evol. 2019, 66, 1543–1556. [Google Scholar]
- Li, H.; Li, D.; Chen, A.; Tang, H.; Li, J.; Huang, S. Characterization of the Kenaf (Hibiscus cannabinus) Global Transcriptome Using Illumina Paired-End Sequencing and Development of EST-SSR Markers. PLoS ONE 2016, 11, e0150548. [Google Scholar] [CrossRef]
- Hui, L.; Li, C.; Huijuan, T.; Mingbao, L.; Gen, P.; Siqi, H.; Hui, J.; Yena, W.; Anguo, C. High-resolution genetic map construction and QTL analysis of important fiber traits in kenaf using RAD-seq. S. Afr. J. Bot. 2023, 153, 46–52. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.; Zhang, X.; Ma, X.; Zhang, L.; Liao, Z.; Zhang, Q.; Wan, X.; Cheng, Y.; Zhang, J. The genome of kenaf (Hibiscus cannabinus L.) provides insights into bast fibre and leaf shape biogenesis. Plant Biotechnol. J. 2020, 18, 1796–1809. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.; Li, Z.; Luo, D.; Wei, F.; Kashif, M.H.; Lu, H.; Hu, Y.; Yue, J.; Huang, Z.; Tan, W. A comprehensive integrated transcriptome and metabolome analyses to reveal key genes and essential metabolic pathways involved in CMS in kenaf. Plant Cell Rep. 2021, 40, 223–236. [Google Scholar] [CrossRef]
- An, X.; Chen, J.; Jin, G. Transcriptome profiling of kenaf (Hibiscus cannabinus L.) under plumbic stress conditions implies the involvement of NAC transcription factors regulating reactive oxygen species-dependent programmed cell death. PeerJ 2020, 8, e8733. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Smith, S.A. Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genom. 2013, 14, 328. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Gong, Y.; Jin, S.; Zhu, Q. Molecular analysis of a UDP-glucose: Flavonoid 3-O-glucosyltransferase (UFGT) gene from purple potato (Solanum tuberosum). Mol. Biol. Rep. 2011, 38, 561–567. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Lin, R.-C.; Rausher, M.D. R2R3-MYB genes control petal pigmentation patterning in Clarkia gracilis ssp. sonomensis (Onagraceae). New Phytol. 2021, 229, 1147–1162. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]



| No. | Retention Time (min) | Identification | Molecular Formula | Negative ion Mass | Ref. | |
|---|---|---|---|---|---|---|
| 1 | 3.9 | Hibiscus acid | C6H5O7 | 207 | [1,2] | |
| 2 | 7.8 | Delphinidin 3-sambubioside (Hibiscin) | C26H27O16 | 595 | [1,2] | |
| 3 | 8.2 | Delphinidin 3-galactoside | C21H19O12 | 463 | [1,2] | |
| 4 | 12.9 | Quercetin 3-glucoside | C21H19O12 | 463 | [2] | |
| TPC 1 | TAC 2 | Hibiscus acid | D3S 3 | D3G 4 | Q3G 5 | |
| Baekma | 63.14 ± 2.95 | 12.60 ± 0.32 | 8.99 ± 0.78 | 8.32 ± 1.58 | 0.00 | 1.29 ± 0.13 |
| Jangdae | 96.81 ± 2.45 ** | 19.90 ± 1.36 ** | 6.13 ± 0.42 ** | 9.75 ± 1.99 | 0.00 | 54.10 ± 0.51 ** |
| Bora | 132.98 ± 12.56 ** | 48.85 ± 6.20 ** | 12.29 ± 0.06 ** | 16.22 ± 1.48 ** | 14.65 ± 2.34 | 50.90 ± 0.54 ** |
| Data | Num. of Transcripts | Length (bp) of Transcripts | Functional Annotation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sum. of bp | Min | Max | Average | N50 | NR Viridiplantae | Interproscan | Total Annotation | ||
| Total transcripts | 81,982 | 127,443,212 | 500 | 15,841 | 1554 | 1882 | 73,666 (89.86%) | 58,963 (71.92%) | 73,808 (90.03%) |
| Representative transcripts | 38,601 | 52,149,684 | 500 | 15,841 | 1350 | 1643 | 32,960 (85.39%) | 25,638 (66.42%) | 33,057 (85.64%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, J.I.; Ryu, J.; Kim, D.-G.; Kim, J.M.; Ahn, J.-W.; Kwon, S.-J.; Kim, S.H.; Kang, S.-Y. Comparative Transcriptome Analysis Identified Potential Genes and Transcription Factors for Flower Coloration in Kenaf (Hibiscus cannabinus L.). Agronomy 2023, 13, 715. https://doi.org/10.3390/agronomy13030715
Lyu JI, Ryu J, Kim D-G, Kim JM, Ahn J-W, Kwon S-J, Kim SH, Kang S-Y. Comparative Transcriptome Analysis Identified Potential Genes and Transcription Factors for Flower Coloration in Kenaf (Hibiscus cannabinus L.). Agronomy. 2023; 13(3):715. https://doi.org/10.3390/agronomy13030715
Chicago/Turabian StyleLyu, Jae Il, Jaihyunk Ryu, Dong-Gun Kim, Jung Min Kim, Joon-Woo Ahn, Soon-Jae Kwon, Sang Hoon Kim, and Si-Yong Kang. 2023. "Comparative Transcriptome Analysis Identified Potential Genes and Transcription Factors for Flower Coloration in Kenaf (Hibiscus cannabinus L.)" Agronomy 13, no. 3: 715. https://doi.org/10.3390/agronomy13030715
APA StyleLyu, J. I., Ryu, J., Kim, D. -G., Kim, J. M., Ahn, J. -W., Kwon, S. -J., Kim, S. H., & Kang, S. -Y. (2023). Comparative Transcriptome Analysis Identified Potential Genes and Transcription Factors for Flower Coloration in Kenaf (Hibiscus cannabinus L.). Agronomy, 13(3), 715. https://doi.org/10.3390/agronomy13030715







