Improved Forage Quality in Alfalfa (Medicago sativa L.) via Selection for Increased Stem Fiber Digestibility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Development of Selected Populations
2.2. Evaluation of Selected Populations
2.3. Statistical Analyses
2.4. Environmental Stability Analysis
3. Results
3.1. Response to Selection for IVNDFD
3.2. Genetic Gain by Cycle for IVNDFD and Fiber Digestibility
3.3. Heritability and Genetic Gain of IVNDFD and Detergent Fiber Components
3.4. Effects of Plant Maturity and Harvest on IVNDFD
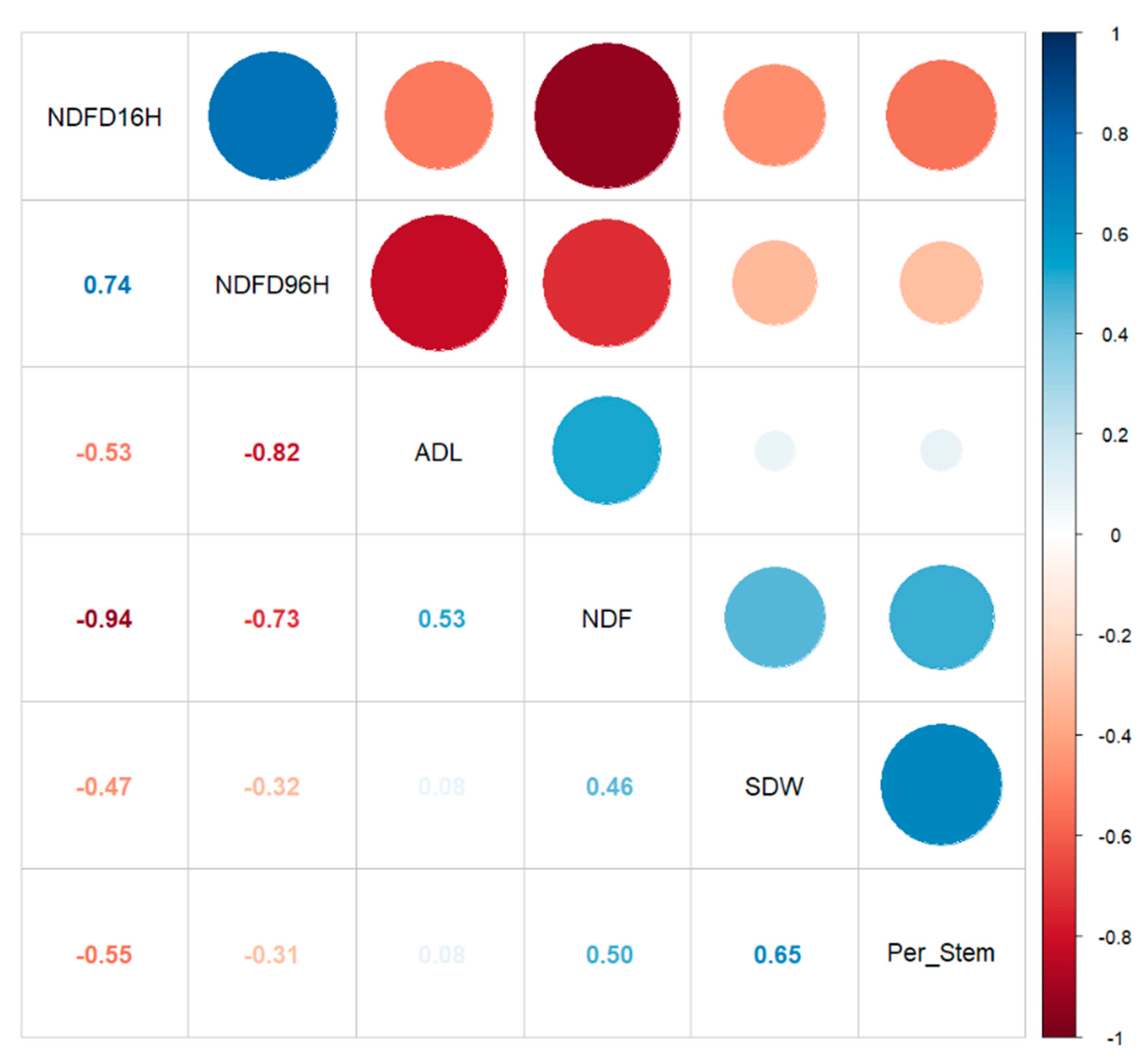
3.5. Additive Gene Effects May Control Alfalfa Stem Digestibility
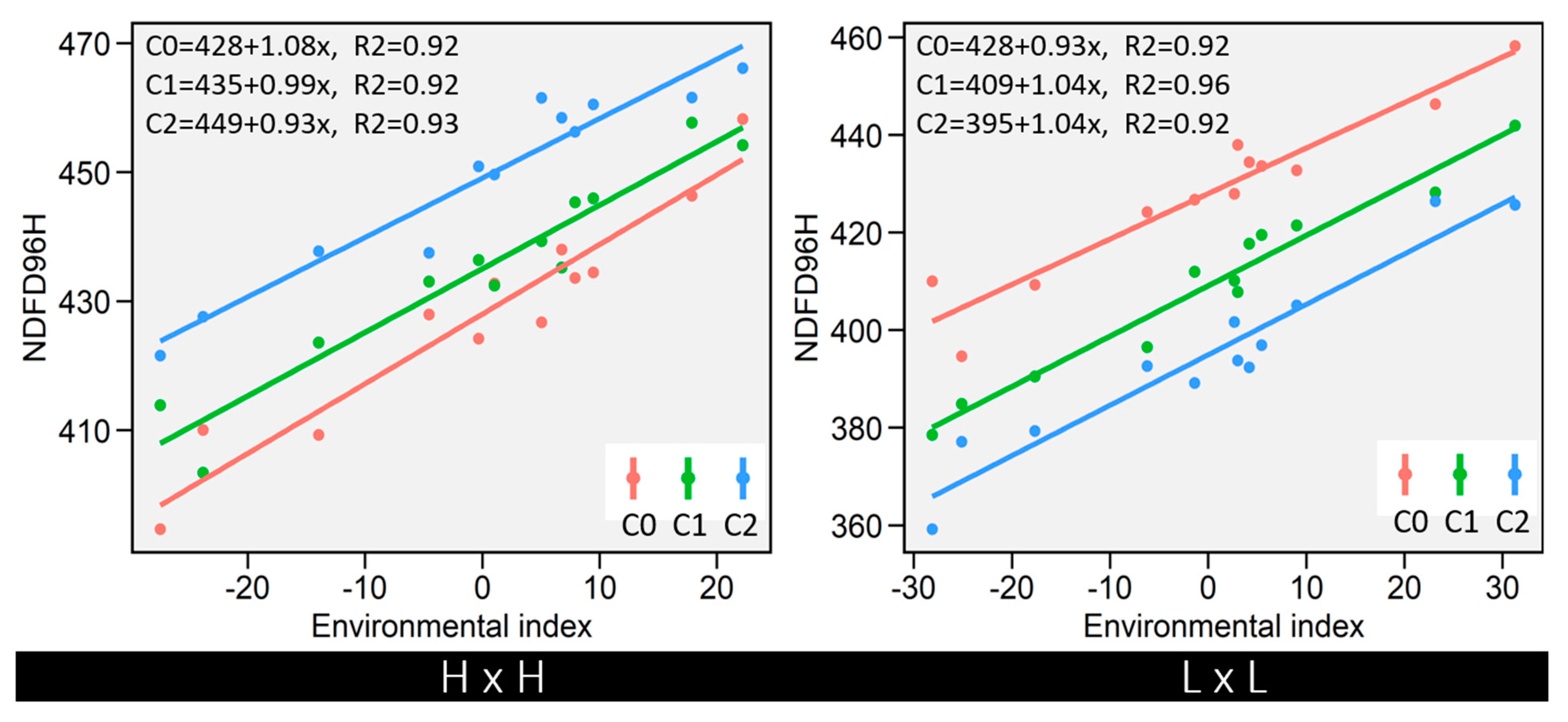
3.6. Stability of IVNDFD with Cycles of Selection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
EOE Statement
References
- Putnam, D.; Meccage, E. Profitable Alfalfa Production Sustains the Environment. Available online: https://calhaysymposium.com/wp-content/uploads/2022/11/PROFITABLE-ALFALFA-PRODUCTION-SUSTAINS-THE-ENVIRONMENT-Dan-Putnam.pdf (accessed on 2 March 2023).
- Fernandez, A.; Sheaffer, C.; Tautges, N.; Putnam, D.; Hunter, M. Alfalfa, Wildlife & the Environment; National Alfalfa and Forage Alliance: St. Paul, MN, USA, 2019. [Google Scholar]
- De Ondarza, M.; Tricarico, J. Advantages and limitations of dairy efficiency measures and the effects of nutrition and feeding management interventions. Prof. Anim. Sci. 2017, 33, 393–400. [Google Scholar] [CrossRef]
- Jung, H.G.; Allen, M.S. Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J. Anim. Sci. 1995, 73, 2774–2790. [Google Scholar] [CrossRef] [PubMed]
- Oba, M.; Allen, M. Evaluation of the importance of the digestibility of neutral detergent fiber from forage: Effects on dry matter intake and milk yield of dairy cows. J. Dairy Sci. 1999, 82, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Wilman, D.; Altimimi, M.A. The in-vitro digestibility and chemical composition of plant parts in white clover, red clover and lucerne during primary growth. J. Sci. Food Agric. 1984, 35, 133–138. [Google Scholar] [CrossRef]
- Engels, F.; Jung, H. Alfalfa stem tissues: Cell-wall development and lignification. Ann. Bot. 1998, 82, 561–568. [Google Scholar] [CrossRef] [Green Version]
- Sheaffer, C.C.; Martin, N.P.; Lamb, J.F.; Cuomo, G.R.; Jewett, J.G.; Quering, S.R. Leaf and stem properties of alfalfa entries. Agron. J. 2000, 92, 733–739. [Google Scholar] [CrossRef]
- Hatfield, R.; Ralph, J.; Grabber, J. Cell wall structural foundations: Molecular basis for improving forage digestibilities. Crop Sci. 1999, 39, 27–37. [Google Scholar] [CrossRef]
- Lamb, J.F.S.; Jung, H.J.G.; Samac, D.A. Environmental Variability and/or Stability of Stem Fiber Content and Digestibility in Alfalfa. Crop Sci. 2014, 54, 2854–2863. [Google Scholar] [CrossRef]
- Buxton, D.R.; Redfearn, D.D. Plant limitations to fiber digestion and utilization. J. Nutr. 1997, 127, 814S–818S. [Google Scholar] [CrossRef] [Green Version]
- Buxton, D.; Marten, G.; Hornstein, J. Genetic variation for forage quality of alfalfa stems. Can. J. Plant Sci. 1987, 67, 1057–1067. [Google Scholar] [CrossRef]
- Buxton, D.R. Quality-related characteristics of forages as influenced by plant environment and agronomic factors. Anim. Feed Sci. Technol. 1996, 59, 37–49. [Google Scholar] [CrossRef]
- Albrecht, K.A.; Wedin, W.F.; Buxton, D.R. Cell-wall composition and digestibility of alfalfa stems and leaves. Crop Sci. 1987, 27, 735–741. [Google Scholar] [CrossRef]
- Wang, Z.; Qiang, H.; Zhao, H.; Xu, R.; Zhang, Z.; Gao, H.; Wang, X.; Liu, G.; Zhang, Y. Association Mapping for Fiber-Related Traits and Digestibility in Alfalfa (Medicago sativa). Front. Plant Sci. 2016, 7, 331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.; Sheaffer, C.; Barnes, D.; Halgerson, J. Forage quality variation in the US alfalfa core collection. Crop Sci. 1997, 37, 1361–1366. [Google Scholar] [CrossRef]
- Bertrand, A.; Claessens, A.; Thivierge, M.-N.; Rocher, S.; Lajeunesse, J.; Castonguay, Y.; Seguin, P. Field Assessment of Alfalfa Populations Recurrently Selected for Stem Cell Wall Digestibility. Crop Sci. 2018, 58, 1632–1643. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.G.; Lamb, J.F.S. Stem Morphological and Cell Wall Traits Associated with Divergent In Vitro Neutral Detergent Fiber Digestibility in Alfalfa Clones. Crop Sci. 2006, 46, 2054–2061. [Google Scholar] [CrossRef] [Green Version]
- Laboski, C.; Lamb, J.; Dowdy, R.; Baker, J.; Wright, J. Irrigation scheduling for a sandy soil using mobile frequency domain reflectometry with a checkbook method. J. Soil Water Conserv. 2001, 56, 97–100. [Google Scholar]
- Kaiser, D.E.; Lamb, J.A.; Eliason, R. Fertilizer Guidelines for Agronomic Crops in Minnesota. Available online: https://conservancy.umn.edu/bitstream/handle/11299/198924/Fertilizer%20Guidelines%20for%20Agronomic%20Crops%20in%20Minnesota.pdf?sequence=1 (accessed on 2 September 2022).
- Van Soest, P.v.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Jung, H.-J.G.; Lamb, J.F. Identification of lucerne stem cell wall traits related to in vitro neutral detergent fibre digestibility. Anim. Feed Sci. Technol. 2003, 110, 17–29. [Google Scholar] [CrossRef]
- McDougall, E. Studies on ruminant saliva. 1. The composition and output of sheep’s saliva. Biochem. J. 1948, 43, 99. [Google Scholar] [CrossRef] [Green Version]
- Vogel, K.P.; Pedersen, J.F.; Masterson, S.D.; Toy, J.J. Evaluation of a filter bag system for NDF, ADF, and IVDMD forage analysis. Crop Sci. 1999, 39, 276–279. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Mendiburu, F.D.; Yaseen, M. Agricolae: Statistical procedures for agricultural research. R package version 1.4. 0. 2020. Available online: https://cran.r-project.org/web/packages/agricolae/index.html (accessed on 2 March 2023).
- Wei, T.; Simko, V.; Levy, M. Package “Corrplot”: Visualization of a Correlation Matrix. 2017. Version 0.84 2021., R package, online resources. Available online: https://cran.microsoft.com/snapshot/2016-08-01/web/packages/corrplot/vignettes/corrplot-intro.html#:~:text=The%20corrplot%20package%20is%20a%20graphical%20display%20of,choosing%20color%2C%20text%20labels%2C%20color%20labels%2C%20layout%2C%20etc. (accessed on 2 March 2023).
- Warton, D.I.; Duursma, R.A.; Falster, D.S.; Taskinen, S. smatr 3– an R package for estimation and inference about allometric lines. Methods Ecol. Evol. 2012, 3, 257–259. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics, 4th ed.; Longman: Harlow, UK, 1996. [Google Scholar]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Kassambara, A.; Kassambara, M.A. Package ‘Ggpubr’. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 31 January 2023).
- Olivoto, T.; Lúcio, A.D.C. metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef] [Green Version]
- Kephart, K.D.; Buxton, D.R.; Hill, R.R., Jr. Digestibility and cell-wall components of alfalfa following selection for divergent herbage lignin concentration. Crop Sci. 1990, 30, 207–212. [Google Scholar] [CrossRef]
- Kephart, K.D.; Buxton, D.; Hill, R., Jr. Morphology of alfalfa divergently selected for herbage lignin concentration. Crop Sci. 1989, 29, 778–782. [Google Scholar] [CrossRef]
- Damiran, D.; Biligetu, B.; Pearce, L.; Lardner, H. PSXI-15 Evaluation of low-lignin alfalfa ‘Hi-Gest® 360′ on the Canadian prairies: Productivity, nutrient profile, and rumen degradation kinetics. J. Anim. Sci. 2021, 99, 348. [Google Scholar] [CrossRef]
- Jungers, J.; Cherney, J.; Martinson, K.; Jaqueth, A.; Sheaffer, C. Forage nutritive value of modern alfalfa cultivars. Crop Forage Turfgrass Manag. 2020, 6, e20076. [Google Scholar] [CrossRef]
- Grev, A.M.; Wells, M.S.; Samac, D.A.; Martinson, K.L.; Sheaffer, C.C. Forage Accumulation and Nutritive Value of Reduced Lignin and Reference Alfalfa Cultivars. Agron. J. 2017, 109, 2749–2761. [Google Scholar] [CrossRef] [Green Version]
- Grev, A.M.; Wells, M.S.; Catalano, D.N.; Martinson, K.L.; Jungers, J.M.; Sheaffer, C.C. Stem and leaf forage nutritive value and morphology of reduced lignin alfalfa. Agron. J. 2020, 112, 406–417. [Google Scholar] [CrossRef]
- Arnold, A.M.; Cassida, K.A.; Albrecht, K.A.; Hall, M.H.; Min, D.; Xu, X.; Orloff, S.; Undersander, D.J.; Santen, E.; Sulc, R.M. Multistate Evaluation of Reduced-Lignin Alfalfa Harvested at Different Intervals. Crop Sci. 2019, 59, 1799–1807. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.; Engels, F. Alfalfa stem tissues: Cell wall deposition, composition, and degradability. Crop Sci. 2002, 42, 524–534. [Google Scholar] [CrossRef]
- Lin, S.; Medina, C.A.; Boge, B.; Hu, J.; Fransen, S.; Norberg, S.; Yu, L.-X. Identification of genetic loci associated with forage quality in response to water deficit in autotetraploid alfalfa (Medicago sativa L.). BMC Plant Biol. 2020, 20, 1–18. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Nazzicari, N.; Li, X.; Wei, Y.; Pecetti, L.; Brummer, E.C. Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genom. 2015, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Marten, G.; Buxton, D.; Barnes, R. Feeding value (forage quality). Alfalfa Alfalfa Improv. 1988, 29, 463–491. [Google Scholar]
- Biazzi, E.; Nazzicari, N.; Pecetti, L.; Brummer, E.C.; Palmonari, A.; Tava, A.; Annicchiarico, P. Genome-wide association mapping and genomic selection for alfalfa (Medicago sativa) forage quality traits. PLoS ONE 2017, 12, e0169234. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.; Zhao, F.; Wang, X.; Han, J.; Zhao, H.; Liu, G.; Wang, Z. Genomic prediction for 25 agronomic and quality traits in alfalfa (Medicago sativa). Front. Plant Sci. 2018, 9, 1220. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.H.; Williams, S.M. Epistasis and its implications for personal genetics. Am. J. Hum. Genet. 2009, 85, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Howard, R.; Carriquiry, A.L.; Beavis, W.D. Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3 Genes Genomes Genet. 2014, 4, 1027–1046. [Google Scholar] [CrossRef] [Green Version]
- Gianola, D.; de Los Campos, G. Inferring genetic values for quantitative traits non-parametrically. Genet. Res. 2008, 90, 525–540. [Google Scholar] [CrossRef]
- De los Campos, G.; Gianola, D.; Rosa, G.J.; Weigel, K.A.; Crossa, J. Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet. Res. 2010, 92, 295–308. [Google Scholar] [CrossRef] [Green Version]


| Population Names | Population Combinations | Cycle Number | Number of Plants Intermated |
|---|---|---|---|
| UMN3097 | Parental | C0 | |
| UMN3355 | H16 × H96 | C1 | 117 |
| UMN3356 | H16 × L96 | C1 | 28 |
| UMN3357 | L16 × H96 | C1 | 26 |
| UMN3358 | L16 × L96 | C1 | 33 |
| UMN4016 | H16 × H96 | C2 | 60 |
| UMN4017 | H16 × L96 | C2 | ~30 |
| UMN4018 | L16 × H96 | C2 | ~30 |
| UMN4019 | L16 × L96 | C2 | ~30 |
| Term | NDFD16 g kg−1 | NDFD96 g kg−1 | ADL g kg−1 NDF | NDF g kg−1 DM | SDW g plant−1 | Per_stem % |
|---|---|---|---|---|---|---|
| Y | 61,131 *** | 37,685 *** | 3868 *** | 79,412 *** | 851 * | 1471 *** |
| M | 136,873 *** | 292,141 *** | 20,875 *** | 789,443 *** | 71,707 *** | 21,300 *** |
| L | 11,393 *** | 191,234 *** | 49,474 *** | 12,798 *** | 3510 *** | 996 *** |
| H | 20,023 *** | 39,845 *** | 10,591 *** | 62,982 *** | 106,104 *** | 33,054 *** |
| G | 5023 *** | 51,724 *** | 5257 *** | 27,206 *** | 2587 *** | 201 *** |
| H:G | 173 *** | 846 *** | 31 | 919 *** | 317 * | 24 |
| L:G | 89 | 768 * | 100 *** | 689 * | 464 * | 15 |
| M:G | 136 ** | 1609 *** | 75 ** | 823 *** | 714 *** | 33 |
| Y:G | 99 | 893** | 61 | 599 | 118 | 18 |
| L:H:G | 66 | 312 | 24 | 320 | 109 | 25 |
| L:M:G | 137 ** | 756 ** | 50 | 478 | 914 *** | 38 * |
| M:H:G | 70 | 256 | 19 | 372 | 128 | 25 |
| Y:H:G | 89 | 274 | 32 | 383 | 55 | 17 |
| Y:L:G | 142 * | 1258 *** | 128 *** | 505 | 164 | 15 |
| Y:M:G | 38 | 337 | 53 | 270 | 236 | 18 |
| L:M:H:G | 47 | 278 | 24 | 268 | 273 | 22 |
| Y:L:H:G | 50 | 184 | 20 | 270 | 71 | 18 |
| Y:L:M:G | 66 | 430 | 37 | 386 | 193 | 34 |
| Y:M:H:G | 60 | 282 | 17 | 219 | 114 | 24 |
| Y:L:M:H:G | 40 | 120 | 15 | 201 | 110 | 14 |
| Trait | Heritability | GG | Slope | p-Value |
|---|---|---|---|---|
| ADL (g kg−1 NDF) | 0.75 | −3.98 | −3.42 | 1.17 × 10−9 |
| NDFD96H (g kg−1) | 0.70 | 4.54 | 10.13 | 2.50 × 10−9 |
| NDF (g kg−1 DM) | 0.47 | −3.15 | −9.79 | 4.60 × 10−6 |
| NDFD16H (g kg−1) | 0.46 | 4.27 | 4.18 | 1.84 × 10−5 |
| SDW_gm (g plant−1) | 0.15 | −16.1 | −1.31 | 8.34 × 10−2 |
| Per_Stem (%) | 0.11 | −2.02 | −0.01 | 2.62 × 10−1 |
| Maturity | Harvest | Cycle | NDFD 16H (g kg−1) | LSD 16H | %/cycle 16H | NDFD 96H (g kg−1) | LSD 96H | %/Cycle 96H |
|---|---|---|---|---|---|---|---|---|
| EB | 1 | C2 | 218 | a | 2.25 | 485 | a | 2.03 |
| EB | 1 | C1 | 213 | a | 3.02 | 476 | a | 0.25 |
| EB | 1 | C0 | 207 | a | 475 | a | ||
| EB | 2 | C2 | 210 | a | 1.39 | 468 | a | 1.27 |
| EB | 2 | C1 | 207 | ab | 0.89 | 462 | a | 1.55 |
| EB | 2 | C0 | 205 | b | 455 | a | ||
| EB | 3 | C2 | 192 | a | 3.44 | 440 | a | 3.84 |
| EB | 3 | C1 | 186 | a | −0.41 | 424 | a | −1.28 |
| EB | 3 | C0 | 187 | a | 430 | a | ||
| LF | 1 | C2 | 182 | a | 0.54 | 442 | a | 2.47 |
| LF | 1 | C1 | 181 | a | 3.81 | 431 | a | 4.11 |
| LF | 1 | C0 | 174 | b | 414 | b | ||
| LF | 2 | C2 | 188 | a | 1.50 | 456 | a | 3.86 |
| LF | 2 | C1 | 186 | ab | 3.05 | 439 | b | 2.86 |
| LF | 2 | C0 | 180 | b | 427 | c | ||
| LF | 3 | C2 | 197 | a | 4.27 | 445 | a | 5.33 |
| LF | 3 | C1 | 189 | ab | 1.43 | 422 | b | 1.21 |
| LF | 3 | C0 | 186 | b | 417 | b | ||
| GP | 1 | C2 | 178 | a | 1.66 | 417 | a | 3.22 |
| GP | 1 | C1 | 175 | ab | 1.29 | 404 | b | 1.42 |
| GP | 1 | C0 | 173 | b | 398 | b | ||
| GP | 2 | C2 | 191 | a | 4.75 | 450 | a | 4.08 |
| GP | 2 | C1 | 182 | b | 2.81 | 433 | b | 3.65 |
| GP | 2 | C0 | 177 | b | 417 | c | ||
| GP | 3 | C2 | 199 | a | 4.11 | 452 | a | 5.09 |
| GP | 3 | C1 | 191 | b | 1.91 | 430 | b | 2.41 |
| GP | 3 | C0 | 188 | b | 420 | b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Heuschele, D.J.; Lamb, J.F.S.; Jung, H.-J.G.; Samac, D.A. Improved Forage Quality in Alfalfa (Medicago sativa L.) via Selection for Increased Stem Fiber Digestibility. Agronomy 2023, 13, 770. https://doi.org/10.3390/agronomy13030770
Xu Z, Heuschele DJ, Lamb JFS, Jung H-JG, Samac DA. Improved Forage Quality in Alfalfa (Medicago sativa L.) via Selection for Increased Stem Fiber Digestibility. Agronomy. 2023; 13(3):770. https://doi.org/10.3390/agronomy13030770
Chicago/Turabian StyleXu, Zhanyou, Deborah J. Heuschele, JoAnn F. S. Lamb, Hans-Joachim G. Jung, and Deborah A. Samac. 2023. "Improved Forage Quality in Alfalfa (Medicago sativa L.) via Selection for Increased Stem Fiber Digestibility" Agronomy 13, no. 3: 770. https://doi.org/10.3390/agronomy13030770
APA StyleXu, Z., Heuschele, D. J., Lamb, J. F. S., Jung, H.-J. G., & Samac, D. A. (2023). Improved Forage Quality in Alfalfa (Medicago sativa L.) via Selection for Increased Stem Fiber Digestibility. Agronomy, 13(3), 770. https://doi.org/10.3390/agronomy13030770







