Removing Harmful Pericarp Character of Weedy Rice as the First Step of Domestication towards Direct-Seeding Rice Using CRISPR/Cas9-Targeted Mutagenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Construction of CRISPR/Cas9-Rc Mutant Vector
2.3. Agrobacterium-Mediated Genetic Transformation of Weedy Rice
2.4. Molecular Identification of Rc Mutants
2.5. Phenotype Traits
2.6. Drought Tolerance in Seed Germination
2.7. Data Analysis
3. Results
3.1. Construction of CRISPR/Cas9-Rc Mutant Vector and Agrobacterium-Mediated Genetic Transformation
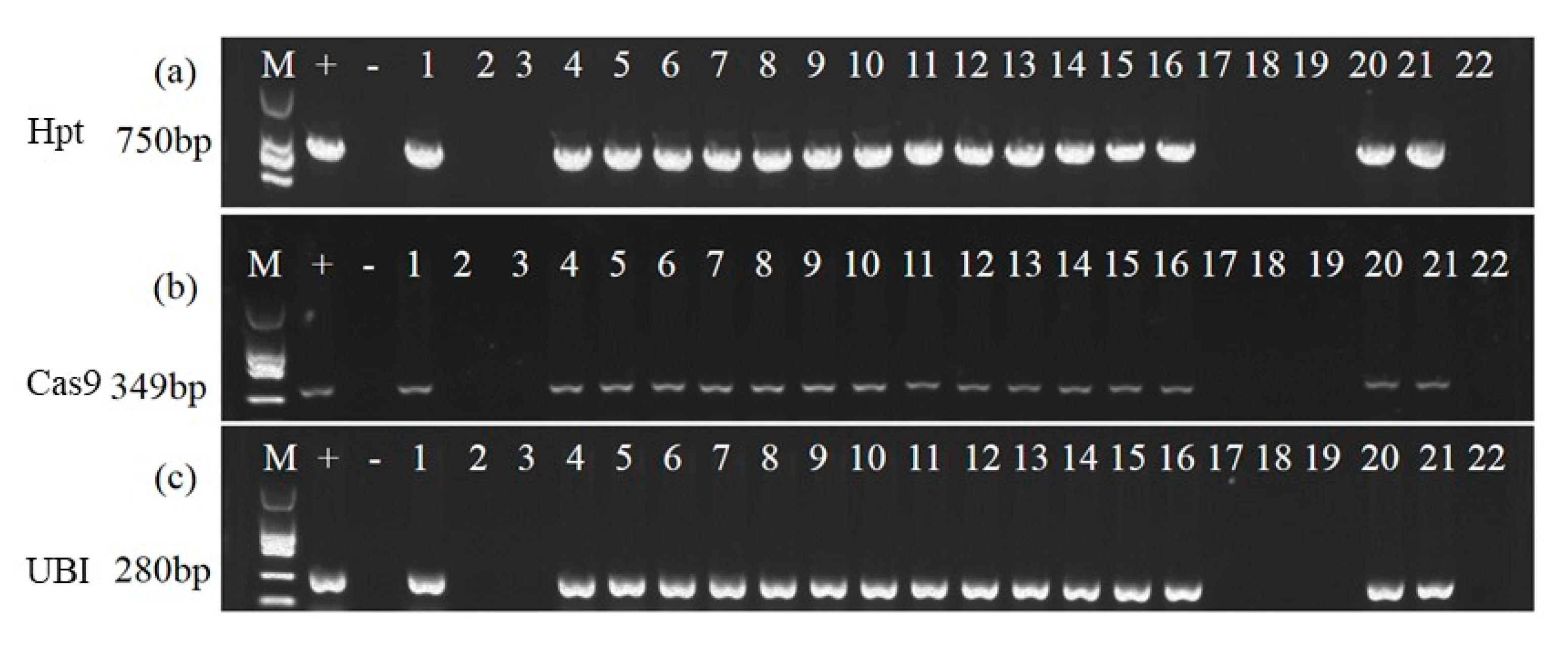
3.2. Molecular Identification of T1-T3 Generation Plantlets
3.3. Predicted Amino Acid Sequence of Rc Gene Mutant
3.4. Off-Target Detection of Rc Mutant
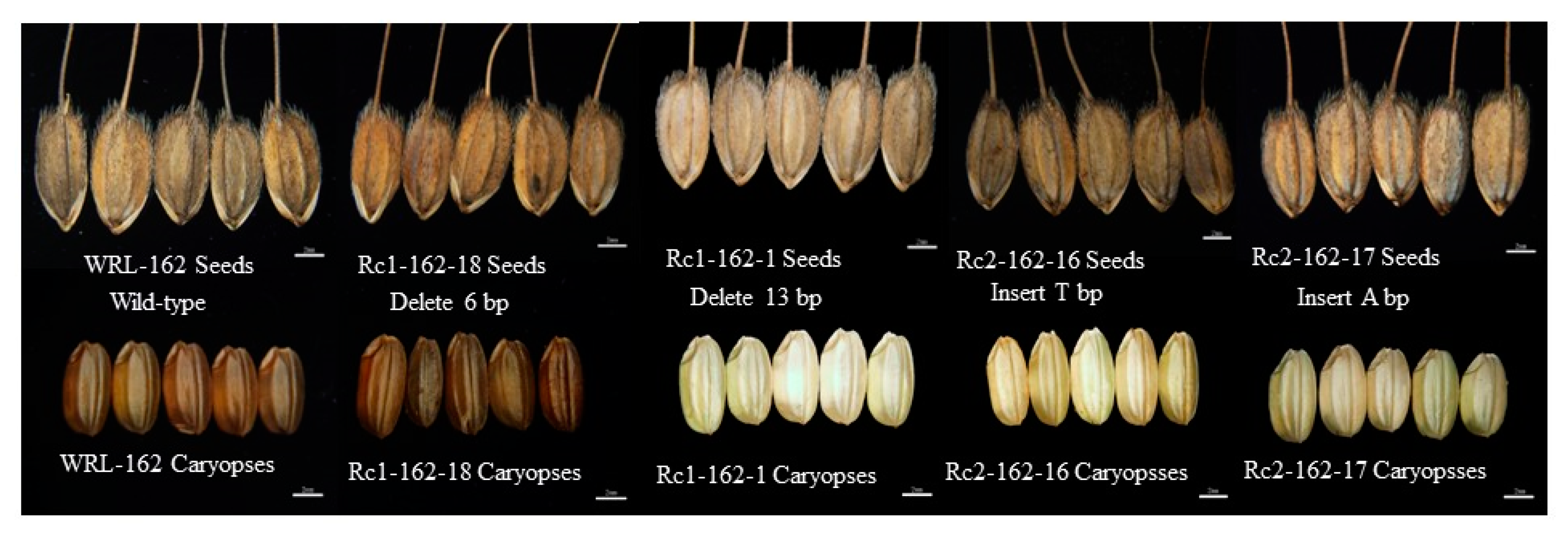
3.5. Drought Tolerance in Seed Germination and Phenotype Traits of Wild Type and T3 Mutant of Weedy Rice
4. Discussion
4.1. The bHLH Region of the Rc Gene Plays a Critical Role in Determining the Pericarp Color
4.2. Domestication of Weedy Rice into Direct-Seeding Rice Might Be Achieved by Precisely Editing a Few Unfavorable Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delouche, J.C.; Burgos, N.R.; Gealy, D.R.; Zorrillad, S.M.G.; Labrada, R.; Larinde, M.; Rosell, C. Weedy Rices: Origin, Biology, Ecology and Control; FAO Plant Production & Protection Paper; FAO: Rome, Italy, 2007. [Google Scholar]
- Estorninos, L.E.; Gealy, D.R.; Gbur, E.E.; Talbert, R.E.; McClelland, M.R. Rice and red rice interference. II. Rice response to population densities of three red rice (Oryza sativa) ecotypes. Weed Sci. 2005, 53, 683–689. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2005, 144, 31. [Google Scholar] [CrossRef]
- Chauhan, B.S. Strategies to manage weedy rice in Asia. Crop Prot. 2013, 48, 51–56. [Google Scholar] [CrossRef]
- Hill, J.E.; Smith, R.J.; Bayer, D.E. Rice weed control: Current technology and emerging issues in temperate rice. Aust. J. Exp. Agric. 1994, 34, 1021–1029. [Google Scholar] [CrossRef]
- Ziska, L.H.; Gealy, D.R.; Burgos, N.; Caicedo, A.L.; Gressel, J.; Lawton-Rauh, A.L.; Avila, L.A.; Theisen, G.; Norsworthy, J.; Ferrero, A.; et al. Chapter Three—Weedy (red) rice: An emerging constraint to global rice production. Adv. Agron. 2015, 129, 181–228. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; Mccouch, S.R. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, M.T.; Thomson, M.J.; Cho, Y.G.; Park, Y.J.; Williamson, S.H.; Bustamante, C.D.; McCouch, S.R. Global Dissemination of a Single Mutation Conferring White Pericarp in Rice. PLoS Genet. 2007, 3, 1418–1424. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Zhou, Y.; Mao, L.; Ye, C.; Wang, W.; Zhang, J.; Yu, Y.; Fu, F.; Wang, Y.; Qian, F.; et al. Genomic variation associated with local adaptation of weedy rice during dedomestication. Nat. Commun. 2017, 8, 15323. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Jia, Y.; Caicedo, A.L.; Olsen, K.M. Signatures of adaptation in the weedy rice genome. Nat. Genet. 2017, 49, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mo, S.D.; Kong, M.Y.; Chao, J.; Chen, X.F.; Yang, J.L.; Yan, Y.J.; Shi, Z.H.; Qiang, S.; Song, X.L.; et al. Better performance of germination in hyperosmotic solutions in conspecific weedy rice than cultivated rice. J. Syst. Evol. 2019, 57, 519–529. [Google Scholar] [CrossRef]
- Zhang, L.J.; Dai, W.M.; Wu, C.; Song, X.L.; Qiang, S. Genetic diversity and origin of Japonica- and Indica-like rice biotypes of weedy rice in the Guangdong and Liaoning provinces of China. Genet. Resour. Crop Evol. 2012, 59, 399–410. [Google Scholar] [CrossRef]
- Wang, H.Q.; Dai, W.M.; Zhang, Z.X.; Li, M.S.; Meng, L.C.; Zhang, Z.; Lu, H.; Song, X.L.; Qiang, S. Occurrence pattern and morphological polymorphism of weedy rice in China. J. Integr. Agric. 2023, 22, 2–22. [Google Scholar] [CrossRef]
- Wu, C.; Dai, W.M.; Song, X.L.; Qiang, S. Diversity of plant traits of weedy rice in Liaoning and Jiangsu provinces. Biodivers. Sci. 2010, 18, 29–36. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef]
- Andres, A.; Fogliatto, S.; Ferrero, A.; Vidotto, F. Growth variability of Italian weedy rice populations grown with or without cultivated rice. Crop Sci. 2015, 55, 394–402. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, Y.J.; Sun, X.X.; He, X.T.; Yang, J.L.; Chen, X.F.; Shi, Z.H.; Song, X.L.; Qiang, S.; Dai, W.M. Weedy rice de-domesticated from cultivated rice has evolved strong resistance to seed ageing. Weed Res. 2021, 61, 396–405. [Google Scholar] [CrossRef]
- Furukawa, T.; Maekawa, M.; Oki, T.; Suda, I.; Iida, S.; Shimada, H.; Takamure, I.; Kadowaki, K.I. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 2007, 49, 91–102. [Google Scholar] [CrossRef]
- Buck, M.J.; Atchley, W.R. Phylogenetic Analysis of Plant Basic Helix-Loop-Helix Proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Bai, M.; Kim, J.; Wang, T.; Oh, E.; Chen, L.; Park, C.H.; Son, S.; Kim, S.; Mudgett, M.B.; et al. The bHLH Transcription Factor HBI1 Mediates the Trade-Off between Growth and Pathogen-Associated Molecular Pattern-Triggered Immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [Green Version]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [Green Version]
- Brooks, S.A.; Yan, W.; Jackson, A.K.; Deren, C.W. A natural mutation in rc reverts white-rice-pericarp to red and results in a new, dominant, wild-type allele: Rc-g. Theor Appl Genet. 2008, 117, 575–580. [Google Scholar] [CrossRef]
- Lee, D.; Lupotto, E.; Powell, W. G-string slippage turns white rice red. Genome 2009, 52, 490–493. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep. 2015, 5, 11491. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Lin, Y.; Chen, S.; Liu, H.; Chen, Z.; Fan, M.; Hu, T.; Mei, F.; Chen, J.; Chen, L.; et al. CRISPR/Cas9-mediated functional recovery of the recessive rc allele to develop red rice. Plant Biotechnol. J. 2019, 17, 2096–2105. [Google Scholar] [CrossRef] [Green Version]
- Li, X.Y.; Qiang, S.; Song, X.; Cai, K.; Dai, W.M. Haplotype analysis of Rc gene for weedy rice in Jiangsu Province. Chin. J. Rice Sci. 2014, 28, 304–313. [Google Scholar]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.Z.; Wei, X.H. Descriptors and Data Standard for Rice (Oryza sativa L.); China Agriculture Press: Beijing, China, 2006. [Google Scholar]
- International Rice Research Institute. Standard Evaluation System for Rice; International Rice Research Institute: Manila, Philippines, 2002. [Google Scholar]
- Xia, H.B.; Xia, H.; Ellstrand, N.C.; Yang, C.; Lu, B.R. Rapid evolutionary divergence and ecotypic diversification of germination behavior in weedy rice populations. New Phytol. 2011, 191, 1119–1127. [Google Scholar] [CrossRef]
- Basu, S.; Roychoudhury, A.; Saha, P.P.; Sengupta, D.N. Differential antioxidative responses of indica rice cultivars to drought stress. Plant Growth Regul. 2009, 60, 51–59. [Google Scholar] [CrossRef]
- Qian, B.; Li, X.; Liu, X.; Wang, M. Improved oxidative tolerance in suspension-cultured cells of C4-pepctransgenic rice by H2O2 and Ca2+under PEG-6000. J. Integr. Plant Biol. 2015, 57, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Baskin, J.M.; Baskin, C.C. Seeds; Academic Press: New York, NY, USA, 1998. [Google Scholar]
- Woffelman, C. DNAMAN for Windows, version 5.2.10.; Lynon Biosoft: San Ramon, CA, USA, 2004. [Google Scholar]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- Gu, X.Y.; Foley, M.E.; Horvath, D.P.; Anderson, J.V.; Feng, J.; Zhang, L.; Mowry, C.R.; Ye, H.; Suttle, J.C.; Kadowaki, K.; et al. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 2011, 189, 1515–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zande, P.V.; Hill, M.S.; Wittkopp, P.J. Pleiotropic effects of trans-regulatory mutations on fitness and gene expression. Science 2022, 377, 105–109. [Google Scholar] [CrossRef]
- Zsögön, A.; Cermak, T.; Voytas, D.; Peres, L.E.P. Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: Case study in tomato. Plant Sci. 2017, 256, 120–130. [Google Scholar] [CrossRef]
- Li, T.D.; Yang, X.P.; Yu, Y.; Si, X.M.; Zhai, X.W.; Zhang, H.W.; Dong, W.X.; Gao, C.X.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [Green Version]



| Positive Regenerated Plants | Mutation Type | Mutation Sequence | Phenotype of Pericarp Color | |
|---|---|---|---|---|
| Wrl-162 | Wild type | ACCTGAATCAAGGGGCGGGAAAGGCGCAAG | Red | |
| TGGAACGCGAAAAGTCGGTGCCATCCAAG | ||||
| Rc 1-162-18 | −6 bp | −6 bp | ACCTGAATCAAGGGGCGGGAA−−−−−−AAGT | Red |
| GGAACGCGAAAAGTCGGTGCCATCCAAG | ||||
| Rc 1-162-1 | −13 bp | −13 bp | ACCTGAATCAAGGGGCGGGAAAGGCG−−−−−−−−−−−−−AAAAGTCGGTGCCATCCAAGGTGATTTCA | White |
| Rc 2-162-16 | +T | +1 bp | ACCTGAATCAAGGGGCGGGAAAGGCGCTAAG | White |
| TGGAACGCGAAAAGTCGGTGCCATCCAAG | ||||
| Rc 2-162-17 | +A | +1 bp | ACGCGAAAAGTCGGTGCCATCCAAGGTGA | White |
| TTTCAGTGCCAACCATGTGCTGAAAAGAG | ||||
| Samples | bHLH Region | Pericarp Color |
|---|---|---|
| WRL-162 | ESRGGKGASGTRKVGAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| Rc 1-162-T1-18 | ESRGGKSGTRKVGAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| Rc 1-162-T1-1 | ESRGGKGEKSVPSKVISVPTMC*KRGEEERSSMRSS*FCDLWYLS*QRWTRRRY*ATRSST*SS*GTAYKSSSRRRRRHE | White |
| Rc 2-162-T1-16 | ESRGGKGAKWNAKSRCHPRFQCQPCAEREEKKREAQEVHNSAIFGTFHDKDGQGVDTRRHDRVREAAKEPHTR | White |
| Rc 2-162-T1-17 | ESRGGKGASGTRKVGAIQGDFSANHVLKREEKKREAQ.EVHNSAIFGTFHDKDGQGVDTRRHDRVREAAKEPHT | White |
| Rc-r (Perla) | EQKHLNQGAGKAQVDAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| Rc (H75) | ESRGGKGASGTRKVGAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| Rc (Oryza rufipogon) | ESRGGKGASGTRKVGAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| Rc g (WELL mutant) | ESRAGKAQVDAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| Rc-s (Surjamkuhi) | ESRGGKGASGTRKVGAIQGDFSANHVLKERRRREKLNEKFIILRSLVPFMTKMDKASILGDTIEYVKQLRNRIQELE | Red |
| rc (Nipponbare) | ESRGGKGASGCHPRFQCQPCAEREEKKREAQEVHNSAIFGTFHDKDGQGVDTRRHDRVREAAKEPHTRARV | White |
| rc (Jefferson) | ESRGGKGASGCHPRFQCQPCAEREEKKREAQEVHNSAIFGTFHDKDGQGVDTRRHDRVREAAKEPHTRARV | White |
| rc (WELL) | ESRGGKGASGCHPRFQCQPCAEREEKKREAQEVHNSAIFGTFHDKDGQGVDTRRHDRVREAAKEPHTRARV | White |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, M.; He, X.; Yin, Z.; Chen, X.; Zhang, Y.; Shi, Z.; Song, X.; Qiang, S.; Dai, W. Removing Harmful Pericarp Character of Weedy Rice as the First Step of Domestication towards Direct-Seeding Rice Using CRISPR/Cas9-Targeted Mutagenesis. Agronomy 2023, 13, 1130. https://doi.org/10.3390/agronomy13041130
Kong M, He X, Yin Z, Chen X, Zhang Y, Shi Z, Song X, Qiang S, Dai W. Removing Harmful Pericarp Character of Weedy Rice as the First Step of Domestication towards Direct-Seeding Rice Using CRISPR/Cas9-Targeted Mutagenesis. Agronomy. 2023; 13(4):1130. https://doi.org/10.3390/agronomy13041130
Chicago/Turabian StyleKong, Mengyao, Xiaotong He, Zhendong Yin, Xianshu Chen, Yujie Zhang, Zhihua Shi, Xiaoling Song, Sheng Qiang, and Weimin Dai. 2023. "Removing Harmful Pericarp Character of Weedy Rice as the First Step of Domestication towards Direct-Seeding Rice Using CRISPR/Cas9-Targeted Mutagenesis" Agronomy 13, no. 4: 1130. https://doi.org/10.3390/agronomy13041130
APA StyleKong, M., He, X., Yin, Z., Chen, X., Zhang, Y., Shi, Z., Song, X., Qiang, S., & Dai, W. (2023). Removing Harmful Pericarp Character of Weedy Rice as the First Step of Domestication towards Direct-Seeding Rice Using CRISPR/Cas9-Targeted Mutagenesis. Agronomy, 13(4), 1130. https://doi.org/10.3390/agronomy13041130







