Nontarget Site-Based Resistance to Fenoxaprop-P-ethyl and Candidate Genes Involved in Alopecurus japonicus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. RNA Extraction, Library Construction, and Illumina Sequencing
2.3. Identification of Differentially Expressed Genes (DEGs) and Functional Annotation
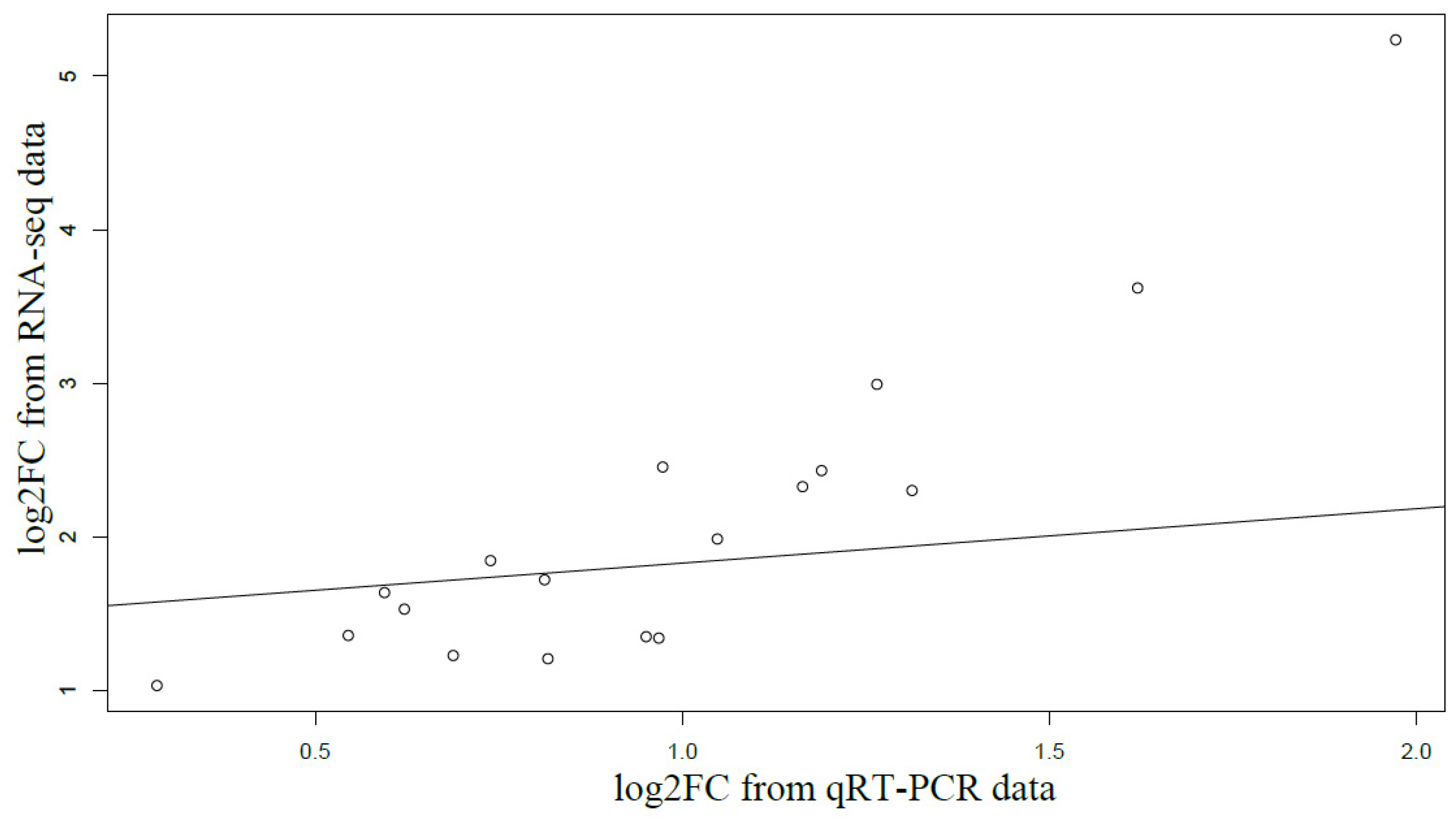
2.4. Quantitative Real-Time PCR (qRT-PCR) Validation
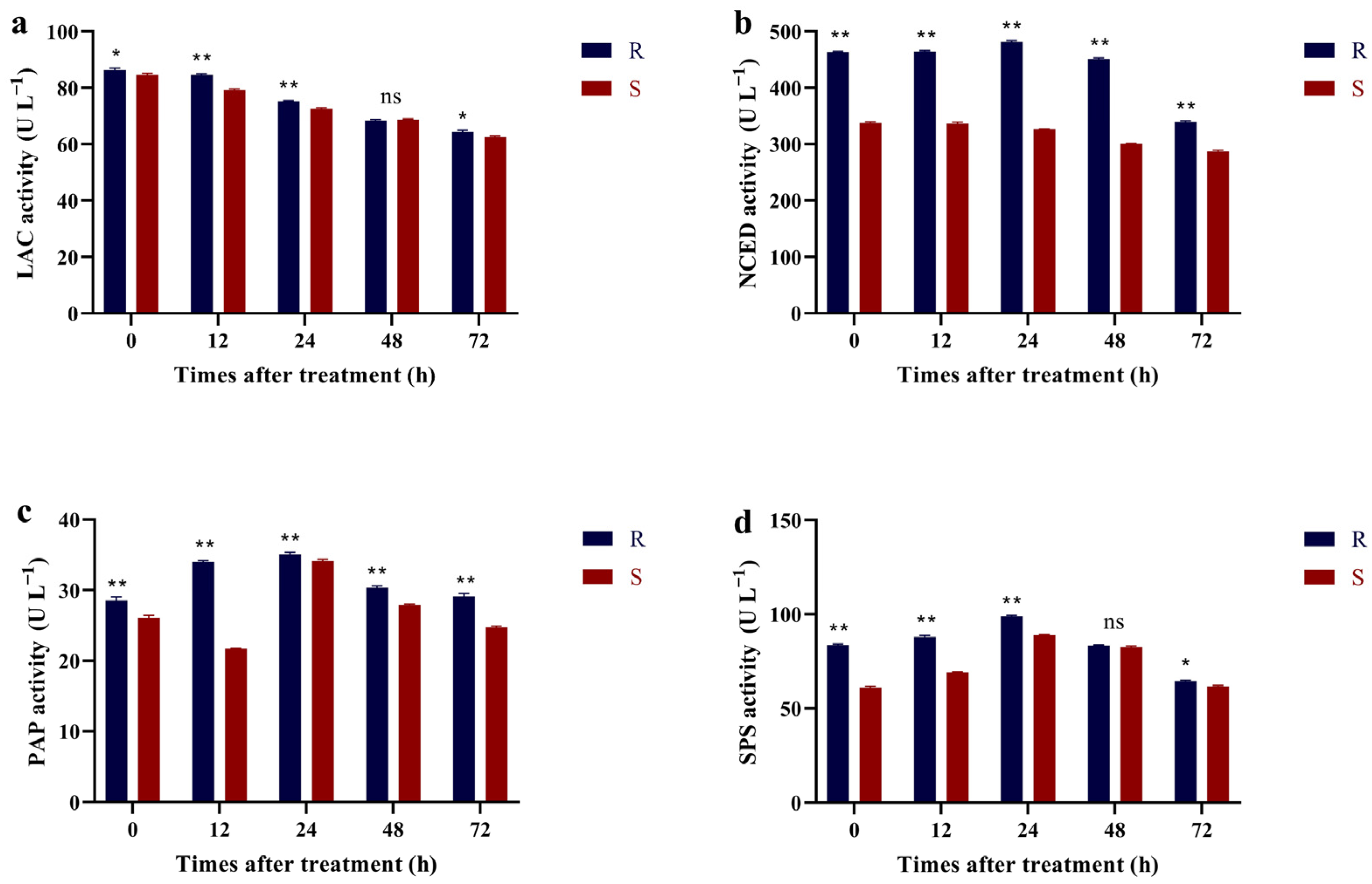
2.5. Activity Assay of Related Metabolic Enzymes
3. Results
3.1. Transcriptome Sequencing and Assembly
3.2. Differential Gene Expression and Functional Annotation
3.3. Candidate NTSR Genes and qPT-PCR Validation
3.4. Related Metabolic Enzyme Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.W.; Wang, J.; DiTommaso, A.; Zhang, C.X.; Zheng, G.P.; Liang, W.; Islam, F.; Yang, C.; Chen, X.X.; Zhou, W.J. Weed research status, challenges, and opportunities in China. Crop Prot. 2020, 134, 104449. [Google Scholar] [CrossRef]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef]
- Comont, D.; Lowe, C.; Hull, R.; Crook, L.; Hicks, H.L.; Onkokesung, N.; Beffa, R.; Childs, D.Z.; Edwards, R.; Freckleton, R.P.; et al. Evolution of generalist resistance to herbicide mixtures reveals a trade-off in resistance management. Nat. Commun. 2020, 11, 3086. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.P.; He, Z.Z.; Liu, T.T.; Li, J.; Dong, L.Y. A novel mutation Asp-2078-Glu in ACCase confers resistance to ACCase herbicides in barnyardgrass (Echinochloa crus-galli). Pestic. Biochem. Physiol. 2020, 168, 104634. [Google Scholar] [CrossRef]
- Ghanizadeh, H.; Harrington, K.C. Non-target site mechanisms of resistance to herbicides. Crit. Rev. Plant Sci. 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef]
- Chen, W.; Wu, L.M.; Wang, J.Z.; Yu, Q.; Bai, L.Y.; Pan, L. Quizalofop-p-ethyl resistance in Polypogon fugax involves glutathione S-transferases. Pest Manag. Sci. 2020, 76, 3800–3805. [Google Scholar] [CrossRef]
- Pan, L.; Yu, Q.; Wang, J.Z.; Han, H.P.; Mao, L.F.; Nyporko, A.; Maguza, A.; Fan, L.J.; Bai, L.Y.; Powles, S. An ABCC-type transporter endowing glyphosate resistance in plants. Proc. Natl. Acad. Sci. USA 2021, 118, 33846264. [Google Scholar] [CrossRef] [PubMed]
- Von, R.U.; Hüttl, R.; Lottspeich, F.; Gierl, A.; Frey, M. Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 2001, 28, 633–642. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.Y.; Wang, H.Z.; Bai, S.; Wang, Q.; Liu, W.T.; Wang, J.X. Acetolactate synthase overexpression in mesosulfuron-methyl-resistant shortawn foxtail (Alopecurus aequalis Sobol.): Reference gene selection and herbicide target gene expression analysis. J. Agric. Food Chem. 2018, 66, 9624–9634. [Google Scholar] [CrossRef]
- Bi, Y.L.; Liu, W.T.; Guo, W.L.; Li, L.X.; Yuan, G.H.; Du, L.; Wang, J.X. Molecular basis of multiple resistance to ACCase- and ALS-inhibiting herbicides in Alopecurus japonicus from China. Pestic. Biochem. Physiol. 2016, 126, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Q.; Xu, H.L.; Zhang, T.; Bai, C.Q.; Dong, L.Y. Fenoxaprop-P-ethyl resistance conferred by cytochrome P450s and target site mutation in Alopecurus japonicus. Pest Manag. Sci. 2018, 74, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, I.A.; Li, R.; You, Z.; Li, Z. Japanese foxtail (Alopecurus japonicus) resistance to fenoxaprop and pinoxaden in China. Weed Sci. 2012, 60, 167–171. [Google Scholar] [CrossRef]
- Tang, H.; Li, J.; Dong, L.Y.; Dong, A.B.; Lü, B.; Zhu, X.D. Molecular bases for resistance to acetyl-coenzyme A carboxylase inhibitor in Japanese foxtail (Alopecurus japonicus). Pest Manag. Sci. 2012, 68, 1241–1247. [Google Scholar] [CrossRef]
- Xu, H.L.; Zhu, X.D.; Wang, H.C.; Li, J.; Dong, L.Y. Mechanism of resistance to fenoxaprop in Japanese foxtail (Alopecurus japonicus) from China. Pestic. Biochem. Physiol. 2013, 107, 25–31. [Google Scholar] [CrossRef]
- Xu, H.L.; Li, J.; Zhang, D.; Cheng, Y.; Jiang, Y.; Dong, L.Y. Mutations at codon position 1999 of acetyl-CoA carboxylase confer resistance to ACCase-inhibiting herbicides in Japanese foxtail (Alopecurus japonicus). Pest Manag. Sci. 2015, 70, 1894–1901. [Google Scholar] [CrossRef]
- Chen, G.Q.; Wang, L.Y.; Xu, H.L.; Wu, X.B.; Pan, L.; Dong, L.Y. Cross-resistance Patterns to Acetyl-CoA Carboxylase Inhibitors Associated with Different Mutations in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2017, 65, 444–451. [Google Scholar] [CrossRef]
- Yu, X.Y.; Tang, W.; Yang, Y.J.; Zhang, J.P.; Lu, Y.L. Comparative transcriptome analysis revealing the different germination process in aryloxyphenoxypropionate-resistant and APP-susceptible Asia minor bluegrass (Polypogon fugax). Plants 2020, 9, 1191. [Google Scholar] [CrossRef]
- Pan, L.; Gao, H.; Xia, W.W.; Zhang, T.; Dong, L.Y. Establishing a herbicide-metabolizing enzyme library in Beckmannia syzigachne to identify genes associated with metabolic resistance. J. Exp. Bot. 2016, 67, 1745–1757. [Google Scholar] [CrossRef]
- Franco, O.S.; Goldberg, C.A.; Walker, A.; Brazier, H.M.; Onkokesung, N.; Edwards, R. Non-target site herbicide resistance is conferred by two distinct mechanisms in black-grass (Alopecurus myosuroides). Front. Plant Sci. 2021, 12, 636652. [Google Scholar] [CrossRef]
- Gaines, T.A.; Lorentz, L.; Figge, A.; Herrmann, J.; Maiwald, F.; Ott, M.; Han, H.P.; Busi, R.; Yu, Q.; Powles, S.B.; et al. RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 2014, 78, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.L.; Li, J.; Wu, R.H.; Su, W.C.; Wu, X.B.; Wang, L.Y.; Dong, L.Y. Identification of reference genes for studying herbicide resistance mechanisms in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2017, 65, 557–566. [Google Scholar] [CrossRef]
- Tan, L.R.; Lu, Y.C.; Zhang, J.J.; Luo, F.; Yang, H. A collection of cytochrome P450 monooxygenase genes involved in modification and detoxification of herbicide atrazine in rice (Oryza sativa) plants. Ecotoxicol. Environ. Saf. 2015, 119, 25–34. [Google Scholar] [CrossRef]
- Xiao, Y.; Wen, J.; Meng, R.; Meng, Y.; Zhou, Q.; Nie, Z.L. The expansion and diversity of the CYP75 gene family in Vitaceae. PeerJ 2021, 9, e12174. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.Y.; Ge, L.A.; Zhu, B.L.; Liu, W.T.; Wang, J.X. Target site mutations and cytochrome P450s confer resistance to fenoxaprop-P-ethyl and mesosulfuron-methyl in Alopecurus aequalis. Pest Manag. Sci. 2018, 75, 204–214. [Google Scholar] [CrossRef]
- Ohkawa, H.; Inui, H. Metabolism of agrochemicals and related environmental chemicals based on cytochrome P450s in mammals and plants. Pest Manag. Sci. 2015, 71, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Guo, Q.S.; Wang, J.Z.; Shi, L.; Yang, X.; Zhou, Y.Y.; Yu, Q.; Bai, L.Y. Cyp81a68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in echinochloa crus-galli. J. Hazard. Mater. 2022, 428, 128225. [Google Scholar] [CrossRef]
- Zhang, X.D.; Zhao, K.X.; Yang, Z.M. Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus. Gene 2018, 664, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Windsor, B.; Roux, S.J.; Lloyd, A. Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 428–433. [Google Scholar] [CrossRef]
- Xi, J.; Xu, P.; Xiang, C.B. Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J. 2012, 69, 782–791. [Google Scholar] [CrossRef]
- Zhao, N.; Li, W.; Bai, S.; Guo, W.L.; Yuan, G.H.; Wang, F.; Liu, W.T.; Wang, J.X. Transcriptome profiling to identify genes involved in mesosulfuron-methyl resistance in Alopecurus aequalis. Front. Plant Sci. 2017, 8, 1391. [Google Scholar] [CrossRef]
- Liu, W.T.; Bai, S.; Zhao, N.; Jia, S.S.; Li, W.; Zhang, L.L.; Wang, J.X. Non-target site-based resistance to tribenuron-methyl and essential involved genes in Myosoton aquaticum (L.). BMC Plant Biol. 2018, 18, 225. [Google Scholar] [CrossRef]
- Zhang, X.L.; Wang, H.Z.; Bei, F.; Wu, C.X.; Zhang, L.L.; Jia, S.S.; Wang, J.X.; Liu, W.T. Investigating the mechanism of metabolic resistance to tribenuron-methyl in Capsella bursa-pastoris (L.) Medik. by full-length transcriptome assembly combined with RNA-Seq. J. Agric. Food Chem. 2021, 69, 3692–3701. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary enzyme based technologies for bioremediation: A review. J. Environ. Manag. 2018, 210, 10–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L.Z.; Wang, X.F.; Chen, B.; Zhao, J.; Cui, J.; Li, Z.K.; Yang, J.; Wu, L.Q.; Wu, J.H.; et al. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2019, 20, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Peng, Q.; Gao, X.; Zhong, S.; Fang, Y.; Yang, X.L.; Ling, Y.; Liu, X.L. Novel fungicide 4-chlorocinnamaldehyde thiosemicarbazide (PMDD) inhibits laccase and controls the causal agent of take-all disease in wheat, Gaeumannomyces graminis var. tritici. J. Agric. Food Chem. 2020, 68, 5318–5326. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.T.; Lu, Y.C.; Zhang, S.; Luo, F.; Yang, H. Rice (Oryza sativa) laccases involved in modification and detoxification of herbicides atrazine and isoproturon residues in plants. J. Agric. Food Chem. 2016, 64, 6397–6406. [Google Scholar] [CrossRef]
- Pan, L.; Zhao, H.W.; Yu, Q.; Bai, L.Y.; Dong, L.Y. miR397/Laccase gene mediated network improves tolerance to fenoxaprop-P-ethyl in Beckmannia syzigachne and Oryza sativa. Front. Plant Sci. 2017, 8, 879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.X.; Ruan, J.H.; Ho, T.H.D.; You, Y.; Yu, T.T.; Quatrano, R.S. Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 2005, 21, 3074–3081. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Alquezar, B.; Zacarías, L. Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J. Exp. Bot. 2006, 57, 633–643. [Google Scholar] [CrossRef]
- Dong, Y.H.; Guo, J.X. Transcriptional analysis of 9-cis-epoxycarotenoid dioxygenase, glucosyltransferase, 8′-hydroxylase and b-glucosidase genes that regulate abscisic acid homeostasis around the onset of grape berry ripening. J. Agric. Sci. Technol. A 2012, 2, 873–881. (In Chinese) [Google Scholar]
- Luo, J.; Chen, S.J.; Cao, S.H.; Zhang, T.; Li, R.R.; Chan, Z.L.; Wang, C.Y. Rose (Rosa hybrida) ethylene responsive factor 3 promotes rose flower senescence via direct activation of the abscisic acid synthesis-related 9-cis-epoxycarotenoid dioxygenase gene. Plant Cell Physiol. 2021, 62, 1030–1043. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, Y.M.; Liu, Y.T.; Zhang, F.; Wang, Z.K.; Wang, H.Y.; Wang, F.; Li, D.P.; Mao, D.D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Wan, X.R.; Li, L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem. Biophys. Res. Commun. 2006, 347, 1030–1038. [Google Scholar] [CrossRef]
- Kraft, M.; Kuglitsch, R.; Kwiatkowski, J.; Frank, M.; Grossmann, K. Indole-3-acetic acid and auxin herbicides up-regulate 9-cis-epoxycarotenoid dioxygenase gene expression and abscisic acid accumulation in cleavers (Galium aparine): Interaction with ethylene. J. Exp. Bot. 2007, 58, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Eliécer, E.M. The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase gene family. Front. Plant Sci. 2015, 6, 341. [Google Scholar] [CrossRef]
- Zhu, X.L.; Lee, S.Y.; Yang, W.T.; Lee, S.W.; Baek, D.W.; Li, M.S.; Kim, D.H. The burholderia pyrrocinia purple acid phosphatase Pap9 mediates phosphate acquisition in plants. J. Exp. Biol. 2019, 62, 342–350. [Google Scholar] [CrossRef]
- Zhu, S.N.; Chen, M.H.; Liang, C.Y.; Xue, Y.B.; Lin, S.L.; Tian, J. Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in Soybean. Front. Plant Sci. 2020, 11, 661. [Google Scholar] [CrossRef]
- Widyasari, K.; Tran, P.T.; Shin, J.; Son, H.; Kim, K.H. Overexpression of a purple acid phosphatase GmPAP2.1 confers soybean mosaic virus resistant in a susceptible soybean cultivar. J. Exp. Bot. 2021, 73, 1623–1642. [Google Scholar] [CrossRef]
- Reddy, C.S.; Kim, K.M.; James, D.; Varakumar, P.; Reddy, M.K. PgPAP18, a heat-inducible novel purple acid phosphatase 18-like gene (PgPAP18-like) from Pennisetum glaucum, may play a crucial role in environmental stress adaptation. Acta Physiol. Plant. 2017, 39, 54. [Google Scholar] [CrossRef]
- Taneja, D.; Das, N. Molecular cloning, sequence analyses, and expression studies of sucrose-phosphate synthase in the potato (Solanum tuberosum L.) cultivars. Acta Physiol. Plant. 2014, 36, 2253–2269. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.G.; Sapeta, H.; Leitão, A.E.; Ramalho, J.C.; Lam, T.T.; Negrão, S.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017, 40, 1197–1213. [Google Scholar] [CrossRef] [PubMed]
- Nemati, F.; Ghanati, F.; Ahmadi Gavlighi, H.; Sharifi, M. Comparison of sucrose metabolism in wheat seedlings during drought stress and subsequent recovery. Biol. Plant. 2018, 62, 595–599. [Google Scholar] [CrossRef]
- Zhang, W.J.; Wang, J.Q.; Huang, Z.L.; Mi, L.; Xu, K.F.; Wu, J.J.; Fan, Y.H.; Ma, S.Y.; Jiang, D.G. Effects of low temperature at booting stage on sucrose metabolism and endogenous hormone contents in winter wheat spikelet. Front. Plant Sci. 2019, 10, 498. [Google Scholar] [CrossRef] [PubMed]
- María Gloria, S.G.; Gerardo, A.A.; José, L.B.; León Francisco, R.H.; Joel Edmundo, L.M.; Lenin, S.C.; Yazmín, C.A.; Miguel, M.T. Arabidopsis thaliana sucrose phosphate synthase (sps) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expression Patterns 2017, 25–26, 92–101. [Google Scholar] [CrossRef]
- Solís-Guzmán, M.G.; Argüello-Astorga, G.; López-Bucio, J.; Ruiz-Herrera, L.F.; López-Meza, J.E.; Sánchez-Calderón, L.; Carreón-Abud, Y.; Martínez-Trujillo, M. Arabidopsis thaliana sucrose phosphate synthase (sps) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expr. Patterns 2017, 25–26, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Liang, C.; Wang, S.K.; Hou, Y.N.; Gao, L.; Liu, L.; He, W.R.; Ma, W.B.; Mo, B.X.; Chen, X.M. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 2018, 9, 5080. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.W.; Liu, W.Z.; Yu, W.Q.; Li, P.; Zhao, C.J.; Yan, F.C.; Wang, G.J.; Zhang, L.G.; Xie, H.P.; Qiu, Z.M. Antitoxin EndoAI can induce disease resistance in tobacco as a protein elicitor. Chem. Biol. Technol. Agric. 2021, 8, 66. [Google Scholar] [CrossRef]
- Xie, X.Z.; Xue, Y.J.; Zhou, J.J.; Zhang, B.; Chang, H.; Takano, M. Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 2011, 4, 688–696. [Google Scholar] [CrossRef]
- Xu, X.; Hayashi, N.; Wang, C.T.; Fukuoka, S.; Kawasaki, S.; Takatsuji, H.; Jiang, C.J. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol. Breed. 2014, 34, 691–700. [Google Scholar] [CrossRef]
- Kim, S.H.; Qi, D.; Ashfield, T.; Helm, M.; Innes, R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 2016, 351, 684–687. [Google Scholar] [CrossRef]
- Wang, J.J.; Chen, J.C.; Li, X.J.; Li, D.; Li, Z.; Cui, H.L. Pro-197-Ser mutation in ALS and high-level GST activities: Multiple resistance to ALS and ACCase inhibitors in Beckmannia syzigachne. Front. Plant Sci. 2020, 11, 572610. [Google Scholar] [CrossRef] [PubMed]




| Gene | Gene Annotation | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|---|
| c80456.graph_c1 | Laccase-15, LAC15 | TCAGGTGAGACAGTTGATGCTTTGG | GAGGCAATAGGCTCGTCAAGTTACC |
| c86897.graph_c0 | Purple acid phosphatase 4, PAP4 | AGCAGCAGCAGCAACACAGTAG | TCATAGCTCCACCAGTCCACATCC |
| c92899.graph_c0 | Purple acid phosphatase 15, PAP15 | GCTACTCCTGCTCATTCGCCAAG | GGTCACTGATCTGTCCACCTTTGC |
| c97785.graph_c0 | 9-cis-epoxycarotenoid dioxygenase NCED5 | CAGAGGTGGAAGCAGAAGCAGTC | CGACTTCGCCATCACCGAGAAC |
| c97938.graph_c1 | Sucrose-phosphate synthase 3, SPS3 | TCCCGTGTGACATTTGCATTAGACC | AGCGTAGCGACTGGACTCTCATC |
| c98820.graph_c0 | Flavonoid 3′-monooxygenase CYP75B4 | TGGTTACTTTGATGCGAGTGCTGAG | TGTGGCTTGATTTGATGGTCTGGAG |
| c97281.graph_c2 | ABC transporter G family member 36, ABCG36 | AGCCATTCCATGTAGTCAAACGA | AAATTGCATTGGCTGCGTTG |
| c94207.graph_c0 | Disease resistance protein RGA3 | TGTGTCACCTTGTTTCAGTGGACTG | CAGCTCATGCCATCTGAACTCGTC |
| c93241.graph_c1 | Late blight resistance protein homolog R1B-17 | TCAGCAACGACAAAGAGGTGAAGAC | GCACTGTAATGGAGCCACGAAGG |
| Sample | Read Number | GC Content | % ≥ Q30 |
|---|---|---|---|
| RCK-1 | 24,623,730 | 55.79% | 95.48% |
| RCK-2 | 27,571,212 | 56.03% | 95.77% |
| RCK-3 | 25,166,323 | 55.38% | 95.42% |
| RCK-4 | 24,346,668 | 55.16% | 95.67% |
| RCK-5 | 24,336,167 | 55.33% | 95.63% |
| SCK-1 | 27,500,747 | 56.57% | 95.81% |
| SCK-2 | 26,315,594 | 54.43% | 95.60% |
| SCK-3 | 26,531,889 | 54.79% | 95.44% |
| SCK-4 | 24,984,973 | 56.16% | 95.99% |
| SCK-5 | 26,884,101 | 55.70% | 95.99% |
| RT-1 | 23,851,499 | 54.61% | 95.78% |
| RT-2 | 27,098,152 | 54.26% | 95.78% |
| RT-3 | 30,950,921 | 53.19% | 95.00% |
| RT-4 | 23,823,583 | 56.63% | 95.91% |
| RT-5 | 27,279,367 | 54.34% | 95.56% |
| ST-1 | 30,872,907 | 55.31% | 95.62% |
| ST-2 | 23,060,205 | 56.20% | 95.93% |
| ST-3 | 25,114,713 | 56.29% | 95.56% |
| ST-4 | 20,558,360 | 55.76% | 95.59% |
| ST-5 | 24,928,922 | 55.83% | 95.84% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Ye, X.; Liang, S.; Cheng, J.; Leng, Q.; Sun, L.; Su, W.; Xue, F.; Dong, L.; Wu, R. Nontarget Site-Based Resistance to Fenoxaprop-P-ethyl and Candidate Genes Involved in Alopecurus japonicus. Agronomy 2023, 13, 1587. https://doi.org/10.3390/agronomy13061587
Xu H, Ye X, Liang S, Cheng J, Leng Q, Sun L, Su W, Xue F, Dong L, Wu R. Nontarget Site-Based Resistance to Fenoxaprop-P-ethyl and Candidate Genes Involved in Alopecurus japonicus. Agronomy. 2023; 13(6):1587. https://doi.org/10.3390/agronomy13061587
Chicago/Turabian StyleXu, Hongle, Xiaofan Ye, Shaoqi Liang, Jingping Cheng, Qiuli Leng, Lanlan Sun, Wangcang Su, Fei Xue, Liyao Dong, and Renhai Wu. 2023. "Nontarget Site-Based Resistance to Fenoxaprop-P-ethyl and Candidate Genes Involved in Alopecurus japonicus" Agronomy 13, no. 6: 1587. https://doi.org/10.3390/agronomy13061587
APA StyleXu, H., Ye, X., Liang, S., Cheng, J., Leng, Q., Sun, L., Su, W., Xue, F., Dong, L., & Wu, R. (2023). Nontarget Site-Based Resistance to Fenoxaprop-P-ethyl and Candidate Genes Involved in Alopecurus japonicus. Agronomy, 13(6), 1587. https://doi.org/10.3390/agronomy13061587






