Abstract
Chestnut blight, caused by Cryphonectria parasitica, is a severe disease that may be biologically controlled by the use of hypovirulent strains, but the diversity of the pathogen population affects biocontrol feasibility. Villuercas–Ibores–Jara, the Jerte Valley and La Vera are the main chestnut production districts in Cáceres (central-western Spain). The purpose of this study is to determine the Vegetative Compatibility Groups (VCGs) and mating types in these districts and to identify hypovirulent isolates to be used in biological control. The VCGs were determined by the merging/barrage response; PCR tests were used for the mating type determination and CHV-1 hypovirus detection. In total, 40 sites were surveyed and 269 isolates were obtained, most of them (227) from the Villuercas–Ibores–Jara district, where EU11 was the predominant VCG (88.1%) with EU1 (6.6%) and EU12 (4.4%) being also present. In the Jerte Valley and La Vera, EU1 (61.9%), EU11 (11.9%) and EU12 (11.9%) were the predominant VCGs. Both mating types were detected (48% MAT-1; 50% MAT-2) and in general, only one mating type was found in each site. The presence of Cryphonectria hypovirus 1 (CHV-1), subtype I, was identified in only one isolate (VCG EU11) from Villuercas–Ibores–Jara. The characteristics of the C. parasitica population in this district and the occurrence of CHV-1 hypovirus support the potential of successful biological control in Villuercas–Ibores–Jara using hypovirulent strains, while in the Jerte Valley and La Vera only preventive measures are recommended.
1. Introduction
Cryphonectria parasitica (Murrill) M. E. Barr. is the agent causing chestnut blight, one of the most severe chestnut diseases. The disease was accidentally introduced from Asia into North America in the early 20th Century and it almost caused the extinction of the American chestnut (Castanea dentata (Marsh.) Borkh) [1]. In Europe, C. parasitica was first recorded in 1938, affecting sweet European chestnut (Castanea sativa Mill.) in Italy and from there, the disease spread into the neighbouring regions of France, Switzerland and Slovenia [2,3,4]. To date, chestnut blight is present in all main chestnut-growing areas in continental Europe [5]. In 2011, an outbreak of the disease was discovered in southern England [6] and in further intensive surveys, the disease was detected in more sites [7,8].
Cryphonectria parasitica is a bark pathogen that infects the branches and stems, causing necrotic lesions (cankers) on the bark. The girdling of branches by the fungus induces wilting and dieback of the distal parts. The disease is readily visible from the dry leaves that remain on the twigs and from the copious epicormics shoots produced below the cankers [2,8]. C. parasitica penetrates the host tissue through wounds or growth crack and both sexual and asexual spores (ascospores and conidia) can cause infections. Typical mycelial fans are formed in the bark and cambium, and necrotic lesions (so-called cankers) are produced on the bark. Sexual (perithecia) and asexual (pycnidia) fruiting bodies develop in the stromata. Conidia are mainly splash-dispersed by rain, while ascospores can be wind-dispersed over long distances [5]. In C. parasitica, mating is controlled by a single mating type (MAT) locus, which contains either the MAT-1 or MAT-2 [9], although some isolates may be sexually compatible with both mating types, and may even produce perithecia on their own [10]. Outcrossing and self-fertilisation occur at variable frequencies [11].
Hypovirulence refers to a viral disease in C. parasitica caused by a double-stranded RNA hypovirus [12]. The hypovirus that has been most frequently studied is CHV-1, which reduces the parasitic growth and sporulation capacity of the pathogen [13]. Hypoviruses can be transmitted from infected to non-infected C. parasitica strains via hyphal anastomosis, but the transmission of the hypovirus among fungal isolates is limited by a vegetative incompatibility (vic) system, which involves at least six loci with two alleles at each locus. Allelic combinations at the six vic loci define a total of 64 vic genotypes, which correspond to the 64 vc types (EU-1 to EU-64) that are genetically characterised [14]. However, the presence of vc types incompatible with these 64 described types suggests that the vegetative compatibility in C. parasitica is most likely regulated by more than six vic loci or that additional alleles exist at the known vic loci [5]. This incompatibility system determines the frequencies of hypovirus transmission between isolates, so that frequencies are high when isolates have the same alleles at all loci (they belong to the same vc type or Vegetative Compatibility Group, VCG), but heteroallelism at one or more vic loci generally restricts the hypovirus transmission between isolates that belong to different VCGs [15].
At least 74 VCGs of C. parasitica have been found across Europe [16,17]. The level of diversity of VCGs is an important factor in the spread of hypovirulence in C. parasitica populations and it is a major determinant in the success of disease control by hypovirulence [2,18,19]. Concerning the vertical transmission, the hypovirus is transmitted into asexual spores (conidia) but not into sexual ascospores and, therefore, sexual reproduction in C. parasitica is a major obstacle for the dissemination of the hypovirus [20]. Moreover, sexual reproduction in C. parasitica can increase and maintain vc type diversity through the recombination of polymorphic vic genes 14]. In Europe, hypovirulence is largely a natural phenomenon [2,13]. In areas where no natural hypovirulence is present or it is low, the hypovirus (CHV-1) can be artificially introduced by field application of hypovirulent strains, which, by hyphal anastomosis, will transmit the hypovirus to the virulent C. parasitica population. Based on the transmission of hypovirulence, biological control programmes for chestnut blight have been implemented in France, Italy, Switzerland, Greece, Spain and Portugal [21,22,23,24,25,26]. In Europe, several molecularly identifiable subtypes of CHV-1 have been described, i.e., subtype I, F1, F2, E, D and G [27,28,29].
Sweet chestnut production is currently increasing in the province of Cáceres (Extremadura, central-western Spain) [30]. Sweet chestnut is a crop of major economic significance for the Villuercas–Ibores–Jara district (south-eastern Cáceres), as well as for the Jerte Valley (northern Cáceres), while in La Vera district (north-eastern Cáceres), it has been recently introduced and occupies only a small area. Chestnut blight was first detected in the Villuercas–Ibores–Jara district at the end of the 90s through a personal communication made by a local forestry officer, although there is no written record of it. Since then, the disease has spread throughout the district, becoming a limiting factor for chestnut cultivation in some areas. The information on its presence in the Jerte Valley and La Vera districts is scarce.
In areas with low or no natural hypovirulence, the human-mediated introduction of hypovirulence by way of the treatment of bark cankers with hypovirus-infected C. parasitica strains is recommended [21,22,23]. In order to develop a chestnut blight control programme based on hypovirulence, knowledge of the VCGs of the pathogen occurring in the particular area to be treated is crucial, as well as having hypovirulent strains that are compatible with the local VCGs [2,19,22]. The purposes of this study are: (i) to confirm the presence of C. parasitica in chestnut trees with chestnut blight symptoms in the production districts in Cáceres; (ii) to determine the VCGs present in the different districts; (iii) to identify the mating types present in the different districts; and (iv) to identify hypovirus-infected C. parasitica isolates for further use in biological controls.
2. Material and Methods
2.1. Survey, Sampling and Isolation of C. parasitica
The main sweet chestnut-growing districts in the province of Cáceres (Extremadura, central-western Spain) were surveyed between September and October 2019 and between May and October 2020, 2021 and 2022. The survey sites with symptoms of the disease were identified by the technicians of the chestnut producers’ associations in three different districts, i.e., Villuercas–Ibores–Jara (south-eastern Cáceres), the Jerte Valley and La Vera (northern Cáceres). The number of trees sampled per site varied depending on the area of the site and the number of diseased trees at each site. Bark samples (5–20 cm2) were removed from the margins of the cankers using a razor or a chisel, depending on the thickness of the bark. The surveyed chestnut trees were geo-referenced and only one canker per tree was sampled.
In the laboratory, samples were examined under a dissecting microscope for the presence of perithecia and/or pycnidia. Fragments (approx. 6 × 6 mm) were cut from the samples, the surface disinfected by immersion in 70% ethanol, dried on absorbent paper and placed on a potato dextrose agar medium (PDA) (39 g/L; VWR). Where possible, conidia were extracted from pycnidia with a needle and placed directly onto PDA. The plates were incubated for 3–7 days at room temperature in dark conditions and the growing colonies were transferred to fresh PDA plates (only one isolate per canker). The morphological characteristics of the colonies were assessed after an incubation period of 7 days at 25 °C in dark conditions and 7 days on a laboratory bench at room temperature. Several characters were considered for this characterisation: growth pattern, growth rate, appearance of the mycelium, colony colour and sporulation density (high, medium, low). All the isolates were preserved in two ways: (i) grown on a PDA medium and maintained at 5 °C, and (ii) grown on filter paper discs placed in paper envelopes and maintained at −20 °C. The isolate harbouring the virus was additionally maintained on a PDA medium at 25 °C and replicated frequently (every 1–1.5 months).
2.2. Vegetative Compatibility Groups
The VCGs of the isolates obtained were determined according to the method based on the merging/barrage response of paired isolates [10]. Pairings were performed on a PDA medium in 90-mm diameter Petri plates, with 4–8 pairings per plate, and the conidial inocula of the paired isolates were arranged 3 mm apart. The plates were incubated for 7 days at 25 °C in dark conditions and 7 days on a laboratory bench at room temperature. The formation of a barrier between the paired isolates with a line of pycnidia in the contact zone indicated that they belonged to different VCGs. However, where the colonies merged and no barrier was formed, the isolates belonged to the same VCG. All the isolates from each surveyed site were paired in all combinations to select the tester isolates from each site. The tester isolates from each site were paired with each other in all combinations, as well as with the European vc-type testers representing the most frequent VCGs in Spain and Portugal (EU1, EU2, EU9, EU11, EU12, EU13 and EU28), which were kindly provided by P. Cortesi (Università degli Studi di Milano-Bicocca). Each test was repeated at least twice.
The diversity of VCGs in each population was assessed with Shannon and Wiener diversity index H’ calculated as H’ = −∑ pi ln pi, where pi is the frequency of the ith VCG in each population. When all individuals have the same phenotype (belong to the same VCG), H’ = 0, and when all have different phenotypes (belong to different VCGs), H’ has its highest value [1].
2.3. Mating Types
C. parasitica isolates were grown in potato dextrose broth for 10 days and mycelial mats were removed and washed with sterile distilled water. The DNA was extracted using the SpeedTools Plant DNA extraction kit (Biotools B&M Labs S.A.) according to the manufacturer’s protocol. Previously, the mycelial samples were placed in steel bead tubes and crushed in a SPEX CertiPrep tissue homogeniser (model 1600 miniG®) for 5 min. Mating types were determined by PCR amplification using primers M1-GS1n and M1-GS3-rev for MAT-1 and primers M2-GS3 and gsl-d-1 [31].
Both primer pairs were used for each isolate. A PCR test was carried out on a final volume of 19 μL containing 4 μL of each sample DNA and the reaction mixture (15 μL) consisted of DreamTaq Green Buffer 10× (20 mM), 200 μM of each dNTP, 0.2 µM each primer and DNA polymerase DreamTaq (5 U/μL). A PCR test analysis was performed using a Bio-Rad thermal cycler (model T100) and consisted of an initial denaturation step at 95 °C for 30 s; then 35 cycles of 30 s at 95 °C; 1 min at 66 °C and 4 min at 72 °C, followed by a final extension at 72 °C for 4 min. The PCR products were visualised by electrophoresis in a 1% agarose gel, 1× TBE buffer, 95 V for 55 min.
2.4. Cryphonectria Hypovirus 1 (CHV-1) Detection, Sequencing and Phylogenetic Analysis
C. parasitica isolates showing white-beige colony colour and low sporulation were analysed for CHV-1 detection (Figure 1). This selection was made on the basis of previous reports [5,10,13] that associate the loss of colony pigmentation and the reduced sporulation with mycovirus infection (hypovirulence).
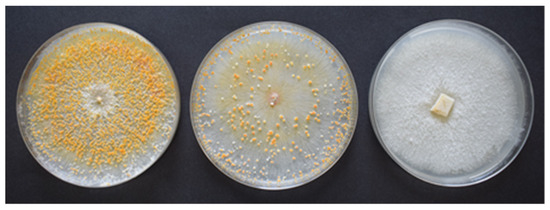
Figure 1.
Colonies of Cryphonectria parasitica with different sporulation density: high (left), not analysed for CHV-1 detection; low (centre) and no sporulation (right), analysed for CHV-1 detection.
C. parasitica isolates were grown in a potato dextrose broth for 10 days and mycelial mats were removed, washed with sterile distilled water and frozen. The frozen mycelium was ground to a fine powder using liquid nitrogen. The total RNA was extracted from the mycelial powder (approx. 50 mg) using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s recommendation. Complementary DNA (cDNA) was generated using the High capacity cDNA Reverse Transcription Kit (Thermo Fisher) according to the manufacturer’s protocol. A PCR test was conducted using primers EP 713-5 and R2280 [26] to amplify a 1439 bp region of open reading frame A (ORF-A) and primers ORF B-12F and ORF B-12R to amplify a 780 bp region of open reading frame B (ORF B) [32]. A PCR test was performed on a final volume of 20 µL, containing 1 µL of the cDNA, 0.2 µL each primer (10 pmol) and 2X PCR SuperMix (Genedirex). Cycling conditions were 2 min at 94 °C followed by 40 cycles of 1 min at 94 °C, 1.5 min at 55 °C, 2.0 min at 72 °C and a final extension of 8 min at 72 °C. The PCR products were visualised by agarose gel electrophoresis on a 1.5% gel stained with GelRed® Nucleic Acid Gel Stain (Biotium, Inc) under UV illumination.
For the CHV1 subtype determination of isolates, the specific region of ORF-A was sequenced using primers hvep1/hvep2 [28] by StabVida laboratories (Caparica, Portugal). The assembled sequences were aligned using Clustal W [33]. The sequences of previously reported subtypes I, F1, F2, D and E deposited in the GenBank (Accession numbers: AF082191, M57938, MF421717, MF431594 and MF431593, respectively) as well as those of CHV-1 subtype I haplotypes [8,28,34] deposited in the GenBank were included for comparison purposes. The alignments were used to reconstruct the phylogenetic trees using the neighbour-joining method with nucleotide identity distances in program MEGA 11.0 [35]. Bootstrap analyses with 1000 replicates were performed to estimate the support for inferred phylogenies [36].
3. Results
3.1. Sampling and Isolation
Forty sweet chestnut growing sites affected by chestnut blight were surveyed. The Villuercas–Ibores–Jara district is the largest of the three surveyed and the disease is highly widespread there. Therefore, most of the surveyed sites (26) were located in this region. Ten and four sites were sampled in the Jerte Valley and La Vera districts, respectively. In adult chestnut trees, the presence of dry branches and/or crown dieback were the most visible symptoms, as well as the presence of mycelial fans when debarking. On the young trees of new plantations, active cankers with orange-reddish colouring were easily found on the branches and stems. Pycnidia were frequently identified on both adult and young trees. Perithecia were not identified in any of the samples examined in the laboratory or in any of the trees surveyed in the field.
The presence of C. parasitica was confirmed in 37 sites, i.e., all the sites surveyed in Villuercas–Ibores–Jara (26) and 11 out of 14 surveyed sites in the Jerte Valley and La Vera (Figure 2). A total of 269 isolates were obtained, most of them (227 isolates, 84.4%) from the Villuercas–Ibores–Jara district, where the number of trees with symptoms of the disease and evident C. parasitica infection in each surveyed site were very high. However, in the Jerte Valley (32 isolates, 11.9%) and La Vera (10 isolates, 3.7%), the distribution of the disease was limited and the number of symptomatic trees in each site was very low, except in one site where the disease was widespread.
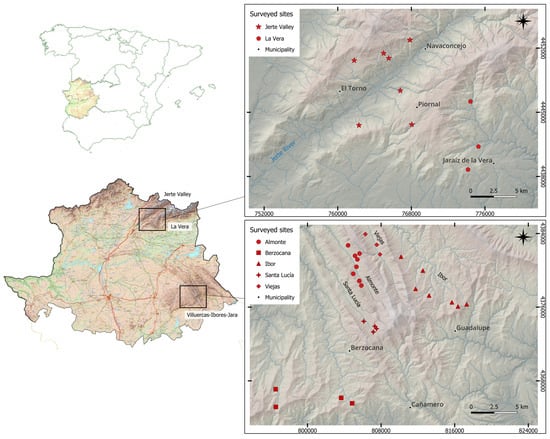
Figure 2.
Map of Spain (on the left) where the area of the Autonomous Region of Extremadura and the Province of Cáceres are highlighted. Enlarged representation of the Jerte Valley and La Vera districts (on the upper right) and the Villuercas–Ibores–Jara district (on the lower right) with the identification of the surveyed sites where C. parasitica was detected.
The morphological characteristics of the isolates corresponded to those typical of C. parasitica. Only six of these isolates, showing a white-beige colony colour and reduced sporulation, were analysed for the presence of CHV-1.
3.2. Vegetative Compatibility Groups
The VCGs of 261 out of the 269 isolates obtained were determined (Figure 3). A total of eight isolates, distributed in three VCGs (named CC1, CC2 and CC3), were incompatible with the European vc type testers used in this study. In the Villuercas–Ibores–Jara district, the predominant VCG was EU11 (200 out of 227 isolates). The topography of this region is characterised by a succession of mountain ranges and parallel valleys (Ibor, Viejas, Almonte, Santa Lucía and Berzocana) with a NW-SE orientation. EU11 was the only VCG present in the Viejas (60 isolates) and Santa Lucía (25 isolates) valleys and it was also the predominant VCG in the Ibor, Almonte and Berzocana valleys (71, 23 and 21 isolates respectively). EU1 was identified (15 isolates) in one site in the Ibor valley and EU12 was only found (10 isolates) on the shady slope of the Almonte valley, while on the sunny slope, only EU11 was present. CC1 was only present in the Berzocana valley (two islolates). In the Jerte Valley and La Vera districts, EU1 was the predominant VCG (26 out of 42 isolates) and EU11, EU12, CC2 and CC3 were identified in lower numbers (5, 5, 4 and 2 isolates, respectively).
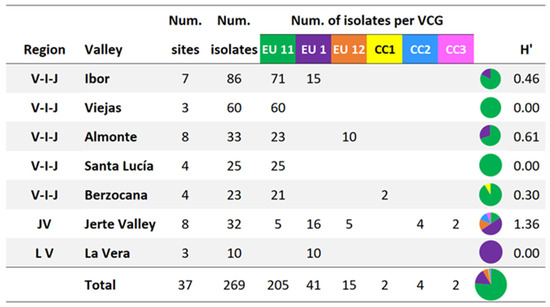
Figure 3.
Vegetative Compatibility Groups (VCGs) and Shannon diversity index (H’) of isolates obtained from surveyed sites in the different valleys of the Villuercas–Ibores–Jara (V-I-J) district and in the Jerte Valley (JV) and La Vera (LV) districts.
The H’-index values (Figure 3) indicate that the greatest diversity in terms of VCGs was found in the Jerte Valley (H’ = 1.36). In the Villuercas–Ibores–Jara district, the diversity was lower in general, with the highest H’-index value corresponding to the Almonte valley (H’ = 0.61).
3.3. Mating Type
The mating type was determined for 133 of the isolates obtained, which were selected to represent the sites sampled and the different VCGs identified (Figure 4). The two MAT idiomorphs were found in almost 1:1 ratio when considering the total number of isolates (64 isolates were identified as MAT-1 and 67 isolates as MAT-2). Two isolates from Villuercas–Ibores–Jara showed an amplification of both MAT-1 and MAT-2 idiomorphs. In this area, the distribution of the mating types varied from one valley to another. In the Santa Lucía and Berzocana valleys, only MAT-2 was identified, whereas in the Viejas and Almonte valleys, MAT-1 was the predominant mating type (92.3% and 91.7%, respectively). In the Ibor valley, 42.1% of the isolates were identified as MAT-1 and 55.3% of them as MAT-2, whereas MAT-1 was the predominant mating type in the Jerte Valley (64.7%) and MAT-2 in La Vera (60.0%).
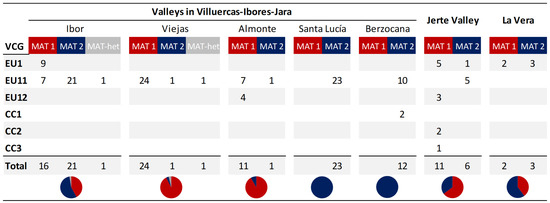
Figure 4.
Distribution of Cryphonectria parasitica mating types (MAT-1, MAT-2 and MAT heterokaryon (MAT-het)) among the Vegetative Compatibility Groups (VCGs) detected in the surveyed sites.
3.4. Cryphonectria Hypovirus 1 (CHV-1) Detection and Subtype Determination
The presence of CHV-1 was detected in only one out of the six isolates analysed, which amplified a PCR product of the expected size with the two primer pairs used. This isolate was obtained from the Villuercas–Ibores–Jara district and belongs to EU11.
The specific region of ORF-A of CHV-1 detected was sequenced (GenBank accession number OQ835557). The first phylogenetic analysis of partial ORF-A sequences, including the sequences of reference isolates from different subtypes (I, F1, F2, D, E), grouped the hypovirus from the Villuercas–Ibores–Jara district into one cluster together with the reference isolates of the CHV-1 subtype I. The second phylogenetic analysis of partial ORF-A sequences, including the sequences of reference isolates from different subtypes (I, F1, F2, D, E) and from the CHV-1 subtype I haplotypes, grouped the hypovirus from the Villuercas–Ibores–Jara district into one cluster together with CHV-1 subtype I isolates from Catalonia (LL0559, LL1783, LL1095, LL1492, LL1383, LL1385) (Figure 5).
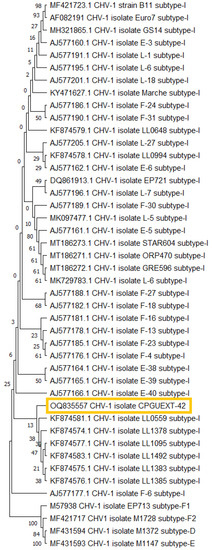
Figure 5.
UPGMA tree derived from ORF A sequences of Cryphonectria hypovirus (CHV-1), showing the sequence generated in the present study (marked in a yellow box) grouping together with other subtype I haplotypes. Sequences of reference isolates from different subtypes (I, F1, F2, D, E) and from CHV-1 subtype I haplotypes were taken from GenBank.
4. Discussion
This study is the first performed on C. parasitica populations and the distribution of the chestnut blight disease in Cáceres (Extremadura).
In the Villuercas–Ibores–Jara district, four VCGs have been identified, with EU11 being the most widespread (88.1% of the isolates), followed by EU1 (6.6%) and EU12 (4.4%); CC1 was present in a lower proportion (0.9%). EU11 has been reported to be present in low proportions in Italy [37], France [17], Croatia [3] and Great Britain [8]. However, EU11 is the predominant VCG in Portugal [38,39] and one of the most common VCGs in Castile and Leon (western Spain) [40], both areas on the border with Extremadura.
The presence of EU1 was limited to one plot in the Ibor valley, with a single owner. The introduction of this VCG may be related to the variety used in the plantation, imported from southern Spain and different from the usual variety in the area (var. Verata). EU12 has only been identified on the shady slope of the Almonte valley, in several plots with different owners, but where pruning and grafting operations were carried out by the same operators. EU12 is the most frequent VCG in south-eastern Europe [3,41,42,43], but its presence is limited in western Europe, where it had only been reported in Portugal and Castile and Leon [38,40].
The situation encountered in the Villuercas–Ibores–Jara district is therefore similar to the situation described in Portugal, where the population of C. parasitica is essentially clonal and dominated by the vc type EU11, although other VCGs (EU1, EU2, EU12 and EU66) are also present and sometimes represented by a few or single isolates [26,38,39]. On the other hand, the distribution of the mating types in this area, associated with its mountainous geography, is favourable for the clonality of the C. parasitica population. The presence of only one mating type in each valley, or the predominance of one of them, implys a very low probability of sexual reproduction. Only in the Ibor valley, both mating types were present in a ratio close to 1:1 (16:21), but they did not occur in the same sites, i.e., the MAT-2 haplotype was present in the sites sampled at the head of the valley, while MAT-1 was present in the remaining sites. The presence of both mating types in a region does not always imply sexual reproduction, as physical proximity between both mating type strains is also important for sexual reproduction to occur [44]. In two of the surveyed sites, two isolates (both EU11) that were amplified for both mating types were detected, although no perithecia were observed in the samples from those sites. C. parasitica has a mixed mating system, with outcrossing and self-fertilisation occurring at variable frequencies [11] and most self-fertile isolates are heterokaryotic for mating type [31]. In the absence of perithecia, we could presume that natural dispersion occurred by conidia, which are mainly splash-dispersed by rain over short distances (a few metres), or washed down the stem and branches [45]. The usual planting frames of chestnut trees in this area are wide (12 × 12 m–15 × 15 m), so the distance between trees could limit the spread of the pathogen by conidia. The wide distribution of the disease in this area suggests a case of human-assisted pathogen dispersion, probably associated with the introduction of infected planting material and agronomic practices without the use of phytosanitary precautions, such as pruning and grafting. It is known that graft unions are especially susceptible to attack by C. parasitica [2].
The situation encountered in the Jerte Valley and La Vera districts was quite different from that of the Villuercas–Ibores–Jara district. The Shannon and Wiener H’-index values indicate that the diversity (in terms of VCGs) in the Jerte Valley was higher than in the Villuercas–Ibores–Jara district. Five VCGs have been found in the Jerte Valley and La Vera, where EU1 was the most widespread VCG (61.9% of the isolates), followed by EU11 (11.9%) and EU12 (11.9%), CC2 (9.5%) and CC3 (4.8%). EU1 has been also detected in other Spanish regions, such as Castile and Leon, Galicia and Asturias [40,46,47] and this VCG is, together with EU2, one of the predominant vc types in Croatia, France, northern Italy, eastern Spain and southern Switzerland [3,35,48]. Both mating types have been found in the Jerte Valley and La Vera, but in general only one mating type was found in each site. Only in one site in La Vera both mating types were found together, and it is known that the second one (MAT 1) had been recently introduced with new chestnut saplings. After being informed, the owner of this plantation decided to remove the new sapling to avoid the risk of perithecia formation. The absence of perithecia and the limited spread of the disease suggest that also in these areas the natural spread occurs only by the action of conidia, although the risk of human-assisted pathogen dispersion is high. Considering the Jerte Valley and La Vera as a whole, the scattered distribution of disease outbreaks, the small number of affected trees in each site, the diversity of VCG and the presence of the two mating types suggest several recent introductions of the pathogen from different sources, probably linked to the trade in plant material, which is very active in this area.
The low proportion of CHV-1-infected isolates found in Cáceres is in line with the low prevalence of the hypovirus reported in other regions of Spain, such as Asturias [49], Galicia [46], Catalonia [34], and Castile and Leon [40], Portugal [38], Bulgaria [50] and Romania [51]. In Europe, natural hypovirulence began to appear and reduce the severity of chestnut blight outbreaks some 10–25 years after the establishment of a new C. parasitica [13]. In Cáceres, the probable date of the initial detection of chestnut blight was the late 90s, and the date of initial detection of the hypovirus is as reported in this paper. The time lapse between the two dates (23 years) corresponds to the predictions based on the correlation found between the year of first identification of chestnut blight and the appearance of healing cankers [2], indicating an initial stage of virus spread. Phylogenetic analysis of sequences indicated that the hypovirus from this study belongs to the Italian subtype (I) of CHV1 and it shows high homology with CHV-1 subtype I isolates from Catalonia (Spain) [34]. However, in Castile and Leon, an area bordering Cáceres, the subtype found is F1 [40]. Subtype I is the most widespread in Europe, which relates to its effective dispersal and establishment capacity compared to other subtypes with low ecological fitness [3,22,27,52]. It is present in Italy, Switzerland, Croatia, Hungary, Greece [27], Macedonia [43], Bosnia-Herzegovina [3], Slovenia [53], France [22], Spain [47], Great Britain [7], Portugal [26] and also in Turkey [54]. Subtypes F1, F2, E and D have been found in France [26], Spain [39,46], Germany [55] and Turkey [54].
Our study reveals differences in C. parasitica populations and distribution patterns of the disease between the Villuercas–Ibores–Jara district and the Jerte Valley and La Vera districts. These differences require the adoption of different disease control strategies in each area. In the Jerte Valley and La Vera districts, preventive measures should be taken to avoid the spread of the disease from existing outbreaks. The use of healthy plant material for planting and grafting, monitoring of new seedlings for early detection of the disease, avoiding wounds in the bark (especially during spore production periods), disinfection of tools, selection of less invasive grafting techniques and extreme care in grafting operations are some of the recommended measures [5]. All these measures are also applicable to the Villuercas–Ibores–Jara district, but in addition, the characteristics of the pathogen populations in the area (the predominance of one VCG (EU11), the absence of the perithecial stage, the geographical distribution of the mating types) and the occurrence of the CHV-1 hypovirus would support the feasibility of successful application of biological control measures using hypovirulent strains.
5. Conclusions
This study is the first on chestnut blight disease in Cáceres (Extremadura) and it reveals differences in C. parasitica populations between the chestnut production districts.
In the Villuercas–Ibores–Jara district, the disease is highly widespread and the characteristics of C. parasitica population are favourable for biological control with hypovirulent strains: there is a predominant VCG (EU11, 88.1%); both mating types are detected, but only one mating type is present in each site; and one CHV-1-infected isolate (belonging to EU-11) is available. Biological control with hypovirulent strains is feasible in this district.
In the Jerte Valley and La Vera districts, the distribution of the disease is limited and EU1 is the most widespread VCG (61.9%), but the diversity (in terms of VCGs) is higher than in the Villuercas–Ibores–Jara district. In general, only one mating type was found in each site. No CHV-1-infected isolate was detected. Preventive measures should be taken to avoid the spread of the disease in this district.
Author Contributions
Conceptualization, M.d.C.R.-M., P.S.-P. and M.D.O.; data curation, M.d.C.R.-M., and M.B.G.-G.; formal analysis, M.d.C.R.-M. and M.D.O.; funding acquisition, M.d.C.R.-M., P.S-P. and M.D.O.; investigation, M.d.C.R.-M., P.S.-P., M.D.O., M.B.G.-G. and E.G.; methodology, M.d.C.R.-M., M.B.G.-G. and E.G.; project administration M.d.C.R.-M.; resources, M.d.C.R.-M. and P.S.-P.; supervision, M.d.C.R.-M.; visualisation, M.d.C.R.-M., P.S.-P. and M.D.O.; writing—original draft M.d.C.R.-M.; writing—review and editing, M.d.C.R.-M., P.S.-P., M.D.O., M.B.G.-G. and E.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Junta de Extremadura and co-funded by the European Regional Development Fund (ERDF) (AGA001 (GR18196); MIPEX and CCESAGROS projects) and the European Agricultural Fund for Rural Development (EAFRD) (Operative Group CASTANEA (PGOF/35/2019)). M.B. García-García was the recipient of a PhD grant from INIA (PRE2019-087630).
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA sequences obtained in this study were deposited in GenBank (accession number: OQ835557).
Acknowledgments
The authors thank Efrén Martín (APCV), Ana Carrón (C.C. Navaconcejo) and Ángel Valderas (ACVJ) for their technical assistance in the survey work and Elena Morales for her technical assistance in the laboratory.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- Heiniger, U.; Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Krajačić, M.; Ćurković-Perica, M. Chestnut blight fungus in Croatia: Diversity of vegetative compatibility types, mating types and genetic variability of associated Cryphonectria hypovirus 1. Plant Pathol. 2008, 57, 1086–1096. [Google Scholar] [CrossRef]
- Prospero, S.; Rigling, D. Invasion genetics of the chestnut blight fungus Cryphonectria parasitica in Switzerland. Phytopathology 2012, 102, 73–82. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef]
- Hunter, G.C.; Wylder, B.; Webber, J.F. First finding of Cryphonectria parasitica causing chestnut blight on Castanea sativa trees in England. New Dis. Rep. 2013, 27, 1. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.; Romón-Ochoa, P.; Gorton, C.; Lewis, A.; Rees, H.; van der Linde, S.; Webber, J. High vegetative compatibility diversity of Cryphonectria parasitica infecting sweet chestnut (Castanea sativa) in Britain indicates multiple pathogen introductions. Plant Pathol. 2019, 68, 727–737. [Google Scholar] [CrossRef]
- Romón-Ochoa, P.; Orlovic, J.K.; Gorton, C.; Lewis, A.; van der Linde, S.; Pérez-Sierra, A. New detections of chestnut bligth in Great Britain during 2019-2020 reveal high Cryphonectria parasitica diversity and limited spread of the disease. Plant Pathol. 2021, 71, 793–804. [Google Scholar] [CrossRef]
- Marra, R.E.; Milgroom, M.G. The mating system of the fungus Cryphonectria parasitica: Selfing and self-incompatibility. Heredity 2001, 86, 134–143. [Google Scholar] [CrossRef]
- Bissegger, M.; Rigling, D.; Heiniger, U. Population structure and disease development of Cryphonectria parasitica in European chestnut forests in the presence of natural hypovirulence. Phytopathology 1997, 87, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Marra, R.E.; Cortesi, P.; Bissegger, M.; Milgroom, M.G. Mixed mating in natural populations of the chestnut blight fungus, Cryphonectria parasitica. Heredity 2004, 93, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.H.; Nuss, D.L. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 1992, 257, 800–803. [Google Scholar] [PubMed]
- Milgroom, M.G.; Cortesi, P. Biological control of chestnut blight with hypovirulence: A critical analysis. Annu. Rev. Phytopathol. 2004, 42, 311–338. [Google Scholar] [CrossRef] [PubMed]
- Cortesi, P.; Milgroom, M.G. Genetics of vegetative incompatibility in Cryphonectria Parasit. Appl. Environ. Microb. 1998, 64, 2988–2994. [Google Scholar] [CrossRef]
- Liu, Y.C.; Milgroom, M.G. Correlation between hypovirus transmission and the number of vegetative incompatibility (vic) genes different among isolates from a natural populations of Cryphonectria parasitica. Phytopathology 1996, 86, 1344–1451. [Google Scholar] [CrossRef]
- Cortesi, P.; McCulloch, C.E.; Song, H.Y.; Lin, H.Q.; Milgroom, M.G. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria Parasit. Genetics 2001, 159, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Robin, C.; Anziani, C.; Cortesi, P. Relationship between biological control, incidence of hypovirulence, and diversity of vegetative compatibility types of Cryphonectria parasitica in France. Phytopathology 2000, 90, 730–737. [Google Scholar] [CrossRef]
- Anagnostakis, S.L.; Hau, B.; Kranz, J. Diversity of vegetative compatibility groups of Cryphonectria parasitica in Connecticut and Europe. Plant Dis. 1986, 70, 536–538. [Google Scholar] [CrossRef]
- MacDonald, W.L.; Fulbright, D.W. Biological control of chestnut blight use and limitations of transmissible hypovirulence. Plant Dis. 1991, 75, 656–661. [Google Scholar] [CrossRef]
- Prospero, S.; Conedera, M.; Heiniger, U.; Rigling, D. Saprophytic activity and sporulation of Cryphonectria parasitica on dead chestnut wood in forests with naturally established hypovirulence. Phytopathology 2006, 96, 1337–1344. [Google Scholar] [CrossRef]
- Heiniger, U.; Rigling, D. Application of the Cryphonectria Hypovirus (CHV-1) to control the chestnut blight, experience from Switzerland. Acta Hortic. 2009, 815, 233–245. [Google Scholar] [CrossRef]
- Robin, C.; Lanz, S.; Soutrenon, A.; Rigling, D. Dominance of natural over released biological control agents of the chestnut blight fungus Cryphonectria parasitica in southeastern France is associated with fitness-related traits. Biol. Control 2010, 53, 55–61. [Google Scholar] [CrossRef]
- Prospero, S.; Rigling, D. Using molecular markers to assess the establishment and spread of a mycovirus applied as a biological control agent against chestnut blight. Biocontrol 2016, 61, 313–323. [Google Scholar] [CrossRef]
- Diamandis, S. Management of chestnut blight in Greece using hypovirulence and silvicultural interventions. Forests 2018, 9, 492–498. [Google Scholar] [CrossRef]
- de Castilla y León, J. El Chancro Del Castaño: Cryphonectria parasit. 2023. Available online: https://medioambiente.jcyl.es/web/es/medio-natural/chancro-castano-cryphonectria-parasitica.html (accessed on 14 March 2023).
- Coelho, V.; Nunes, L.; Gouveia, E. Short and long term efficacy and prevalence of Cryphonectria parasitica hypovirulent strains released as biocontrol agents of chestnut blight. Eur. J. Plant Pathol. 2021, 159, 769–781. [Google Scholar] [CrossRef]
- Allemann, C.; Hoegger, P.; Heiniger, U.; Rigling, D. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 1999, 8, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Gobbin, D.; Hoegger, P.J.; Heiniger, U.; Rigling, D. Sequence variation and evolution of Cryphonectria hypovirus 1 (CHV-1) in Europe. Virus Res. 2003, 97, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rigling, D.; Borst, N.; Cornejo, C.; Supatashvili, A.; Prospero, S. Genetic and phenotypic characterization of Cryphonectria hypovirus 1 from Eurasian Georgia. Viruses 2018, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- MAPA. Avance de datos de Frutales no cítricos y Frutales secos 2021. In Crop Surface Areas and Annual Production from Ministry of Agriculture, Fisheries and Food (Government of Spain); MAPA: Palacio de Fomento Madrid, Spain, 2022; Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/agricultura/superficies-producciones-anuales-cultivos/ (accessed on 22 March 2023).
- McGuire, I.C.; Marra, R.E.; Milgroom, M.G. Mating-type heterokaryosis and selfing in Cryphonectria parasitica. Fungal Genet. Biol. 2004, 41, 521–533. [Google Scholar] [CrossRef]
- Feau, N.; Dutech, C.; Brusini, J.; Rigling, D.; Robin, C. Multiple introductions and recombination in Cryphonectria hypovirus 1: Perspective of a sustainable biological control of chestnut blight. Evol. Appl. 2014, 9, 580–596. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Bassie, L.; Oliach, D.; Gomez, M.; Medina, V.; Liu, B.; Colinas, C. Cryphonectria hypovirus 1 (CHV-1) survey reveals low occurrence and diversity of subtypes in NE Spain. For. Pathol. 2015, 45, 51–59. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies—An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Cortesi, P.; Milgroom, M.G.; Bisiach, M. Distribution and diversity of vegetative compatibility types in subpopulations of Cryphonectria parasitica in Italy. Mycol. Res. 1996, 100, 1087–1093. [Google Scholar] [CrossRef]
- Bragança, H.; Simões, S.; Onofre, N.; Tenreiro, R.; Rigling, D. Cryphonectria parasitica in Portugal: Diversity of vegetative compatibility types, mating types, and occurrence of hypovirulence. For. Pathol. 2007, 37, 391–402. [Google Scholar] [CrossRef]
- Gouveia, E.; Pereira, E.; Araújo, A.; Coelho, V.; Castro, J.; Bragança, H.; Martins, L. Cancro do Castanheiro em Trás-os-Montes (Portugal): Incidência atual e estudo da estrutura populacional de Cryphonectria parasitica para a introdução da luta biológica por hipovirulência. Gaia Sci. 2016, 10, 75–83. [Google Scholar] [CrossRef]
- Zamora, P.; Martin, A.B.; Rigling, D.; Diez, J.J. Diversity of Cryphonectria parasitica in western Spain and identification of hypovirus-infected isolates. For. Pathol. 2012, 42, 412–419. [Google Scholar] [CrossRef]
- Milgroom, M.G.; Sotirovski, K.; Spica, D.; Davis, J.E.; Brewer, M.T.; Milev, M.; Cortesi, P. Clonal population structure of the chestnut blight fungus in expanding ranges in southeastern Europe. Mol. Ecol. 2008, 17, 4446–4458. [Google Scholar] [CrossRef]
- Perlerou, C.; Diamandis, S. Identification and geographic distribution of vegetative compatibility types of Cryphonectria parasitica and occurrence of hypovirulence in Greece. For. Pathol. 2006, 36, 413–421. [Google Scholar] [CrossRef]
- Sotirovski, K.; Milgroom, M.G.; Rigling, D.; Heiniger, U. Occurrence of Cryphonectria hypovirus 1 in the chestnut blight fungus in Macedonia. For. Pathol. 2006, 36, 136–143. [Google Scholar] [CrossRef]
- Ahmad, F.; Baric, S. Genetic diversity of Cryphonectria parasitica causing chestnut blight in South Tyrol (northern Italy). Eur. J. Plant Pathol. 2022, 162, 621–635. [Google Scholar] [CrossRef]
- Griffin, G.J. Chestnut blight and its control. Hortic. Rev. 1986, 8, 291–336. [Google Scholar]
- Montenegro, D.; Aguin, O.; Sainz, M.J.; Hermida, M.; Mansilla, J.P. Diversity of vegetative compatibility types, distribution of mating types and occurrence of hypovirulence of Cryphonectria parasitica in chestnut stands in NW Spain. For. Ecol. Manag. 2008, 256, 973–980. [Google Scholar] [CrossRef]
- González-Varela, G.; González, A.J.; Milgroom, M.G. Clonal population structure and introductions of the chestnut blight fungus, Cryphonectria parasitica, in Asturias, northern Spain. Eur. J. Plant Pathol. 2011, 131, 67–79. [Google Scholar] [CrossRef]
- Robin, C.; Heiniger, U. Chestnut blight in Europe: Diversity of Cryphonectria parasitica, hypovirulence and biocontrol. For. Snow Landsc. Res. 2001, 76, 361–367. [Google Scholar]
- Trapiello, E.; Rigling, D.; González, A.J. Occurrence of hypovirus-infected Cryphonectria parasitica isolates in northern Spain: An encouraging situation for biological control of chestnut blight in Asturian forests. Eur. J. Plant Pathol. 2017, 149, 503–514. [Google Scholar] [CrossRef]
- Risteski, M.; Milev, M.; Rigling, D.; Milgroom, M.G.; Bryner, S.F.; Sotirovski, K. Distribution of chestnut blight and diversity of Cryphonectria parasitica in chestnut forests in Bulgaria. For. Pathol. 2013, 43, 437–443. [Google Scholar] [CrossRef]
- Adamcikova, K.; Ondruskova, E.; Kadasi-Horakova, M.; Botu, M.; Kobza, M.; Achim, G. Distribution and population structure of the chestnut blight fungus in Romania. Plant Protect. Sci. 2015, 51, 141–149. [Google Scholar] [CrossRef]
- Chen, B.; Nuss, D.L. Infectious cDNA clone of hypovirus CHV1-Euro7: A comparative virology approach to investigate virus-mediated hypovirulence of chestnut blight fungus Cryphonectria parasitica. J. Virol. 1999, 73, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Ćurković Perica, M. Diversity of vegetative compatibility types and mating types of Cryphonectria parasitica in Slovenia and occurrence of associated Cryphonectria hypovirus 1. Plant Pathol. 2011, 60, 752–761. [Google Scholar] [CrossRef]
- Akilli, S.; Serce, C.U.; Katircioglu, Y.Z.; Maden, S.; Rigling, D. Characterization of hypovirulent isolates of the chestnut blight fungus, Cryphonectria parasitica from the Marmara and Black Sea regions of Turkey. Eur. J. Plant Pathol. 2013, 135, 323–334. [Google Scholar] [CrossRef]
- Peters, F.S.; Busskamp, J.; Prospero, S.; Rigling, D.; Metzler, B. Genetic diversification of the chestnut blight fungus Cryphonectria parasitica and its associated hypovirus in Germany. Fungal Biol. 2014, 118, 193–210. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).