Comparative Nutritional Assessment and Metabolomics of a WRKY Rice Mutant with Enhanced Germination Rates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
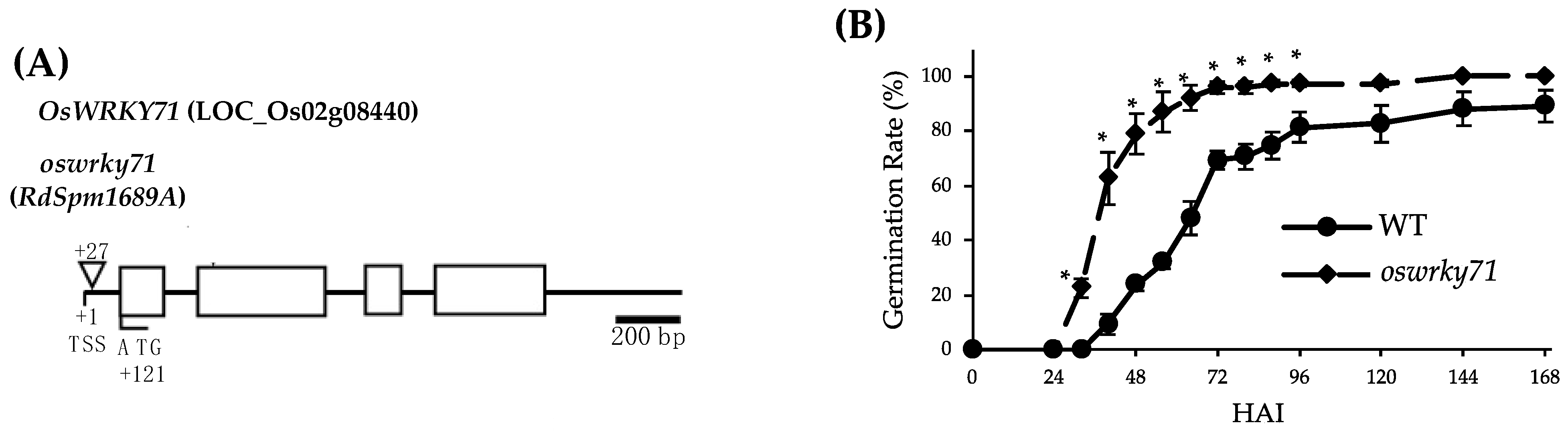
2.2. Genotyping Mutant Lines by PCR
2.3. Germination Assays
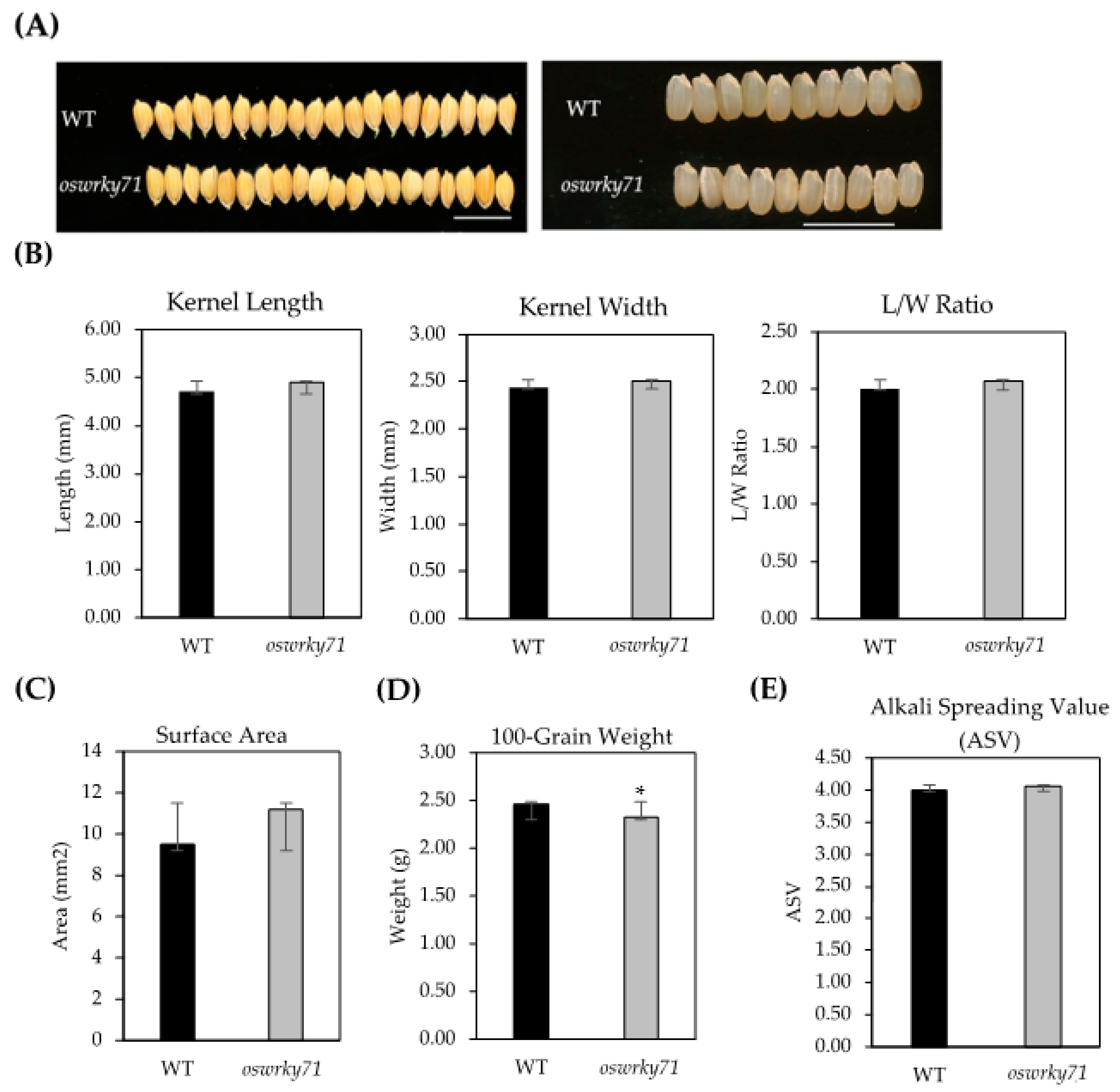
2.4. Kernel Morphology
2.5. Proximate Composition
2.6. Total Protein
2.7. Amino Acids
2.8. B Vitamins
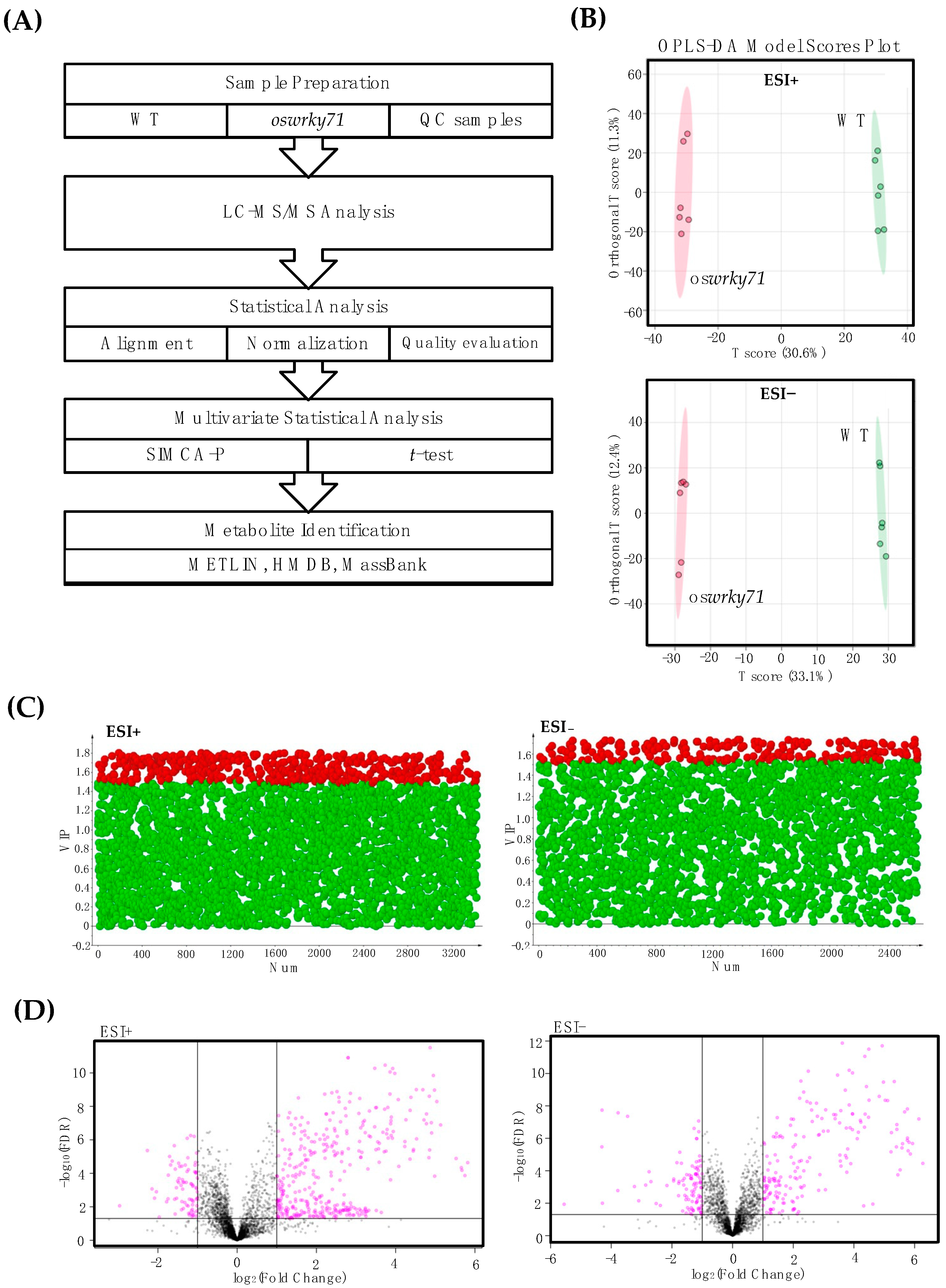
2.9. Untargeted Metabolomics
2.10. Statistical Methods
3. Results
3.1. Transposon-Mediated Mutation in OsWRKY71 Results in Early Germination Phenotype
3.2. Morphological Characterization of oswrky71 Kernel
3.3. Proximate Composition and Mineral Analyses
3.4. Amino Acids Composition Analysis
3.5. Quantification of B Vitamins
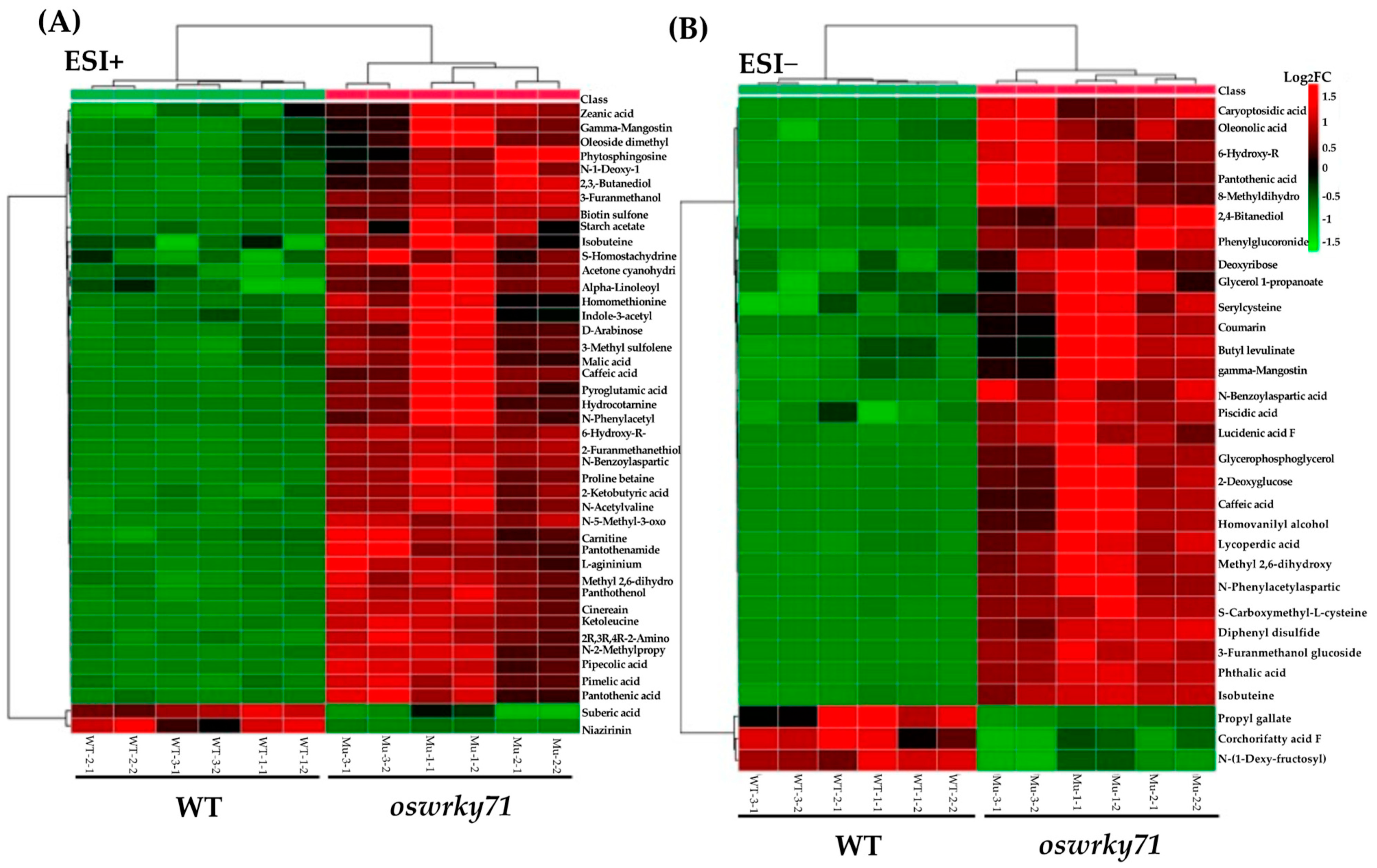
3.6. Identification of Differential Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO, Food and Agriculture Organization of the United Nations. David Lubin Memorial Library. In African Studies Companion Online; Brill: Leiden, The Netherlands, 2022. [Google Scholar]
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for Global Nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Gnanamanickam, S.S. Rice and Its Importance to Human Life. In Biological Control of Rice Diseases; Springer: Amsterdam, The Netherlands, 2009; pp. 1–11. [Google Scholar]
- Ramankutty, N.; Mehrabi, Z.; Waha, K.; Jarvis, L.; Kremen, C.; Herrero, M.; Rieseberg, L.H. Trends in global agricultural land use: Implications for environmental health and food security. Annu. Rev. Plant Biol. 2018, 69, 789–815. [Google Scholar] [CrossRef] [PubMed]
- Pickson, R.B.; He, G.; Boateng, E. Impacts of climate change on rice production: Evidence from 30 Chinese provinces. Environ. Dev. Sustain. 2022, 24, 3907–3925. [Google Scholar] [CrossRef]
- Veronica, N.; Rani, Y.A.; Subrahmanyam, D.; Rao, K.N.; Ahamad, M.; Rani, P.P. Physiological and biochemical responses in rice (Oryza sativa L.) to high temperature: A review. Res. Crops 2016, 17, 21–30. [Google Scholar] [CrossRef]
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef]
- Hussain, S.; Huang, J.; Huang, J.; Ahmad, S.; Nanda, S.; Anwar, S.; Shakoor, A.; Zhu, C.; Zhu, L.; Cao, X. Rice production under climate change: Adaptations and mitigating strategies. In Environment, Climate, Plant and Vegetation Growth; Springer: Cham, Switzerland, 2020; pp. 659–686. [Google Scholar]
- Achary, V.M.M.; Reddy, M.K. CRISPR-Cas9 mediated mutation in GRAIN WIDTH and WEIGHT2 (GW2) locus improves aleurone layer and grain nutritional quality in rice. Sci. Rep. 2021, 11, 21941. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Biol. 2019, 61, 541–563. [Google Scholar] [CrossRef]
- Sohn, S.-I.; Pandian, S.; Kumar, T.S.; Zoclanclounon, Y.A.B.; Muthuramalingam, P.; Shilpha, J.; Satish, L.; Ramesh, M. Seed Dormancy and Pre-Harvest Sprouting in Rice-An Updated Overview. Int. J. Mol. Sci. 2021, 22, 11804. [Google Scholar] [CrossRef]
- Shu, K.; Meng, Y.J.; Shuai, H.W.; Liu, W.G.; Du, J.B.; Liu, J.; Yang, W.Y. Dormancy and germination: How does the crop seed decide? Plant Biol. 2015, 17, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zeng, M.; Wu, Y.; Du, Y.; Liu, J.; Luo, S.; Zeng, Y. Comparative transcriptomic analysis provides insights into the molecular basis underlying pre-harvest sprouting in rice. BMC Genom. 2022, 23, 771. [Google Scholar] [CrossRef]
- Li, C.; Ni, P.; Francki, M.; Hunter, A.; Zhang, Y.; Schibeci, D.; Li, H.; Tarr, A.; Wang, J.; Cakir, M.; et al. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genom. 2004, 4, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Rice Extension. Rice Growing Guidelines for Managing Water Usage. Available online: https://riceextension.org.au/documents/2018/3/21/rice-growing-guidelines-for-managing-water-usage (accessed on 7 April 2023).
- de Mey, Y.; Demont, M. 19 Bird Damage to Rice in Africa: Evidence and Control. In Realizing Africa’s Rice Promise; CABI: Wallingford, UK, 2013; pp. 241–249. [Google Scholar]
- Ni, H.; Moody, K.; Robles, R.; Paller, E.; Lales, J. Oryza sativa plant traits conferring competitive ability against weeds. Weed Sci. 2000, 48, 200–204. [Google Scholar] [CrossRef]
- Chao, S.; Mitchell, J.; Fukai, S. Factors Determining Genotypic Variation in the Speed of Rice Germination. Agronomy 2021, 11, 1614. [Google Scholar] [CrossRef]
- Farooq, M.; Siddique, K.H.; Rehman, H.; Aziz, T.; Lee, D.-J.; Wahid, A. Rice direct seeding: Experiences, challenges and opportunities. Soil Tillage Res. 2011, 111, 87–98. [Google Scholar] [CrossRef]
- Kumar, C.S.; Wing, R.A.; Sundaresan, V. Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J. 2005, 44, 879–892. [Google Scholar] [CrossRef]
- Villacastin, A.J.; Adams, K.S.; Boonjue, R.; Rushton, P.J.; Han, M.; Shen, J.Q. Dynamic differential evolution schemes of WRKY transcription factors in domesticated and wild rice. Sci. Rep. 2021, 11, 14887. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.-L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Zhang, Z.-L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.-H.D.; Shen, Q.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Yang, G.; Komatsu, S.; Shen, Q.J. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006, 46, 231–242. [Google Scholar] [CrossRef]
- Matsumoto, T.; Wu, J.; Itoh, T.; Numa, H.; Antonio, B.; Sasaki, T. The Nipponbare genome and the next-generation of rice genomics research in Japan. Rice 2016, 9, 33. [Google Scholar] [CrossRef]
- Farooq, M.; Basra, S.; Ahmad, N.; Hafeez, K. Thermal hardening: A new seed vigor enhancement tool in rice. J. Integr. Plant Biol. 2005, 47, 187–193. [Google Scholar] [CrossRef]
- IRRI, International Rice Research Institute. Standard Evaluation System for Rice; IRRI, International Rice Research Institute: Los Baños, Philippines, 1996. [Google Scholar]
- Collins, T.J. ImageJ for microscopy. Biotechniques 2007, 43, S25–S30. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35 (Suppl. S1), D521–D526. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E. METLIN: A technology platform for identifying knowns and unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.S.; Chang, J.L. Does gibberellic acid stimulate seed germination via amylase synthesis? Plant Physiol. 1972, 49, 441. [Google Scholar] [CrossRef]
- Kaneko, M.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M. The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 2002, 128, 1264–1270. [Google Scholar] [CrossRef]
- Gong, D.; He, F.; Liu, J.; Zhang, C.; Wang, Y.; Tian, S.; Sun, C.; Zhang, X. Understanding of Hormonal Regulation in Rice Seed Germination. Life 2022, 12, 1021. [Google Scholar] [CrossRef]
- Farooq, M.; Basra, S.; Saleem, B.; Nafees, M.; Chishti, S. Enhancement of tomato seed germination and seedling vigor by osmopriming. Pak. J. Agric. Sci. 2005, 42, 3–4. [Google Scholar]
- Pang, Y.; Ali, J.; Wang, X.; Franje, N.J.; Revilleza, J.E.; Xu, J.; Li, Z. Relationship of rice grain amylose, gelatinization temperature and pasting properties for breeding better eating and cooking quality of rice varieties. PLoS ONE 2016, 11, e0168483. [Google Scholar] [CrossRef]
- Levine, H.; Slade, L. Interpreting the behavior of low-moisture foods. In Water and Food Quality; Elsevier: Amsterdam, The Netherlands, 1989; pp. 71–134. [Google Scholar]
- Laca, A.; Mousia, Z.; Díaz, M.; Webb, C.; Pandiella, S.S. Distribution of microbial contamination within cereal grains. J. Food Eng. 2006, 72, 332–338. [Google Scholar] [CrossRef]
- Adair, C.; Bollich, C.; Bowman, D.; Joson, N.; Johnston, T.; Webb, B.; Atkins, J. Rice breeding and testing methods in the United States. USDA Agric. Handb. 1973, 289, 22–75. [Google Scholar]
- Cogburn, R.R. Susceptibility of varieties of stored rough rice to losses caused by storage insects. J. Stored Prod. Res. 1977, 13, 29–34. [Google Scholar] [CrossRef]
- Goa, K.L.; Brogden, R.N. L-carnitine. Drugs 1987, 34, 1–24. [Google Scholar] [CrossRef]
- Evans, A.M.; Fornasini, G. Pharmacokinetics of L-carnitine. Clin. Pharmacokinet. 2003, 42, 941–967. [Google Scholar] [CrossRef] [PubMed]
- Galland, M.; He, D.; Lounifi, I.; Arc, E.; Clément, G.; Balzergue, S.; Huguet, S.; Cueff, G.; Godin, B.; Collet, B. An integrated “multi-omics” comparison of embryo and endosperm tissue-specific features and their impact on rice seed quality. Front. Plant Sci. 2017, 8, 1984. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. The KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chagoyen, M.; Pazos, F. MBRole: Enrichment analysis of metabolomic data. Bioinformatics 2011, 27, 730–731. [Google Scholar] [CrossRef]
- Bedair, M.; Glenn, K.C. Evaluation of the use of untargeted metabolomics in the safety assessment of genetically modified crops. Metabolomics 2020, 16, 111. [Google Scholar] [CrossRef]
- Codex. Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants (CAC/GL 45-2003). 2003. Available online: http://files.foodmate.com/2013/files_1782.html (accessed on 17 November 2022).
- OECD. Safety Evaluation of Foods Derived by Modern Biotechnology: Concepts and Principles; OECD: Paris, France, 1993; Volume 93, p. 1. [Google Scholar]
- Swamy, B.M.; Samia, M.; Boncodin, R.; Marundan, S.; Rebong, D.B.; Ordonio, R.L.; Miranda, R.T.; Rebong, A.T.; Alibuyog, A.Y.; Adeva, C.C. Compositional analysis of genetically engineered GR2E “Golden Rice” in comparison to that of conventional rice. J. Agric. Food Chem. 2019, 67, 7986–7994. [Google Scholar] [CrossRef] [PubMed]
- Kessler, D.A.; Taylor, M.R.; Maryanski, J.H.; Flamm, E.L.; Kahl, L.S. The safety of foods developed by biotechnology. Science 1992, 256, 1747–1749. [Google Scholar] [CrossRef]
- Potrykus, I. Golden rice and beyond. Plant Physiol. 2001, 125, 1157–1161. [Google Scholar] [CrossRef]
- Tang, G.; Qin, J.; Dolnikowski, G.G.; Russell, R.M.; Grusak, M.A. Golden Rice is an effective source of vitamin A. Am. J. Clin. Nutr. 2009, 89, 1776–1783. [Google Scholar] [CrossRef]
- Dipti, S.S.; Bergman, C.; Indrasari, S.D.; Herath, T.; Hall, R.; Lee, H.; Habibi, F.; Bassinello, P.Z.; Graterol, E.; Ferraz, J.P. The potential of rice to offer solutions for malnutrition and chronic diseases. Rice 2012, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, X.; Yao, X.; Yu, R.; Larkin, P.J.; Liu, C.-M. Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains. Proc. Natl. Acad. Sci. USA 2018, 115, 11327–11332. [Google Scholar] [CrossRef] [PubMed]
- Kayahara, H.; Tsukahara, K.; Tatai, T. Flavor, health and nutritional quality of pre-germinated brown rice. In Food Flavors and Chemistry: Advances of the New Millennium, Proceedings of the 10th International Flavor Conference, Paros, Greece, 4–7 July 2000; The Royal Society of Chemistry: London, UK, 2002; pp. 546–551. [Google Scholar]
- Bui, T.N.; Le, T.H.; Nguyen, D.H.; Tran, Q.B.; Nguyen, T.L.; Le, D.T.; Do, V.A.; Vu, A.L.; Aoto, H.; Okuhara, Y. Pre-germinated brown rice reduced both blood glucose concentration and body weight in Vietnamese women with impaired glucose tolerance. J. Nutr. Sci. Vitaminol. 2014, 60, 183–187. [Google Scholar] [CrossRef]
- Hsu, T.-F.; Kise, M.; Wang, M.-F.; Ito, Y.; Yang, M.-D.; Aoto, H.; Yoshihara, R.; Yokoyama, J.; Kunii, D.; Yamamoto, S. Effects of pre-germinated brown rice on blood glucose and lipid levels in free-living patients with impaired fasting glucose or type 2 diabetes. J. Nutr. Sci. Vitaminol. 2008, 54, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055. [Google Scholar] [CrossRef]
- Laza, M.R.C.; Peng, S.; Akita, S.; Saka, H. Effect of panicle size on grain yield of IRRI-released indica rice cultivars in the wet season. Plant Prod. Sci. 2004, 7, 271–276. [Google Scholar] [CrossRef]
- Salo-väänänen, P.P.; Koivistoinen, P.E. Determination of protein in foods: Comparison of net protein and crude protein (N× 6.25) values. Food Chem. 1996, 57, 27–31. [Google Scholar] [CrossRef]
- Ridley, W.P.; Shillito, R.D.; Coats, I.; Steiner, H.-Y.; Shawgo, M.; Phillips, A.; Dussold, P.; Kurtyka, L. Development of the international life sciences institute crop composition database. J. Food Compos. Anal. 2004, 17, 423–438. [Google Scholar] [CrossRef]
- Alba, R.; Phillips, A.; Mackie, S.; Gillikin, N.; Maxwell, C.; Brune, P.; Ridley, W.; Fitzpatrick, J.; Levine, M.; Harris, S. Improvements to the international life sciences institute crop composition database. J. Food Compos. Anal. 2010, 23, 741–748. [Google Scholar] [CrossRef]
- Sult, T.; Barthet, V.J.; Bennett, L.; Edwards, A.; Fast, B.; Gillikin, N.; Launis, K.; New, S.; Rogers-Szuma, K.; Sabbatini, J. Report: Release of the international life sciences institute crop composition database version 5. J. Food Compos. Anal. 2016, 51, 106–111. [Google Scholar] [CrossRef]
- Park, S.-Y.; Kim, J.K.; Jang, J.-S.; Lee, S.Y.; Oh, S.; Lee, S.M.; Yang, C.-I.; Yeo, Y. Comparative analysis of nutritional composition between the disease-resistant rice variety OsCK1 and conventional comparators. Food Sci. Biotechnol. 2015, 24, 225–231. [Google Scholar] [CrossRef]
- Xin, L.; Xiaoyun, H.; Yunbo, L.; Guoying, X.; Xianbin, J.; Kunlun, H. Comparative analysis of nutritional composition between herbicide-tolerant rice with bar gene and its non-transgenic counterpart. J. Food Compos. Anal. 2008, 21, 535–539. [Google Scholar] [CrossRef]
- Nam, K.-H.; Nam, K.J.; An, J.H.; Jeong, S.-C.; Park, K.W.; Kim, H.-B.; Kim, C.-G. Comparative analysis of key nutrient composition between drought-tolerant transgenic rice and its non-transgenic counterpart. Food Sci. Biotechnol. 2013, 22, 1–7. [Google Scholar] [CrossRef]
- Cho, Y.-H.; Puligundla, P.; Oh, S.-D.; Park, H.-M.; Kim, K.-M.; Lee, S.-M.; Ryu, T.-H.; Lee, Y.-T. Comparative evaluation of nutritional compositions between transgenic rice harboring the CaMsrB2 gene and the conventional counterpart. Food Sci. Biotechnol. 2016, 25, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, J.; Aguilera-Gomez, M.; Barrucci, F.; Cocconcelli, P.; Davies, H.; Denslow, N.; Lou Dorne, J.; Grohmann, L.; Herman, L.; Hogstrand, C. EFSA Scientific Colloquium 24–’Omics in Risk Assessment: State of the Art and Next Steps; EFSA: Parma, Italy, 2018; Volume 15. [Google Scholar]
- Christ, B.; Pluskal, T.; Aubry, S.; Weng, J.-K. Contribution of untargeted metabolomics for future assessment of biotech crops. Trends Plant Sci. 2018, 23, 1047–1056. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Agriculture and Natural Resources; Committee on Genetically Engineered Crops: Past Experience and Future Prospects. Genetically Engineered Crops: Experiences and Prospects; National Academies Press (US): Washington, DC, USA, 2016. [Google Scholar] [CrossRef]
- Ricroch, A.E. Assessment of GE food safety using ‘-omics’ techniques and long-term animal feeding studies. New Biotechnol. 2013, 30, 349–354. [Google Scholar] [CrossRef]
- Ricroch, A.E.; Bergé, J.B.; Kuntz, M. Evaluation of genetically engineered crops using transcriptomic, proteomic, and metabolomic profiling techniques. Plant Physiol. 2011, 155, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Ricroch, A.; Bergé, J.B.; Messean, A. Literature review of the dispersal of transgenes from genetically modified maize. Comptes Rendus Biol. 2009, 332, 861–875. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, V.R.; Manickam, S.; Muthurajan, R. A Comparative Metabolomic Analysis Reveals the Nutritional and Therapeutic Potential of Grains of the Traditional Rice Variety Mappillai Samba. Plants 2022, 11, 543. [Google Scholar] [CrossRef]
- Gong, R.; Huang, D.; Chen, Y.; Li, H.; Wang, Z.; Zhou, D.; Zhao, L.; Pan, Y.; Chang, Y.; Xiang, Y. Comparative metabolomics analysis reveals the variations of eating quality among three high-quality rice cultivars. Mol. Breed. 2020, 40, 112. [Google Scholar] [CrossRef]
- Kim, H.; Kim, O.-W.; Ahn, J.-H.; Kim, B.-M.; Oh, J.; Kim, H.-J. Metabolomic analysis of germinated brown rice at different germination stages. Foods 2020, 9, 1130. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Saito, K. Role of metabolomics in crop improvement. J. Plant Biochem. Biotechnol. 2012, 21, 24–31. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, C.; Xu, H.; Yuan, K.; Lu, X.; Zhu, Z.; Wu, Y.; Xu, G. Metabolic profiling of transgenic rice with cryIAc and sck genes: An evaluation of unintended effects at metabolic level by using GC-FID and GC–MS. J. Chromatogr. B 2009, 877, 725–732. [Google Scholar] [CrossRef]
- Jiao, Z.; Si, X.-X.; Li, G.-K.; Zhang, Z.-M.; Xu, X.-P. Unintended compositional changes in transgenic rice seeds (Oryza sativa L.) studied by spectral and chromatographic analysis coupled with chemometrics methods. J. Agric. Food Chem. 2010, 58, 1746–1754. [Google Scholar] [CrossRef]
| Component | WT | oswrky71 | p-Value b | Lit. Range c | ||
|---|---|---|---|---|---|---|
| Mean ± SD a | Range | Mean ± SD a | Range | |||
| Crude protein (% DB) | 13.10 ± 0.2 | 12.9–13.2 | 13.70 ± 0.2 | 13.6–13.9 | 0.01 | 5.9–11.8 |
| True protein (mg) | 13.03 ± 1.8 | 11.1–14.7 | 9.49 ± 1.3 | 8.7–11.0 | 0.05 | Not available |
| Crude fiber (% DB) | 9.77 ± 0.3 | 9.5–10.1 | 9.83 ± 0.4 | 9.5–10.3 | 0.83 | 8.6–18.2 |
| Crude fat (% DB) | 1.60 ± 0.0 | 1.6–1.6 | 1.70 ± 0.1 | 1.6–1.8 | 0.16 | 0.56–3.47 |
| Ash (% DB) | 4.07 ± 0.1 | 4.0–4.1 | 3.70 ± 0.3 | 3.4–3.9 | 0.08 | 3.61–8.6 |
| Carbohydrate (% DB) | 81.23 ± 0.2 | 81.1–81.4 | 80.83 ± 0.4 | 80.5–81.3 | 0.19 | 80.0–86.4 |
| Moisture (% FW) | 11.57 ± 0.3 | 11.4–11.9 | 9.57 ± 0.86 | 8.8–10.5 | 0.02 | 3.5–73.7 |
| Mineral (mg/100 g DB) | WT | oswrky71 | p-Value b | Lit. Range c | ||
|---|---|---|---|---|---|---|
| Mean ± SD a | Range | Mean ± SD a | Range | |||
| Calcium | 70.00 ± 0.0 | 70–70 | 50.00 ± 0.0 | 50–50 | 0.00 | 10–150 |
| Phosphorus | 380.00 ± 0.0 | 380–380 | 416.67 ± 5.8 | 410–420 | 0.00 | 190–470 |
| Magnesium | 160.00 ± 10.0 | 150–170 | 180.00 ± 10.0 | 170–190 | 0.07 | 30–170 |
| Potassium | 706.67 ± 11.5 | 700–720 | 640.00 ± 26.5 | 610–660 | 0.02 | 170–472 |
| Sulfur | 146.67 ± 1.5 | 140–150 | 143.33 ± 5.8 | 140–150 | 0.52 | 30–220 |
| Sodium | 33.67 ± 1.5 | 32–35 | 10.00 ± 0.0 | 10–10 | 0.00 | 0–100 |
| Zinc | 4.17 ± 0.2 | 4.2–4.0 | 6.0 ± 0.1 | 5.9–6.1 | 0.00 | 2.2–5.3 |
| Iron | 8.63 ± 1.4 | 7.7–10.2 | 7.0 ± 0.2 | 6.8–7.1 | 0.11 | 1.6–9.08 |
| Manganese | 6.67 ± 0.2 | 6.5–6.8 | 5.47 ± 0.2 | 5.3–5.7 | 0.00 | 2–11.7 |
| Copper | 1.30 ± 0.1 | 1.2–1.4 | 1.33 ± 0.1 | 1.3–1.4 | 0.64 | 0.2–1.3 |
| Amino Acid (mg/100 g DB) | WT | oswrky71 | p–Value b | Lit. Range c | ||
|---|---|---|---|---|---|---|
| Mean ± SD a | Range | Mean ± SD a | Range | |||
| Glycine | 462.7 ± 21.5 | 443.2–485.8 | 389.0 ± 48.7 | 360.8–445.2 | 0.07 | 290–510 |
| Alanine | 695.7 ± 7.0 | 688.1–701.8 | 622.2 ± 60.5 | 554.6–671.5 | 0.10 | 330–630 |
| Serine | 497.4 ± 19.9 | 481.8–519.8 | 425.2 ± 43.6 | 374.9–450.6 | 0.06 | 230–560 |
| Proline | 524.8 ± 14.9 | 510.2–540.0 | 469.5 ± 44.7 | 417.9–496.8 | 0.11 | 280–540 |
| Valine | 730.0 ± 38.3 | 700.5–773.3 | 612.8 ± 59.9 | 550.0–669.3 | 0.05 | 340–650 |
| Threonine | 382.6 ± 5.7 | 376.5–387.8 | 334.0 ± 42.1 | 285.8–364.1 | 0.12 | 220–410 |
| Cysteine | 125.0 ± 23.4 | 104.4–150.5 | 125.2 ± 16.3 | 114.8–143.9 | 0.99 | 100–260 |
| Isoleucine | 439.6 ± 10.4 | 427.9–447.3 | 378.3 ± 44.5 | 328.3–413.7 | 0.08 | 240–460 |
| Aspartic acid | 1335.8 ± 39.2 | 1301.9–1378.8 | 1143.0 ± 81.5 | 1050.2–1203.1 | 0.02 | 500–990 |
| Glutamic acid | 2752.6 ± 72.0 | 2676.5–2819.7 | 2341.1 ± 291.9 | 2054.0–2647.4 | 0.08 | 890–1990 |
| Methionine | 91.8 ± 59.2 | 24.4–135.1 | 93.9 ± 4.2 | 89.3–97.4 | 0.95 | 130–310 |
| Histidine | 222.6 ± 9.0 | 212.9–230.7 | 197.8 ± 23.5 | 171.7–217.3 | 0.16 | 140–281 |
| Phenylalanine | 499.5 ± 7.2 | 491.1–504.1 | 434.1 ± 29.4 | 401.4–458.5 | 0.02 | 280–620 |
| Arginine | 920.8 ± 105.6 | 830.8–1037.0 | 775.9 ± 51.9 | 717.0–814.6 | 0.10 | 410–850 |
| Tryptophan | 4.7 ± 0.1 | 4.6–4.7 | 5.9 ± 1.7 | 4.8–7.8 | 0.28 | 50–180 |
| Lysine | 736.2 ± 34.1 | 698.7–765.4 | 580.3 ± 72.8 | 526.2–663.1 | 0.03 | 210–430 |
| Tyrosine | 321.5 ± 22.2 | 301.4–345.3 | 279.9 ± 19.5 | 260.5–299.4 | 0.07 | 50–180 |
| Leucine | 937.1 ± 25.1 | 919.9–965.9 | 814.7 ± 80.6 | 727.3–886.1 | 0.07 | 460–920 |
| Vitamin (mg/kg DB) | WT | oswrky71 | p-Value b | Lit. Range c | ||
|---|---|---|---|---|---|---|
| Mean ± SD a | Range | Mean ± SD a | Range | |||
| Choline chloride | 555.33 ± 21.46 | 541–580 | 448.33 ± 23.71 | 423–470 | 0.00 | Not available |
| Calcium pantotenate (Vit B5) | 4.88 ± 0.09 | 4.88–4.96 | 3.65 ± 0.03 | 3.61–3.66 | 0.00 | 7.22–14.0 |
| Riboflavin (Vit B2) | 0.33 ± 0.01 | 0.32–0.34 | 0.36 ± 0.01 | 0.35–0.36 | 0.02 | 0.4–1.4 |
| Thiamine (Vit B1) | 2.72 ± 0.02 | 2.69–2.74 | 2.47 ± 0.03 | 2.44–2.50 | 0.00 | 2.35–6.25 |
| Pathway | p-Value | FDR |
|---|---|---|
| Metabolic pathways | 7.7 × 10−13 | 8.4 × 10−11 |
| Aminoacyl-tRNA biosynthesis | 4.1 × 10−11 | 2.2 × 10−9 |
| Glycine, serine and threonine metabolism | 5.3 × 10−9 | 1.9 × 10−7 |
| Cysteine and methionine metabolism | 2.2 × 10−8 | 6.5 × 10−7 |
| ABC transporters | 3.6 × 10−6 | 7.8 × 10−5 |
| Alanine, aspartate and glutamate metabolism | 10.0 × 10−6 | 1.8 × 10−4 |
| Biosynthesis of plant hormones | 1.5 × 10−5 | 2.3 × 10−4 |
| Glucosinolate biosynthesis | 3.0 × 10−5 | 4.1 × 10−4 |
| Histidine metabolism | 4.2 × 10−5 | 5.1 × 10−4 |
| Biosynthesis of alkaloids derived from ornithine, lysine and nicotinic acid | 9.8 × 10−5 | 1.1 × 10−3 |
| Biosynthesis of phenylpropanoids | 2.6 × 10−4 | 2.5 × 10−3 |
| Valine, leucine and isoleucine biosynthesis | 3.2 × 10−4 | 2.9 × 10−3 |
| Arginine and proline metabolism | 4.1 × 10−4 | 3.4 × 10−3 |
| Lysine biosynthesis | 6.1 × 10−4 | 4.7 × 10−3 |
| Biosynthesis of alkaloids derived from histidine and purine | 9.3 × 10−4 | 6.7 × 10−3 |
| Phenylpropanoid biosynthesis | 1.2 × 10−3 | 8.4 × 10−3 |
| Biotin metabolism | 1.5 × 10−3 | 9.8 × 10−3 |
| Cyanoamino acid metabolism | 1.9 × 10−3 | 1.2 × 10−2 |
| Nicotinate and nicotinamide metabolism | 2.7 × 10−3 | 1.4 × 10−2 |
| Pantothenate and CoA biosynthesis | 2.7 × 10−3 | 1.4 × 10−2 |
| Pathway | p-Value | FDR |
|---|---|---|
| Biosynthesis of unsaturated fatty acids | 2.5 × 10−1 | 3.0 × 10−9 |
| Alanine, aspartate and glutamate metabolism | 4.2 × 10−1 | 2.5 × 10−8 |
| Biosynthesis of phenylpropanoids | 5.7 × 10−9 | 2.2 × 10−7 |
| Aminoacyl-tRNA biosynthesis | 2.3 × 10−8 | 5.5 × 10−7 |
| ABC transporters | 2.1 × 10−8 | 5.5 × 10−7 |
| Metabolic pathways | 4.4 × 10−8 | 8.7 × 10−7 |
| Biosynthesis of alkaloids derived from ornithine, lysine and nicotinic acid | 7.1 × 10−8 | 1.2 × 10−6 |
| Arginine and proline metabolism | 4.9 × 10−6 | 7.3 × 10−5 |
| Biosynthesis of plant hormones | 7.6 × 10−6 | 1.0 × 10−4 |
| Reductive carboxylate cycle (CO2 fixation) | 1.2 × 10−5 | 1.2 × 10−4 |
| Glycine, serine and threonine metabolism | 5.0 × 10−5 | 5.4 × 10−4 |
| Pyrimidine metabolism | 1.7 × 10−4 | 1.7 × 10−3 |
| Glyoxylate and dicarboxylate metabolism | 2.3 × 10−4 | 2.1 × 10−3 |
| Two-component system | 2.6 × 10−4 | 2.2 × 10−3 |
| Proximal tubule bicarbonate reclamation | 3.2 × 10−4 | 2.4 × 10−3 |
| Phenylalanine metabolism | 3.0 × 10−4 | 2.4 × 10−3 |
| Biosynthesis of alkaloids derived from shikimate pathway | 4.4 × 10−4 | 3.1 × 10−3 |
| Citrate cycle (TCA cycle) | 6.1 × 10−4 | 4.0 × 10−3 |
| Biosynthesis of alkaloids derived from histidine and purine | 6.4 × 10−4 | 4.0 × 10−3 |
| Propanoate metabolism | 7.3 × 10−4 | 4.3 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bataller, S.; Villacastin, A.J.; Shen, Q.J.; Bergman, C. Comparative Nutritional Assessment and Metabolomics of a WRKY Rice Mutant with Enhanced Germination Rates. Agronomy 2023, 13, 1149. https://doi.org/10.3390/agronomy13041149
Bataller S, Villacastin AJ, Shen QJ, Bergman C. Comparative Nutritional Assessment and Metabolomics of a WRKY Rice Mutant with Enhanced Germination Rates. Agronomy. 2023; 13(4):1149. https://doi.org/10.3390/agronomy13041149
Chicago/Turabian StyleBataller, Santiago, Anne J. Villacastin, Qingxi J. Shen, and Christine Bergman. 2023. "Comparative Nutritional Assessment and Metabolomics of a WRKY Rice Mutant with Enhanced Germination Rates" Agronomy 13, no. 4: 1149. https://doi.org/10.3390/agronomy13041149
APA StyleBataller, S., Villacastin, A. J., Shen, Q. J., & Bergman, C. (2023). Comparative Nutritional Assessment and Metabolomics of a WRKY Rice Mutant with Enhanced Germination Rates. Agronomy, 13(4), 1149. https://doi.org/10.3390/agronomy13041149






