Citrus Disease Image Generation and Classification Based on Improved FastGAN and EfficientNet-B5
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset and Test Environment Setup
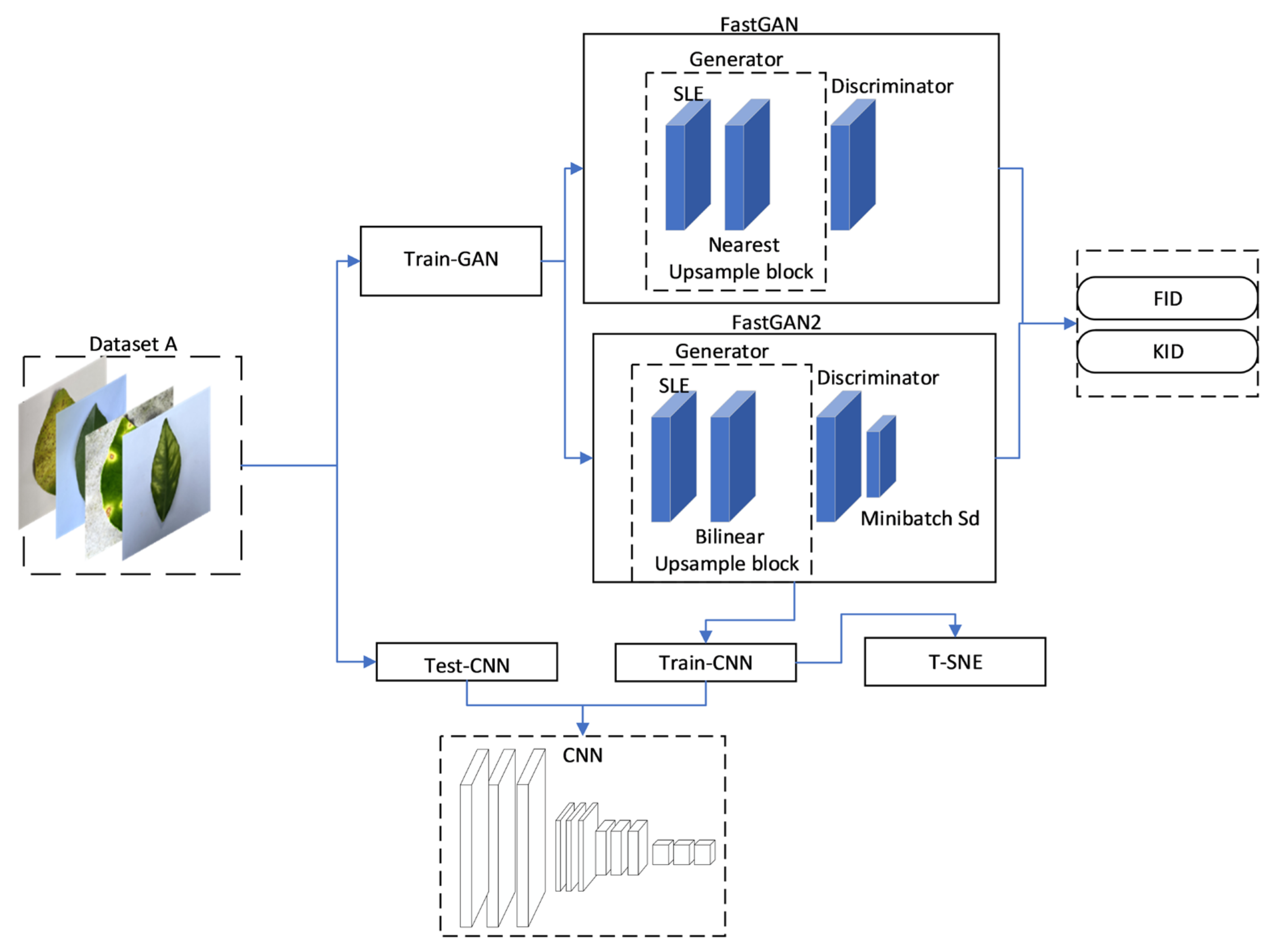
2.2. FastGAN Model
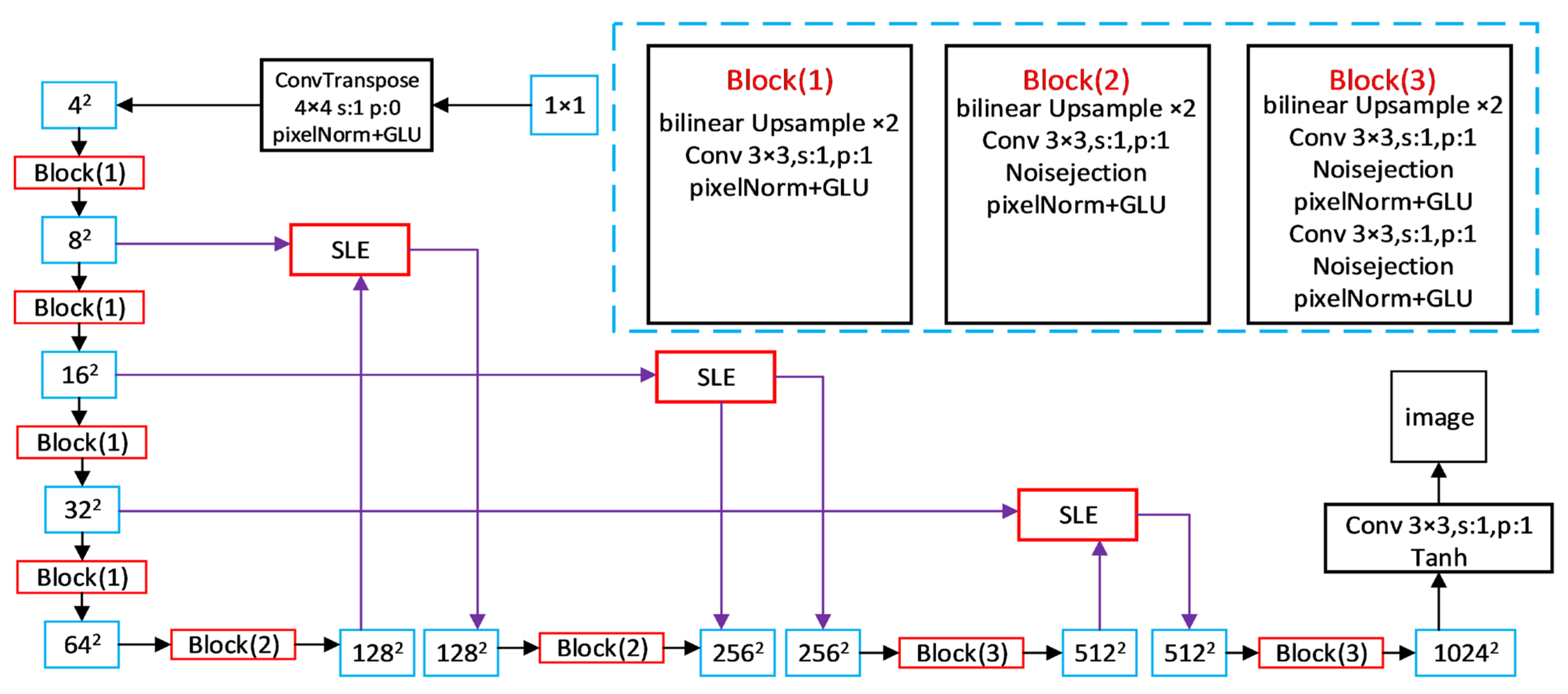
2.3. Improved FastGAN Model
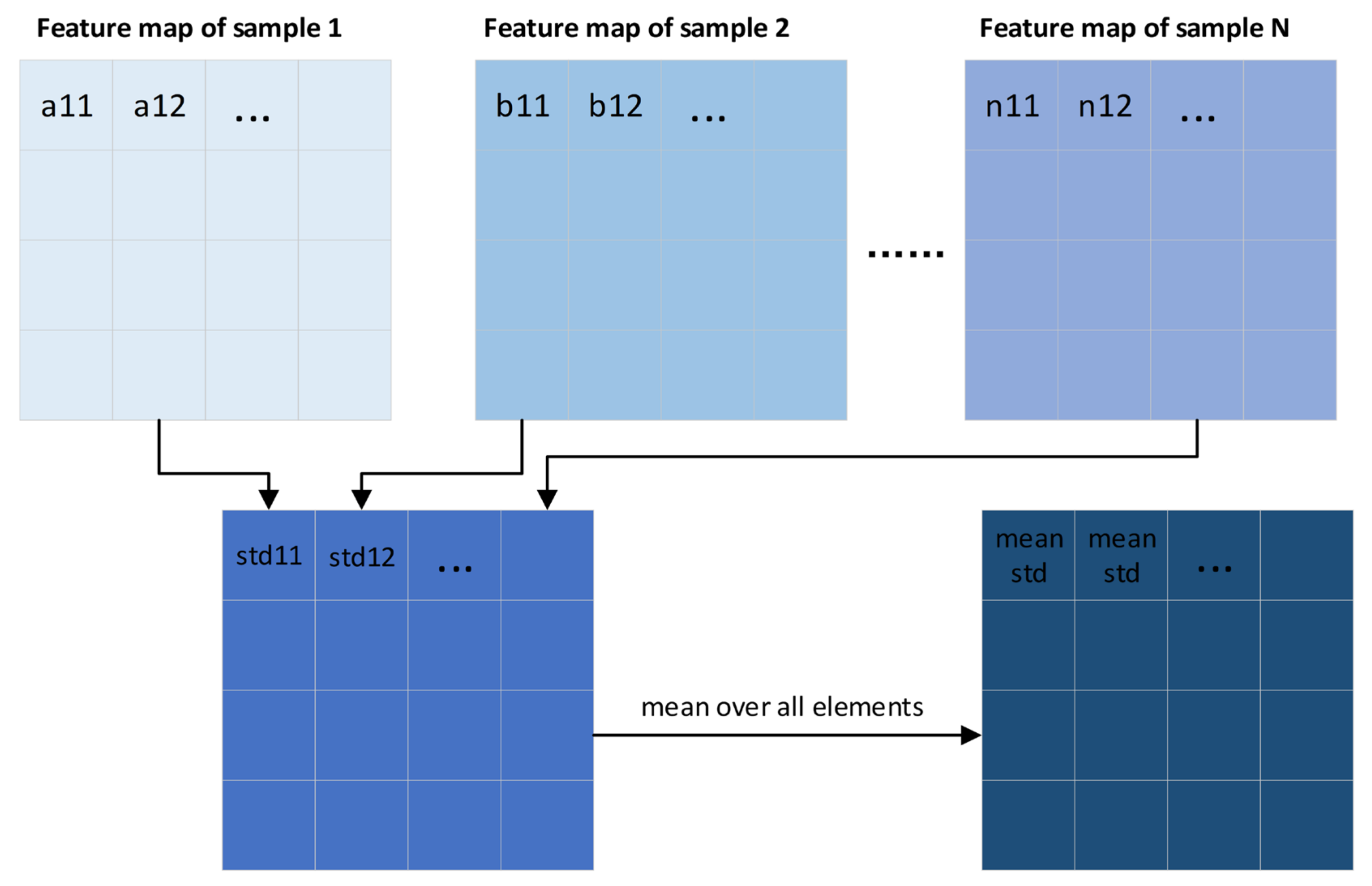
2.4. Improved EfficientNet-B5
3. Results and Discussion
3.1. Generate Image Quality Ratings
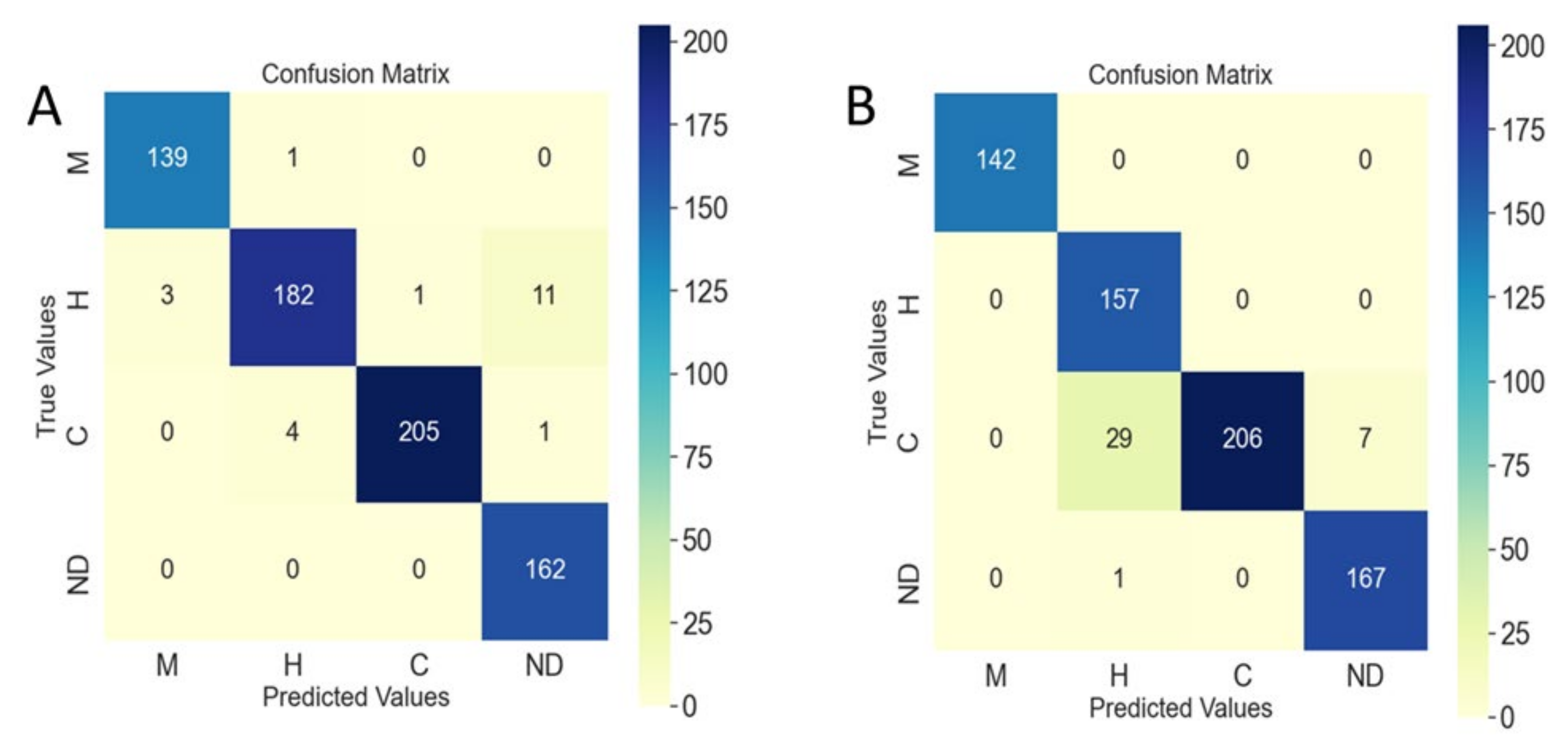
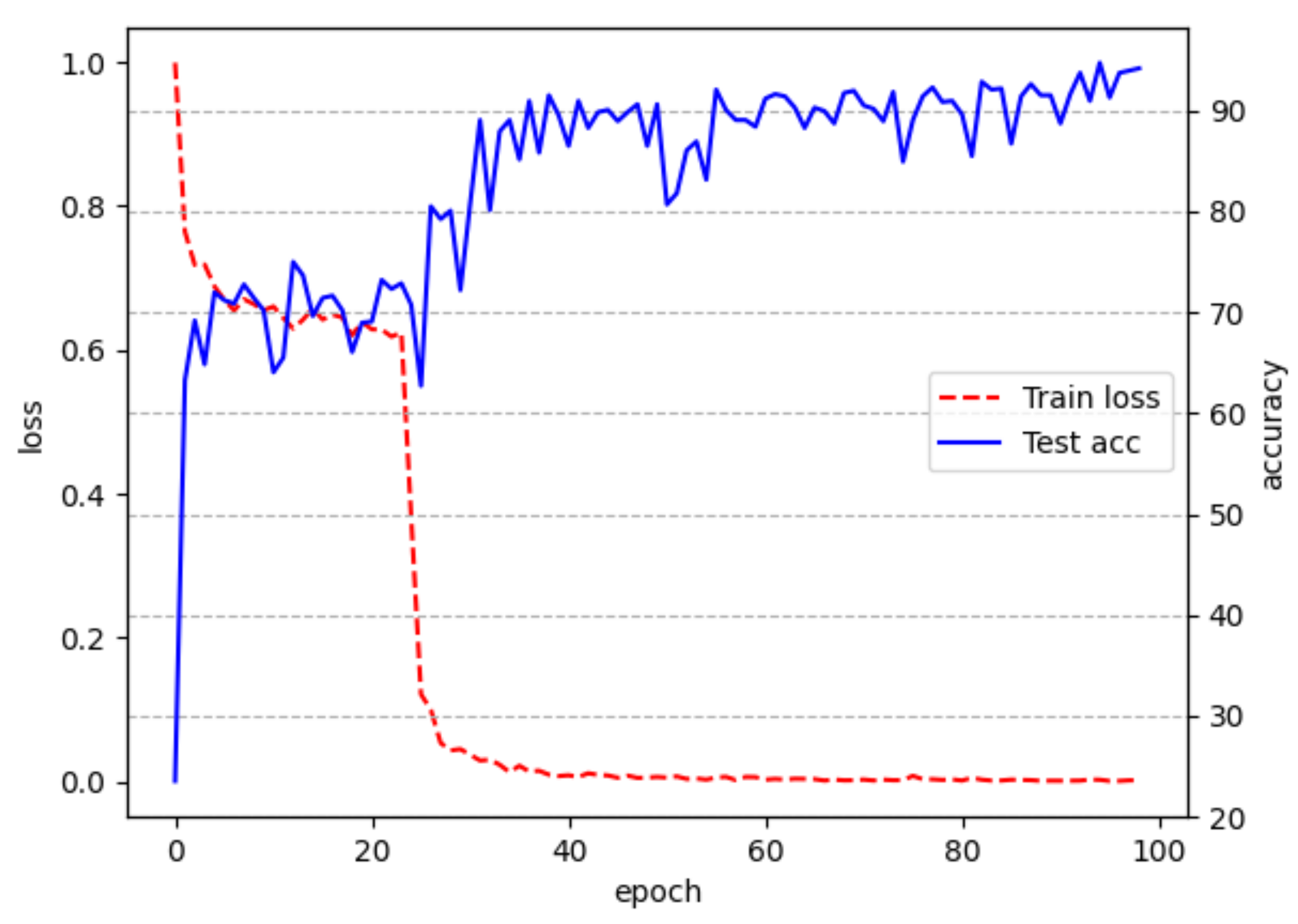
3.2. Classified Network Performance Evaluation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Arcface | Adaptive angular margin |
| FID | Fréchet inception distance |
| KID | Kernel inception distance |
| GAN | Generative adversarial network |
| G | Generator |
| SLE | Skip-layer excitation |
| DNN | Deep neural network |
| WLL | Weight loss landscape |
| AP | Average precision |
| CNN | Convolutional neural network |
| AWP | Adversarial weight perturbation |
| Adam | Adaptive moment estimation |
References
- Iqbal, Z.; Khan, M.A.; Sharif, M.; Shah, J.H.; Rehman, M.H.U.; Javed, K. An automated detection and classification of citrus plant diseases using image processing techniques. Comput. Electron. Agric. 2018, 153, 12–32. [Google Scholar] [CrossRef]
- Gulzar, Y. Fruit Image Classification Model Based on MobileNetV2 with Deep Transfer Learning Technique. Sustainability 2023, 15, 1906. [Google Scholar] [CrossRef]
- Palei, S.; Behera, S.K.; Sethy, P.K. A Systematic Review of Citrus Disease Perceptions and Fruit Grading Using Machine Vision. Procedia Comput. Sci. 2023, 218, 2504–2519. [Google Scholar] [CrossRef]
- Mamat, N.; Othman, M.F.; Abdulghafor, R.; Alwan, A.A.; Gulzar, Y. Enhancing Image Annotation Technique of Fruit Classification Using a Deep Learning Approach. Sustainability 2023, 15, 901. [Google Scholar] [CrossRef]
- Wu, F.Y.; Duan, J.L.; Ai, P.Y.; Chen, Z.Y.; Yang, Z.; Zou, X.J. Rachis detection and three-dimensional localization of cut off point for vision-based banana robot. Comput. Electron. Agric. 2022, 198, 107079. [Google Scholar] [CrossRef]
- Wu, F.Y.; Duan, J.L.; Chen, S.Y.; Ye, Y.X.; Ai, P.Y.; Yang, Z. Multi-Target Recognition of Bananas and Automatic Positioning for the Inflorescence Axis Cutting Point. Front. Plant. Sci. 2021, 12, 705021. [Google Scholar] [CrossRef]
- Tang, Y.C.; Zhou, H.; Wang, H.J.; Zhang, Y.Q. Fruit detection and positioning technology for a Camellia oleifera C. Abel orchard based on improved YOLOv4-tiny model and binocular stereo vision. Expert Syst. Appl. 2023, 211, 118573. [Google Scholar] [CrossRef]
- Da Silva, J.C.F.; Silva, M.C.; Luz, E.J.S.; Delabrida, S.; Oliveira, R.A.R. Using Mobile Edge AI to Detect and Map Diseases in Citrus Orchards. Sensors 2023, 23, 2165. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldu, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Zhang, S.W.; Wu, X.W.; You, Z.H.; Zhang, L.Q. Leaf image based cucumber disease recognition using sparse representation classification. Comput. Electron. Agric. 2017, 134, 135–141. [Google Scholar] [CrossRef]
- Liu, H.Y.; Jiao, L.; Wang, R.J.; Xie, C.J.; Du, J.M.; Chen, H.B.; Li, R. WSRD-Net: A Convolutional Neural Network-Based Arbitrary-Oriented Wheat Stripe Rust Detection Method. Front. Plant. Sci. 2022, 13, 876069. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zhao, M. Research on deep learning in apple leaf disease recognition. Comput. Electron. Agric. 2020, 168, 105146. [Google Scholar] [CrossRef]
- Yao, N.; Ni, F.; Wang, Z.; Luo, J.; Sung, W.K.; Luo, C.; Li, G. L2MXception: An improved Xception network for classification of peach diseases. Plant Methods 2021, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions in Computer Vision and Pattern Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Janarthan, S.; Thuseethan, S.; Rajasegarar, S.; Lyu, Q.; Zheng, Y.Q.; Yearwood, J. Deep Metric Learning Based Citrus Disease Classification with Sparse Data. IEEE Access 2020, 8, 162588–162600. [Google Scholar] [CrossRef]
- Salman, S.; Liu, X. Overfitting Mechanism and Avoidance in Deep Neural Networks. arXiv 2019, arXiv:1901.06566v1. [Google Scholar]
- Li, W.; Chen, C.; Zhang, M.M.; Li, H.C.; Du, Q. Data Augmentation for Hyperspectral Image Classification with Deep CNN. IEEE Geosci. Remote Sens. Lett. 2019, 16, 593–597. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the 28th Conference on Neural Information Processing Systems, Montréal, QC, Canada, 8–13 August 2014. [Google Scholar]
- You, C.; Li, G.; Zhang, Y.; Zhang, X.; Shan, H.; Li, M.; Ju, S.; Zhao, Z.; Zhang, Z.; Cong, W.; et al. CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). IEEE Trans. Med. Imaging 2020, 39, 188–203. [Google Scholar] [CrossRef]
- Lin, Y.; Li, Y.; Cui, H.; Feng, Z. WeaGAN: Generative Adversarial Network for Weather Translation of Image among Multi-domain. In Proceedings of the International Conference on Behavioral, Economic and Socio-Cultural Computing, Beijing, China, 28–30 October 2019. [Google Scholar]
- Niu, S.L.; Li, B.; Wang, X.G.; Lin, H. Defect Image Sample Generation with GAN for Improving Defect Recognition. IEEE Trans. Automat. Sci. Eng. 2020, 17, 1611–1622. [Google Scholar] [CrossRef]
- Ma, L.; Shuai, R.; Ran, X.; Liu, W.; Ye, C. Combining DC-GAN with ResNet for blood cell image classification. Med. Biol. Eng. Comput. 2020, 58, 1251–1264. [Google Scholar] [CrossRef]
- Cap, Q.H.; Uga, H.; Kagiwada, S.; Iyatomi, H. LeafGAN: An Effective Data Augmentation Method for Practical Plant Disease Diagnosis. IEEE Trans. Automat. Sci. Eng. 2020, 9, 1258–1267. [Google Scholar] [CrossRef]
- Xiao, D.Q.; Zeng, R.L.; Liu, Y.F.; Huang, Y.G.; Liu, J.B.; Feng, J.Z.; Zhang, X.L. Citrus greening disease recognition algorithm based on classification network using TRL-GAN. Comput. Electron. Agric. 2022, 200, 107206. [Google Scholar] [CrossRef]
- Karras, T.; Laine, S.; Aittala, M.; Hellsten, J.; Lehtinen, J.; Aila, T. Analyzing and Improving the Image Quality of StyleGAN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 4217–4228. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhu, Y.; Song, K.; Elgammal, A. Towards Faster and Stabilized GAN Training for High-fidelity Few-shot Image Synthesis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Online, 19–25 June 2021. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathe, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant. Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, X.; Zheng, H.-T.; Sun, J. ShuffleNet V2: Practical Guidelines for Efficient CNN Architecture Design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Tolstikhin, I.; Houlsby, N.; Kolesnikov, A.; Beyer, L.; Zhai, X.; Unterthiner, T.; Yung, J.; Steiner, A.; Keysers, D.; Uszkoreit, J.; et al. MLP-Mixer: An all-MLP Architecture for Vision. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Online, 19–25 June 2021. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Kaggle. CCL’20|Kaggle. Available online: https://www.kaggle.com/datasets/downloader007/ccl20 (accessed on 23 August 2022).
- Loshchilov, I.; Hutter, F. SGDR: Stochastic Gradient Descent with Warm Restarts. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive Growing of GANS for Improved Quality, Stability, and Variation. arXiv 2018, arXiv:1710.10196. [Google Scholar]
- Deng, J.; Guo, J.; Xue, N.; Zafeiriou, S. ArcFace: Additive Angular Margin Loss for Deep Face Recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Xu, Q.; Huang, G.; Yuan, Y.; Guo, C.; Sun, Y.; Wu, F.; Weinberger, K. An empirical study on evaluation metrics of generative adversarial networks. arXiv 2018, arXiv:1806.07755. [Google Scholar]
- Wu, D.; Xia, S.-T.; Wang, Y. Adversarial Weight Perturbation Helps Robust Generalization. In Proceedings of the Advances in Neural Information Processing Systems 33: Annual Conference on Neural Information Processing Systems, Online, 6–12 June 2020. [Google Scholar]
- Arora, S.; Hu, W.; Kothari, P.K. An Analysis of the t-SNE Algorithm for Data Visualization. PlMR 2018, 75, 1455–1462. [Google Scholar]
- Bińkowski, M.; Sutherland, D.J.; Arbel, M.; Gretton, A. Demystifying MMD GANs. arXiv 2018, arXiv:1801.01401. [Google Scholar]
- Luque, A.; Carrasco, A.; Martín, A.; de las Heras, A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019, 91, 216–231. [Google Scholar] [CrossRef]
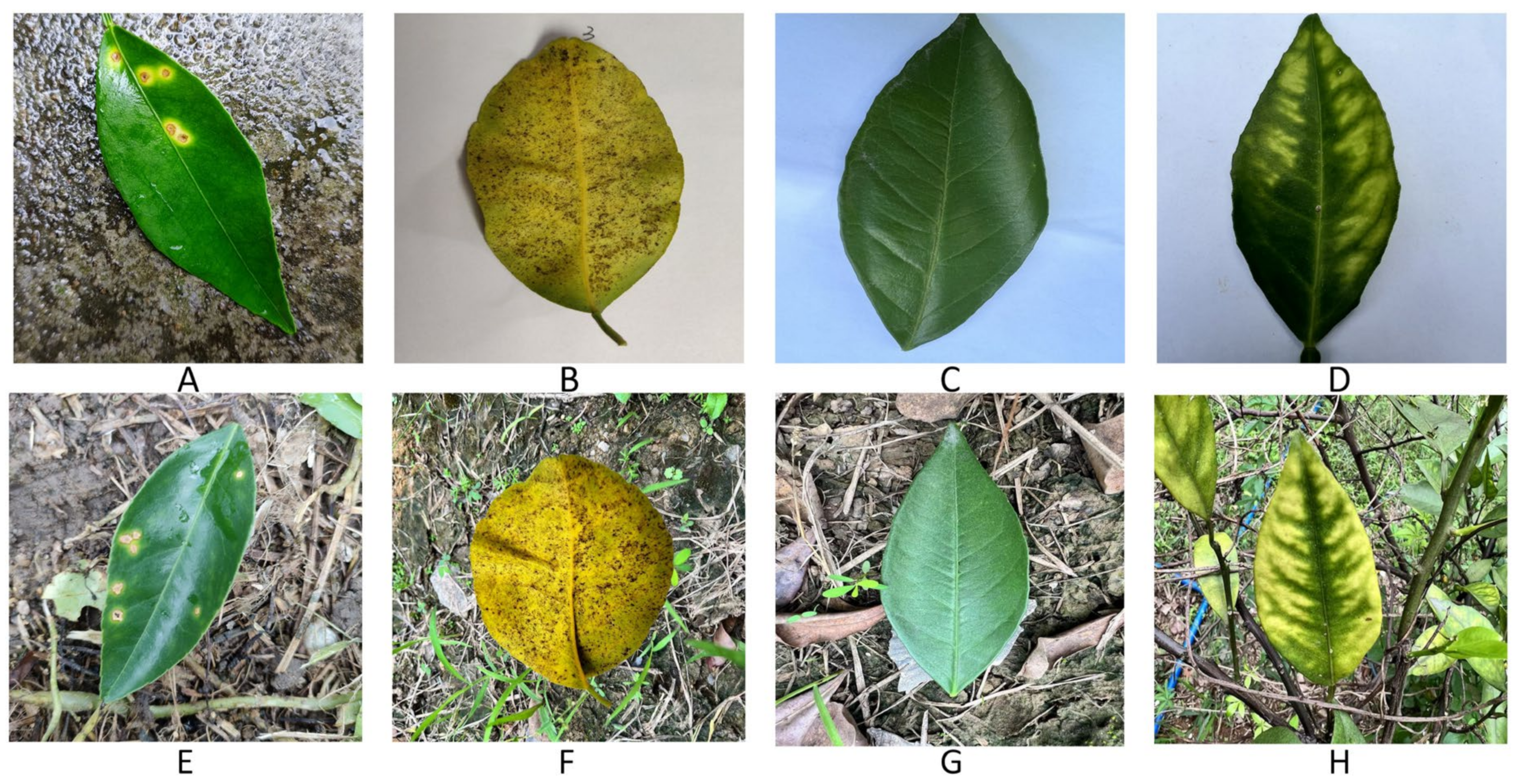

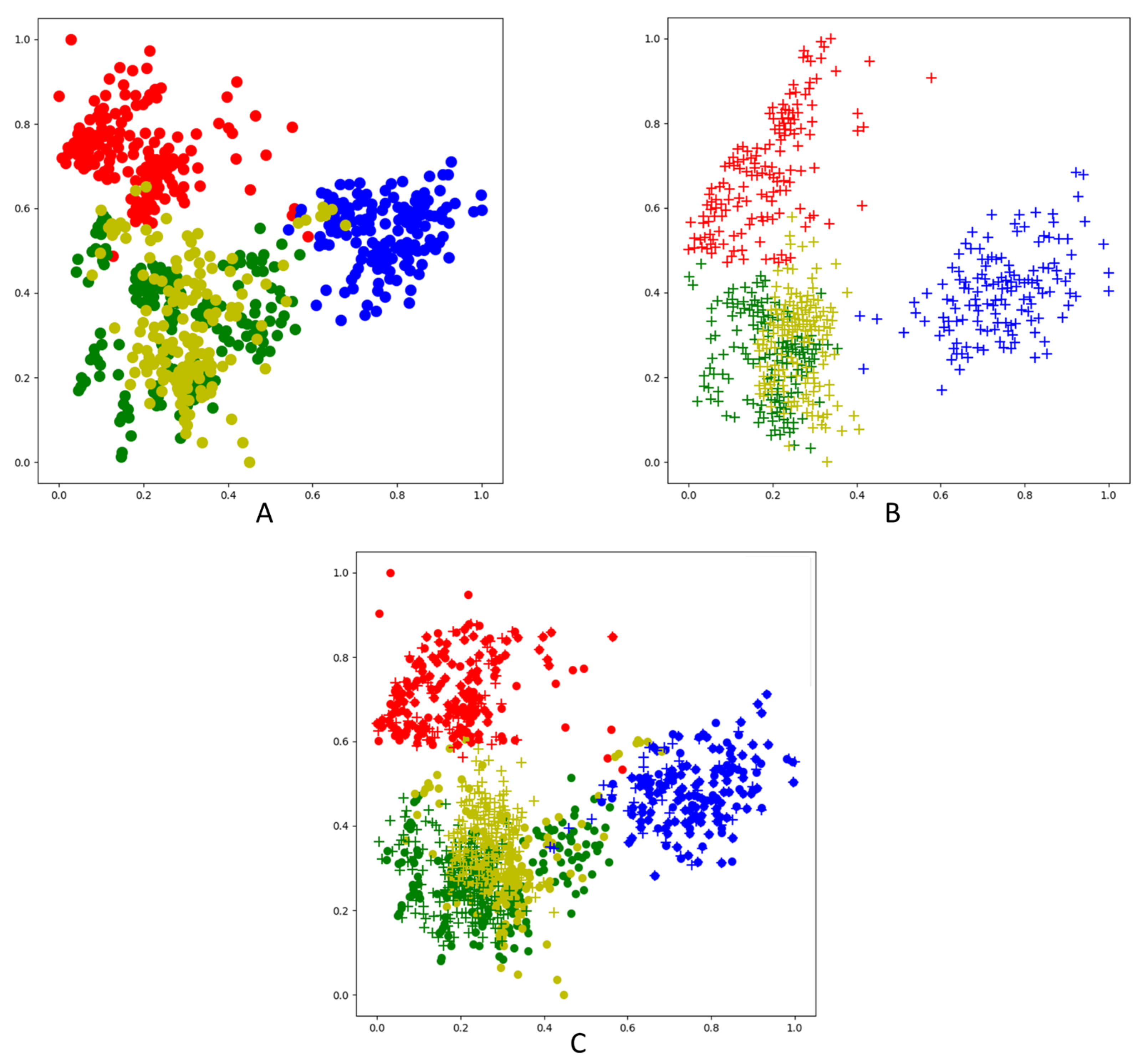




| Leaves | Train-GAN | Train-CNN | Test-CNN |
|---|---|---|---|
| Melanose | 50 | 2000 | 142 |
| Healthy | 50 | 2000 | 187 |
| Canker | 50 | 2000 | 206 |
| Nutritional Deficiency | 50 | 2000 | 174 |
| Leaves | FastGAN2 | FastGAN | ||
|---|---|---|---|---|
| FID | KID | FID | KID | |
| Melanose | 64.73 | 3.01 | 153.52 | 13.79 |
| Healthy | 73.90 | 2.18 | 90.98 | 4.71 |
| Canker | 55.53 | 2.21 | 64.59 | 3.01 |
| Nutritional Deficiency | 53.56 | 2.03 | 54.15 | 2.01 |
| Average | 61.93 | 2.36 | 90.81 | 5.88 |
| Network | Accuracy% | Precision% | Recall% | F1-Score% | ||||
|---|---|---|---|---|---|---|---|---|
| FastGan | FastGan2 | FastGan | FastGan2 | FastGan | FastGan2 | FastGan | FastGan2 | |
| Densenet121 | 82.64 | 90.97 | 88.95 | 93.79 | 74.21 | 91.31 | 72.03 | 91.61 |
| Shufflenetv2 | 80.21 | 94.64 | 83.15 | 94.41 | 70.71 | 94.41 | 70.58 | 94.37 |
| Mlp Mixer | 82.33 | 94.22 | 93.04 | 95.20 | 73.83 | 94.12 | 73.09 | 94.47 |
| MobileNetV3 | 82.29 | 92.81 | 92.63 | 94.13 | 73.87 | 92.49 | 72.98 | 93.06 |
| ResNet50 | 80.56 | 91.26 | 89.99 | 93.65 | 71.70 | 91.56 | 69.33 | 91.86 |
| Vision Transformer | 88.54 | 93.97 | 89.52 | 94.52 | 81.15 | 94.01 | 83.47 | 94.04 |
| Swin Transformer | 88.62 | 93.42 | 73.87 | 95.19 | 71.17 | 93.61 | 71.73 | 93.88 |
| EfficientNet-B3 | 86.11 | 92.10 | 86.05 | 94.12 | 80.33 | 92.34 | 77.15 | 92.63 |
| EfficientNet-B5 | 90.62 | 94.78 | 95.15 | 96.13 | 86.34 | 94.98 | 88.52 | 95.23 |
| EfficientNet-B5-pro | 94.64 | 97.04 | 95.33 | 97.32 | 94.75 | 96.96 | 94.84 | 97.09 |
| Average | 85.66 | 93.52 | 88.76 | 94.86 | 77.80 | 93.58 | 77.37 | 93.82 |
| Leaves | F1-Score% | Precision% | Recall% | Average% |
|---|---|---|---|---|
| Melanose | 98.58 | 99.29 | 97.89 | 98.59 |
| Healthy | 94.79 | 92.39 | 97.33 | 94.84 |
| Canker | 98.56 | 97.62 | 99.51 | 98.56 |
| Zinc Deficiency | 95.92 | 97.92 | 94 | 95.95 |
| Magnesium Deficiency | 96.93 | 99.3 | 94.67 | 96.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Q.; Guo, Y.; Li, Z.; Song, S.; Lyu, S.; Sun, D.; Wang, Y.; Chen, Z. Citrus Disease Image Generation and Classification Based on Improved FastGAN and EfficientNet-B5. Agronomy 2023, 13, 988. https://doi.org/10.3390/agronomy13040988
Dai Q, Guo Y, Li Z, Song S, Lyu S, Sun D, Wang Y, Chen Z. Citrus Disease Image Generation and Classification Based on Improved FastGAN and EfficientNet-B5. Agronomy. 2023; 13(4):988. https://doi.org/10.3390/agronomy13040988
Chicago/Turabian StyleDai, Qiufang, Yuanhang Guo, Zhen Li, Shuran Song, Shilei Lyu, Daozong Sun, Yuan Wang, and Ziwei Chen. 2023. "Citrus Disease Image Generation and Classification Based on Improved FastGAN and EfficientNet-B5" Agronomy 13, no. 4: 988. https://doi.org/10.3390/agronomy13040988
APA StyleDai, Q., Guo, Y., Li, Z., Song, S., Lyu, S., Sun, D., Wang, Y., & Chen, Z. (2023). Citrus Disease Image Generation and Classification Based on Improved FastGAN and EfficientNet-B5. Agronomy, 13(4), 988. https://doi.org/10.3390/agronomy13040988








