Apical Dominance and Branching in Plantlets of Colt Cherry Lines Expressing Different Light and Auxin Signalling Sensitivities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Culture Medium
2.2. Experimental Design and Parameters
2.3. Statistical Analysis
3. Results
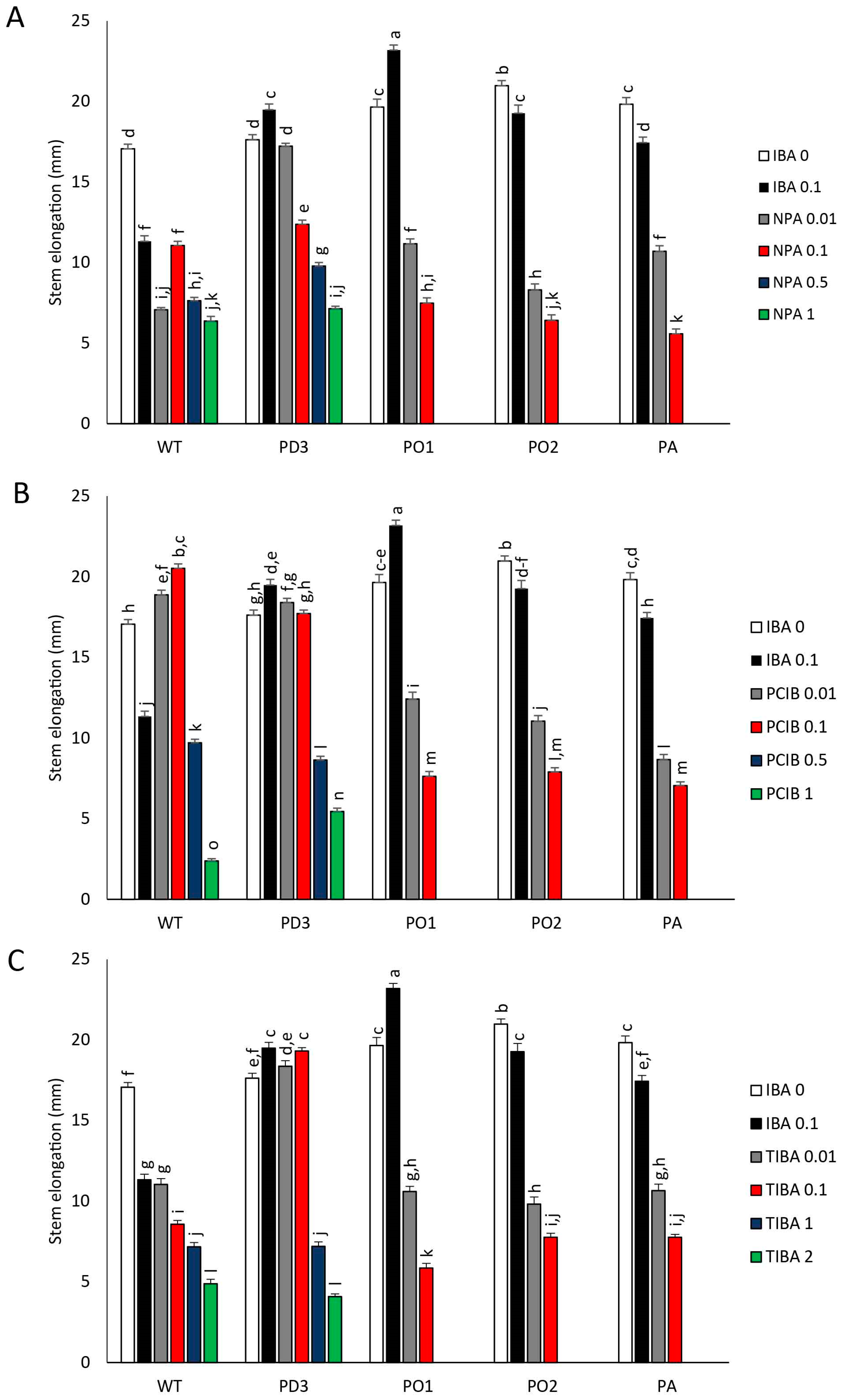
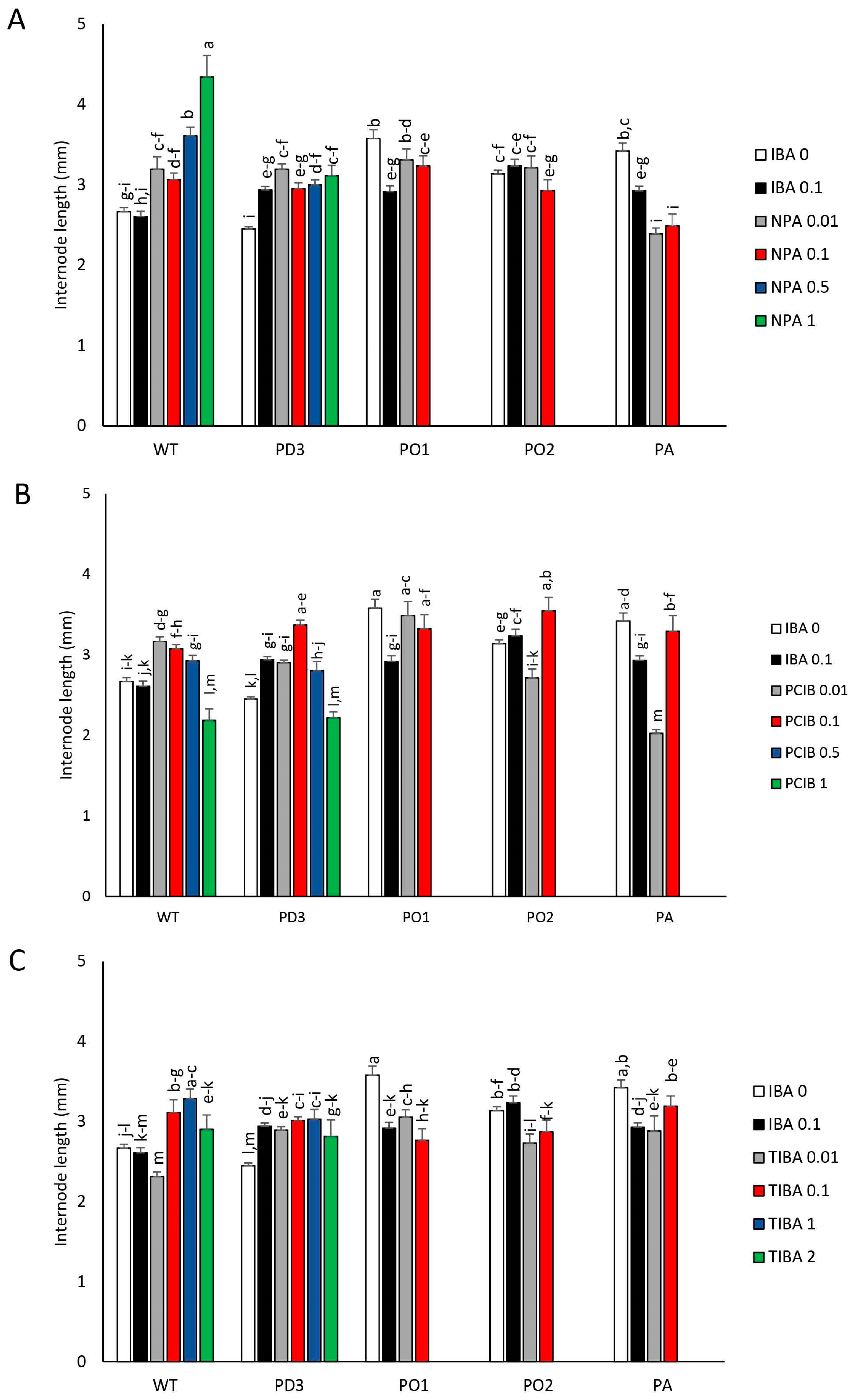
3.1. Plantlet Growth
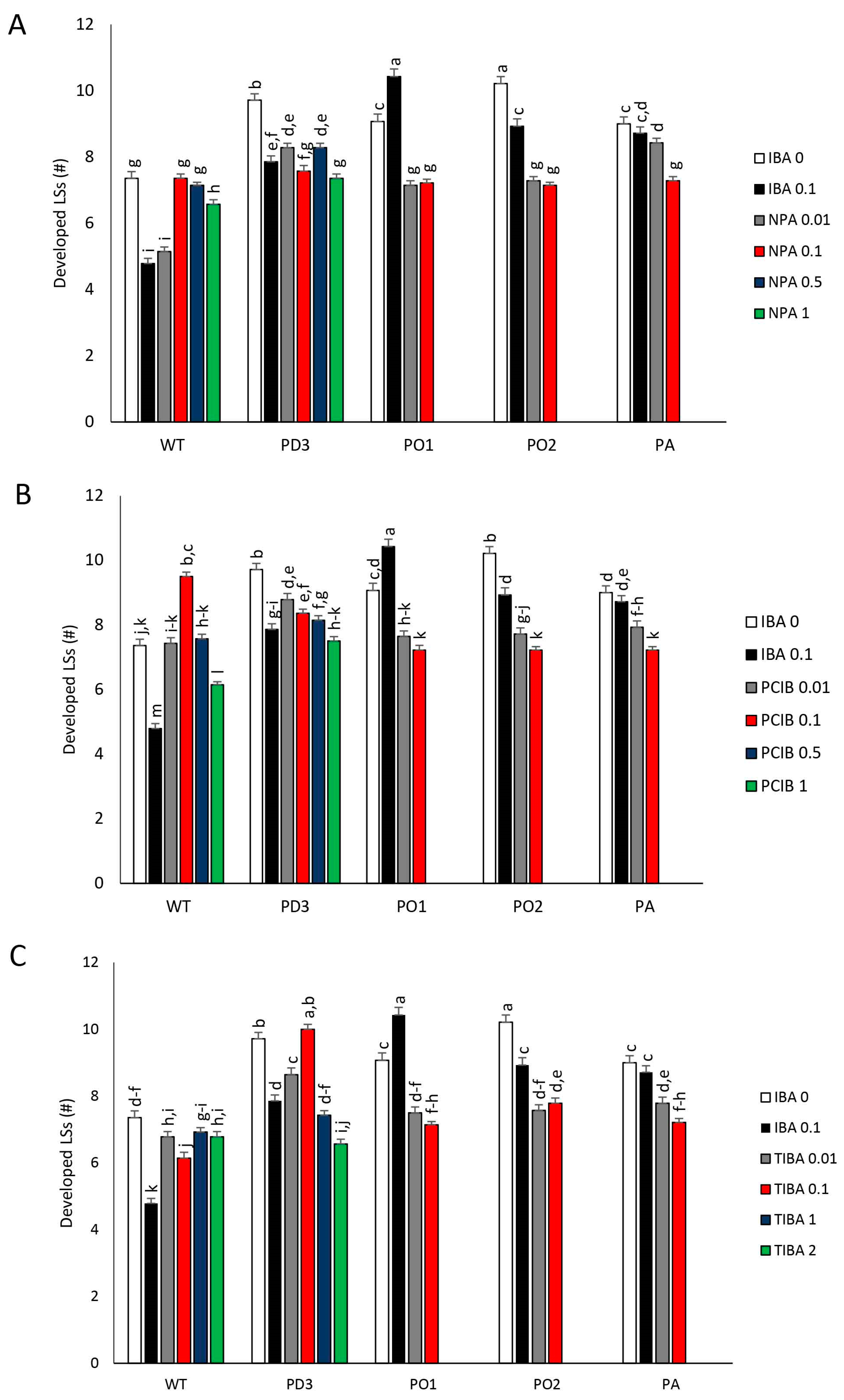
3.2. Development of Nodes (LBs) and Apical Dominance (AD)
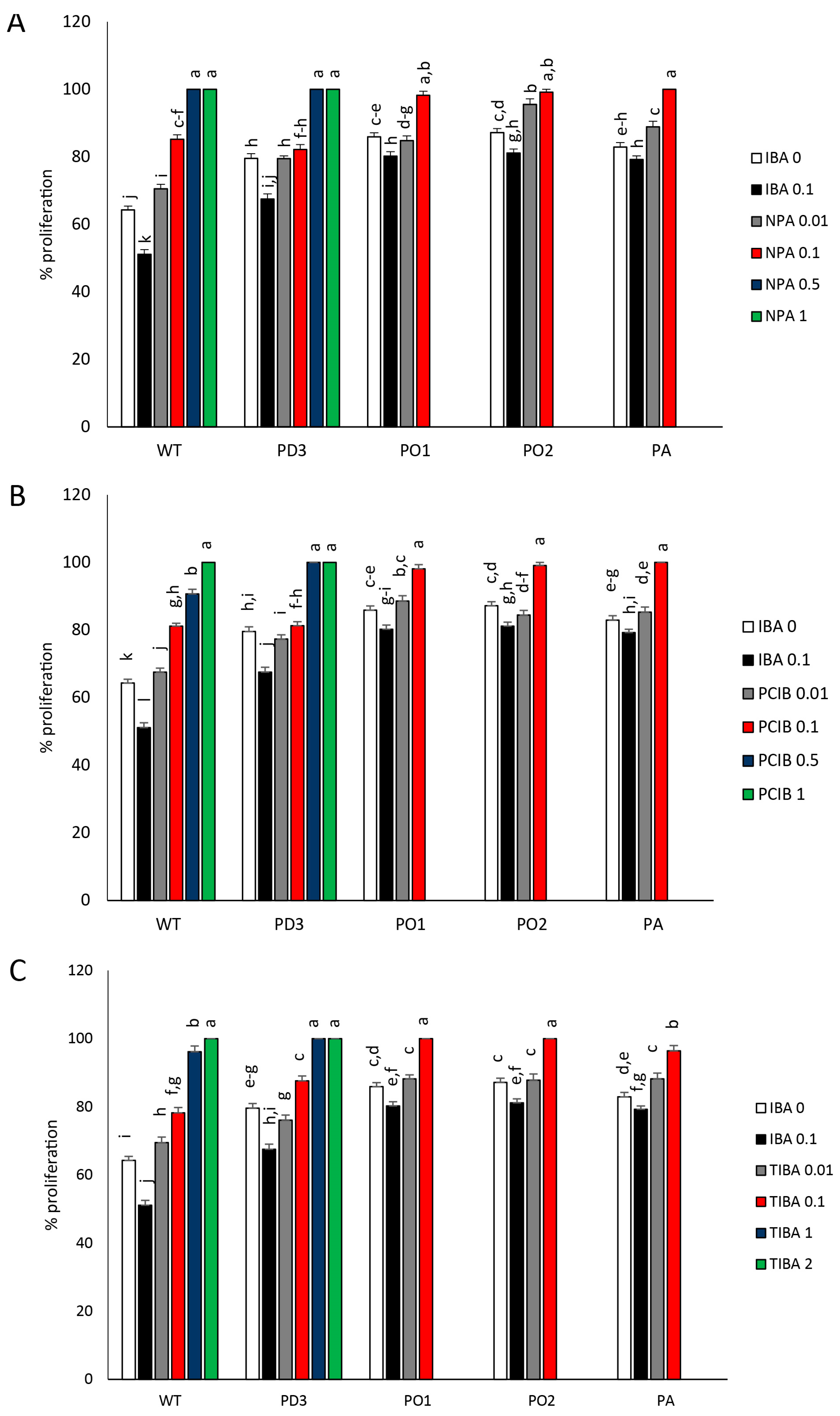
3.3. Shoot Branching (Proliferation Rate) and Dimensions of Developed Lateral Shoots
3.4. Multi-Response Permutation Procedure (MRPP) and Canonical Discriminant Analyses (CDA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Apical dominance |
| ARF | Auxin response factors |
| Aux/IAA | Auxin/Indoleacetic acid |
| IAA | Indole-3-acetic acid |
| IBA | Indole-3-Butyric Acid |
| LB | Lateral bud |
| LS | Lateral shoot |
| NPA | 1-N-naphthylphthalamic acid |
| PCIB | p-Chlorophenoxyisobutyric acid |
| PIN | PIN-FORMED |
| PHYA | PHYTOCHROME A (protein) |
| phyA | phytochrome A (gene) |
| TIBA | 2,3,5,-triiodobenzoic acid |
Appendix A
DNA and RNA Extraction and Southern Blotting
References
- Beveridge, C.A.; Rameau, C.; Wijerathna-Yapa, A. Lessons from a Century of Apical Dominance Research. J. Exp. Bot. 2023, erad137. [Google Scholar] [CrossRef]
- Cline, M. Concepts and Terminology of Apical Dominance. Am. J. Bot. 1997, 84, 1064. [Google Scholar] [CrossRef] [Green Version]
- Petrášek, J.; Friml, J. Auxin Transport Routes in Plant Development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [Green Version]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and Regulation of Auxin Biosynthesis in Arabidopsis Roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef]
- Ljung, K.; Nemhauser, J.L.; Perata, P. New Mechanistic Links between Sugar and Hormone Signalling Networks. Curr. Opin. Plant Biol. 2015, 25, 130–137. [Google Scholar] [CrossRef]
- Di, D.-W.; Zhang, C.; Luo, P.; An, C.-W.; Guo, G.-Q. The Biosynthesis of Auxin: How Many Paths Truly Lead to IAA? Plant Growth Regul 2016, 78, 275–285. [Google Scholar] [CrossRef]
- Baluska, F. Polar Transport of Auxin: Carrier-Mediated Flux across the Plasma Membrane or Neurotransmitter-like Secretion? Trends Cell Biol. 2003, 13, 282–285. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar Demand, Not Auxin, Is the Initial Regulator of Apical Dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef]
- Peer, W.A.; Murphy, A.S. Flavonoids and Auxin Transport: Modulators or Regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in Action: Signalling, Transport and the Control of Plant Growth and Development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Peer, W.A.; Blakeslee, J.J.; Yang, H.; Murphy, A.S. Seven Things We Think We Know about Auxin Transport. Mol. Plant 2011, 4, 487–504. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Peer, W.A.; Murphy, A.S. Auxin Transport. Curr. Opin. Plant Biol. 2005, 8, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, U.; Lee, Y.; Martinoia, E.; et al. Plant ABC Proteins--a Unified Nomenclature and Updated Inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Barbier, F.F.; Dun, E.A.; Beveridge, C.A. Apical Dominance. Curr. Biol. 2017, 27, R864–R865. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, S.B.; Wang, X.-J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA Proteins Are Active Repressors, and Their Stability and Activity Are Modulated by Auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Zazimalova, E.; Murphy, A.S.; Yang, H.; Hoyerova, K.; Hosek, P. Auxin Transporters—Why So Many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef] [Green Version]
- Balla, J.; Medveďová, Z.; Kalousek, P.; Matiješčuková, N.; Friml, J.; Reinöhl, V.; Procházka, S. Auxin Flow-Mediated Competition between Axillary Buds to Restore Apical Dominance. Sci. Rep. 2016, 6, 35955. [Google Scholar] [CrossRef] [Green Version]
- Leonel, L.V.; De Oliveira Reis, F.; De Assis Figueiredo, F.A.M.M.; Ferraz, T.M.; De Oliveira Maia Júnior, S.; Silva, P.C.; De Andrade, J.R. Light Intensity and Hydrogel Soil Amendment Differentially Affect Growth and Photosynthesis of Successional Tree Species. J. For. Res. 2023, 34, 257–268. [Google Scholar] [CrossRef]
- Kazemi, D.; Dehestani-Ardakani, M.; Hatami, M.; Ghorbanpour, M. Research on the Differences in Phenotypic and Photosynthetic Biophysical Parameters of Begonias (Begonia Rex) Cultivars Under Various Light Spectral Compositions. J. Plant Growth Regul. 2023. [Google Scholar] [CrossRef]
- Croce, J.; Badano, E.I.; Trigo, C.B.; Martinez-Galvez, F.; Tálamo, A. Experimental Approaches to Select Tree Species for Forest Restoration: Effects of Light, Water Availability and Interspecific Competition in Degraded Areas. J. For. Res. 2022, 33, 1197–1207. [Google Scholar] [CrossRef]
- Morini, S.; Muleo, R. Effects of Light Quality on Micropropagation of Woody Species. In Micropropagation of Woody Trees and Fruits; Mohan Jain, S., Ishii, K., Eds.; Kluwer Academic Publisher: Dordrecht, The Netherlands; Boston, MA, USA; London, UK; pp. 3–35.
- Cirvilleri, G.; Spina, S.; Iacona, C.; Catara, A.; Muleo, R. Study of Rhizosphere and Phyllosphere Bacterial Community and Resistance to Bacterial Canker in Genetically Engineered Phytochrome A Cherry Plants. J. Plant Physiol. 2008, 165, 1107–1119. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. Plant Hormone Signaling Lightens up: Integrators of Light and Hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef] [PubMed]
- van Gelderen, K.; Kang, C.; Pierik, R. Light Signaling, Root Development, and Plasticity. Plant Physiol. 2018, 176, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Muleo, R.; Iacona, C.; Nicese, F.; Intrieri, M.; Boscherini, G.; Loreti, F.; Buiatti, B.; Thomas, B. Overexpressing PhyA Changes the Cherry Plant Sensibility to the Proximity of Light Signal. In Proceedings of the XLVII Italian Society of Agricoltural Genetics Annual Congress, Barcelona, Spain, 23 June 2003. [Google Scholar]
- Muleo, R.; Iacona, C. Light Perception and Timekeeping Systems in Plants: The Biological Value of the Domain of Time. Riv. Biol. 2007, 100, 16–21. [Google Scholar]
- Sgamma, T.; Forgione, I.; Luziatelli, F.; Iacona, C.; Mancinelli, R.; Thomas, B.; Ruzzi, M.; Muleo, R. Monochromic Radiations Provided by Light Emitted Diode (LED) Modulate Infection and Defense Response to Fire Blight in Pear Trees. Plants 2021, 10, 1886. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Whitelam, G.C. Phytochromes and Shade-Avoidance Responses in Plants. Ann. Bot. 2005, 96, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childs, K.L.; Cordonnier-Pratt, M.-M.; Pratt, L.H.; Morgan, P.W. Genetic Regulation of Development in Sorghum Bicolor: VII. ma(3) Flowering Mutant Lacks a Phytochrome That Predominates in Green Tissue. Plant Physiol. 1992, 99, 765–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childs, K.L.; Miller, F.R.; Cordonnier-Pratt, M.M.; Pratt, L.H.; Morgan, P.W.; Mullet, J.E. The Sorghum Photoperiod Sensitivity Gene, Ma3, Encodes a Phytochrome B. Plant Physiol. 1997, 113, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.W.; Nagpal, P.; Poole, D.S.; Furuya, M.; Chory, J. Mutations in the Gene for the Red/Far-Red Light Receptor Phytochrome B Alter Cell Elongation and Physiological Responses throughout Arabidopsis Development. Plant Cell 1993, 5, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Yanovsky, M.J.; Casal, J.J.; Whitelam, G.C. Phytochrome A, Phytochrome B and HY4 Are Involved in Hypocotyl Growth Responses to Natural Radiation in Arabidopsis: Weak de-Etiolation of the PhyA Mutant under Dense Canopies. Plant Cell Environ. 1995, 18, 788–794. [Google Scholar] [CrossRef]
- Smith, H.; Xu, Y.; Quail, P.H. Antagonistic but Complementary Actions of Phytochromes A and B Allow Optimum Seedling De-Etiolation. Plant Physiol. 1997, 114, 637–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C. Plant Blue-Light Receptors. Trends Plant Sci. 2000, 5, 337–342. [Google Scholar] [CrossRef]
- Ballaré, C.L. Keeping up with the Neighbours: Phytochrome Sensing and Other Signalling Mechanisms. Trends Plant Sci. 1999, 4, 97–102. [Google Scholar] [CrossRef]
- Gilbert, I.R.; Jarvis, P.G.; Smith, H. Proximity Signal and Shade Avoidance Differences between Early and Late Successional Trees. Nature 2001, 411, 792–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muleo, R.; Thomas, B. Effects of Light Quality on Shoot Proliferation of Prunus Cerasifera In Vitro Are the Result of Differential Effects on Bud Induction and Apical Dominance. J. Hortic. Sci. 1997, 72, 483–499. [Google Scholar] [CrossRef]
- Stirk, W.A.; Bálint, P.; Tarkowská, D.; Novák, O.; Maróti, G.; Ljung, K.; Turečková, V.; Strnad, M.; Ördög, V.; van Staden, J. Effect of Light on Growth and Endogenous Hormones in Chlorella Minutissima (Trebouxiophyceae). Plant Physiol. Biochem. 2014, 79, 66–76. [Google Scholar] [CrossRef]
- Roman, H.; Girault, T.; Barbier, F.; Péron, T.; Brouard, N.; Pěnčík, A.; Novák, O.; Vian, A.; Sakr, S.; Lothier, J.; et al. Cytokinins Are Initial Targets of Light in the Control of Bud Outgrowth. Plant Physiol. 2016, 172, 489–509. [Google Scholar] [CrossRef] [Green Version]
- Muleo, R.; Morini, S.; Casano, S. Photoregulation of Growth and Branching of Plum Shoots: Physiological Action of Two Photosystems. In Vitro Cell. Dev. Biol.-Plant 2001, 37, 609–617. [Google Scholar] [CrossRef]
- Muleo, R.; Morini, S. Light Quality Regulates Shoot Cluster Growth and Development of MM106 Apple Genotype in In Vitro Culture. Sci. Hortic. 2006, 108, 364–370. [Google Scholar] [CrossRef]
- Blythe, E.K.; Sibley, J.L.; Tilt, K.M.; Ruter, J.M. Methods of Auxin Application in Cutting Propagation: A Review of 70 Years of Scientific Discovery and Commercial Practice. J. Environ. Hortic. 2007, 25, 166–185. [Google Scholar] [CrossRef]
- Strader, L.C.; Bartel, B. Transport and Metabolism of the Endogenous Auxin Precursor Indole-3-Butyric Acid. Mol. Plant 2011, 4, 477–486. [Google Scholar] [CrossRef] [Green Version]
- Cavallaro, V.; Pellegrino, A.; Muleo, R.; Forgione, I. Light and Plant Growth Regulators on In Vitro Proliferation. Plants 2022, 11, 844. [Google Scholar] [CrossRef]
- Driver, J.A.; Kuniyuki, A. In Vitro Propagation of Paradox Walnut Root Stock. HortScience 1984, 19, 507–509. [Google Scholar] [CrossRef]
- Onofri, A. DSAASTAT a New Excel VBA Macro to Perform Basic Statistical Analyses of Field Trials; Department of Agriculture and Environmental Sciences, University of Perugia: Perugia, Italy, 2010. [Google Scholar]
- Muleo, R.; Morini, S. Physiological Dissection of Blue and Red Light Regulation of Apical Dominance and Branching in M9 Apple Rootstock Growing In Vitro. J. Plant Physiol. 2008, 165, 1838–1846. [Google Scholar] [CrossRef] [PubMed]
- Marcelis-van Acker, C.A.M.; Scholten, H.J. Development of Axillary Buds of Rose In Vitro. Sci. Hortic. 1995, 63, 47–55. [Google Scholar] [CrossRef]
- Dobránszki, J.; Teixeira da Silva, J.A. Micropropagation of Apple—A Review. Biotechnol. Adv. 2010, 28, 462–488. [Google Scholar] [CrossRef] [PubMed]
- Pati, P.K.; Rath, S.P.; Sharma, M.; Sood, A.; Ahuja, P.S. In Vitro Propagation of Rose—A Review. Biotechnol. Adv. 2006, 24, 94–114. [Google Scholar] [CrossRef]
- Kumar, N.; Reddy, M. In Vitro Plant Propagation: A Review. J. Forest Environ. Sci. 2011, 27, 61–72. [Google Scholar]
- Nacheva, L.; Dimitrova, N.; Koleva-Valkova, L.; Stefanova, M.; Ganeva, T.; Nesheva, M.; Tarakanov, I.; Vassilev, A. In Vitro Multiplication and Rooting of Plum Rootstock ‘Saint Julien’ (Prunus domestica Subsp. Insititia) under Fluorescent Light and Different LED Spectra. Plants 2023, 12, 2125. [Google Scholar] [CrossRef]
- Cavallaro, V.; Avola, G.; Fascella, G.; Pellegrino, A.; Ierna, A. Effects of Spectral Quality and Light Quantity of LEDs on In Vitro Shoot Development and Proliferation of Ananas comosus L. Merr. Agronomy 2023, 13, 1072. [Google Scholar] [CrossRef]
- Abdouli, D.; Soufi, S.; Bettaieb, T.; Werbrouck, S.P.O. Effects of Monochromatic Light on Growth and Quality of Pistacia vera L. Plants 2023, 12, 1546. [Google Scholar] [CrossRef] [PubMed]
- Aygun, A.; Dumanoglu, H. In Vitro Shoot Proliferation and In Vitro and Ex Vitro Root Formation of Pyrus elaeagrifolia Pallas. Front. Plant Sci. 2015, 6, 225. [Google Scholar] [CrossRef]
- Sadeghi, F.; Yadollahi, A.; Kermani, M.J.; Eftekhari, M. Optimizing Culture Media for In Vitro Proliferation and Rooting of Tetra (Prunus empyrean 3) Rootstock. J. Genet. Eng. Biotechnol. 2015, 13, 19–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, F.; Moran, R.; Day, M.; Halteman, W.; Zhang, D. In Vitro Shoot Proliferation of Apple Rootstocks ‘B.9′, ‘G.30′, and ‘G.41′ Grown under Red and Blue Light. Horts 2015, 50, 430–433. [Google Scholar] [CrossRef]
- Müller, D.; Leyser, O. Auxin, Cytokinin and the Control of Shoot Branching. Ann. Bot. 2011, 107, 1203–1212. [Google Scholar] [CrossRef] [Green Version]
- Balla, J.; Kalousek, P.; Reinöhl, V.; Friml, J.; Procházka, S. Competitive Canalization of PIN-Dependent Auxin Flow from Axillary Buds Controls Pea Bud Outgrowth: Competitive Canalization of Auxin Flow. Plant J. 2011, 65, 571–577. [Google Scholar] [CrossRef]
- Frick, E.M.; Strader, L.C. Roles for IBA-Derived Auxin in Plant Development. J. Exp. Bot. 2018, 69, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Li, G.; Chen, X.; Xing, L.; Ma, J.; Zhang, D.; Ge, H.; Han, M.; Sha, G.; An, N. Role of Cytokinin, Strigolactone, and Auxin Export on Outgrowth of Axillary Buds in Apple. Front. Plant Sci. 2019, 10, 616. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Janssen, B.J.; Snowden, K.C. The Molecular and Genetic Regulation of Shoot Branching. Plant Physiol. 2021, 187, 1033–1044. [Google Scholar] [CrossRef]
- Fattorini, L.; Veloccia, A.; Della Rovere, F.; D’Angeli, S.; Falasca, G.; Altamura, M.M. Indole-3-Butyric Acid Promotes Adventitious Rooting in Arabidopsis Thaliana Thin Cell Layers by Conversion into Indole-3-Acetic Acid and Stimulation of Anthranilate Synthase Activity. BMC Plant Biol. 2017, 17, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abas, L.; Kolb, M.; Stadlmann, J.; Janacek, D.P.; Lukic, K.; Schwechheimer, C.; Sazanov, L.A.; Mach, L.; Friml, J.; Hammes, U.Z. Naphthylphthalamic Acid Associates with and Inhibits PIN Auxin Transporters. Proc. Natl. Acad. Sci. USA 2021, 118, e2020857118. [Google Scholar] [CrossRef] [PubMed]
- Dhonukshe, P.; Grigoriev, I.; Fischer, R.; Tominaga, M.; Robinson, D.G.; Hašek, J.; Paciorek, T.; Petrášek, J.; Seifertová, D.; Tejos, R.; et al. Auxin Transport Inhibitors Impair Vesicle Motility and Actin Cytoskeleton Dynamics in Diverse Eukaryotes. Proc. Natl. Acad. Sci. USA 2008, 105, 4489–4494. [Google Scholar] [CrossRef]
- Zou, M.; Ren, H.; Li, J. An Auxin Transport Inhibitor Targets Villin-Mediated Actin Dynamics to Regulate Polar Auxin Transport. Plant Physiol. 2019, 181, 161–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oono, Y.; Ooura, C.; Rahman, A.; Aspuria, E.T.; Hayashi, K.; Tanaka, A.; Uchimiya, H. P-Chlorophenoxyisobutyric Acid Impairs Auxin Response in Arabidopsis Root. Plant Physiol. 2003, 133, 1135–1147. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Xie, F.; Jiang, Y.; Li, Z.; Huang, X.; Li, L. Phytochrome A Negatively Regulates the Shade Avoidance Response by Increasing Auxin/Indole Acidic Acid Protein Stability. Dev. Cell 2018, 44, 29–41.e4. [Google Scholar] [CrossRef] [Green Version]
- Iacona, C.; Muleo, R. L’azione Della Qualità Della Luce Nei Diversi Stadi Della Propagazione In Vitro e Post Vitro Del Portinnesto Colt. Italus Hortus 2012, 19, 37–49. [Google Scholar]
- Kay, S.A.; Nagatani, A.; Keith, B.; Deak, M.; Furuya, M.; Chua, N.H. Rice Phytochrome Is Biologically Active in Transgenic Tobacco. Plant Cell 1989, 1, 775–782. [Google Scholar] [CrossRef] [Green Version]
- McCormac, A.C.; Cherry, J.R.; Hershey, H.P.; Vierstra, R.D.; Smith, H. Photoresponses of Transgenic Tobacco Plants Expressing an Oat Phytochrome Gene. Planta 1991, 185, 162–170. [Google Scholar] [CrossRef]
- González-Grandío, E.; Poza-Carrión, C.; Sorzano, C.O.S.; Cubas, P. BRANCHED1 Promotes Axillary Bud Dormancy in Response to Shade in Arabidopsis. Plant Cell 2013, 25, 834–850. [Google Scholar] [CrossRef] [Green Version]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple Pathways Regulate Shoot Branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef]
- Girault, T.; Abidi, F.; Sigogne, M.; Pelleschi-Travier, S.; Boumaza, R.; Sakr, S.; Leduc, N. Sugars Are under Light Control during Bud Burst in Rosa Sp.: Photocontrol of Sugars during Bud Burst. Plant Cell Environ. 2010, 33, 1339–1350. [Google Scholar] [CrossRef]
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.-D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B.; et al. Sucrose Is an Early Modulator of the Key Hormonal Mechanisms Controlling Bud Outgrowth in Rosa Hybrida. J. Exp. Bot. 2015, 66, 2569–2582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, K.A. Shade Avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Kanegae, H.; Shinomura, T.; Miyao, A.; Hirochika, H.; Furuya, M. Isolation and Characterization of Rice Phytochrome A Mutants. Plant Cell 2001, 13, 521–534. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.L.; Murfet, I.C.; Reid, J.B. Pea Mutants with Reduced Sensitivity to Far-Red Light Define an Important Role for Phytochrome A in Day-Length Detection. Plant Physiol. 1997, 114, 1225–1236. [Google Scholar] [CrossRef] [Green Version]
- Kneissl, J.; Shinomura, T.; Furuya, M.; Bolle, C. A Rice Phytochrome A in Arabidopsis: The Role of the N-Terminus under Red and Far-Red Light. Mol. Plant 2008, 1, 84–102. [Google Scholar] [CrossRef]
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome Regulation of Branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927. [Google Scholar] [CrossRef] [Green Version]
- Krishna Reddy, S.; Finlayson, S.A. Phytochrome B Promotes Branching in Arabidopsis by Suppressing Auxin Signaling. Plant Physiol. 2014, 164, 1542–1550. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Zhang, B.; Li, Y.; André, D.; Nilsson, O. Phytochrome B and PHYTOCHROME INTERACTING FACTOR8 Modulate Seasonal Growth in Trees. New Phytol. 2021, 232, 2339–2352. [Google Scholar] [CrossRef] [PubMed]
- Forgione, I.; Wołoszyńska, M.; Pacenza, M.; Chiappetta, A.; Greco, M.; Araniti, F.; Abenavoli, M.R.; Van Lijsebettens, M.; Bitonti, M.B.; Bruno, L. Hypomethylated Drm1 Drm2 Cmt3 Mutant Phenotype of Arabidopsis Thaliana Is Related to Auxin Pathway Impairment. Plant Sci. 2019, 280, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; CSHL Press: Cold Spring Harbor, NY, USA, 1989; Volume 2, ISBN 978-0-87969-309-1. [Google Scholar]
- Glenn, G.; Andreou, L.-V. Analysis of DNA by Southern Blotting. Methods Enzymol. 2013, 529, 47–63. [Google Scholar] [CrossRef] [PubMed]



| Size Classification of Lateral Shoots | |||
|---|---|---|---|
| <5 mm | 5~10 mm | >10 mm | |
| Mean [%] ± SD | Mean [%] ± SD | Mean [%] ± SD | |
| WT IBA 0 | 74 ± 16.2 | 26 ± 16.2 | 0 ± 0 |
| WT IBA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT NPA 0.01 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT NPA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT NPA 0.5 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT NPA 1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 IBA 0 | 62 ± 10.3 | 38 ± 10.3 | 0 ± 0 |
| PD3 IBA 0.1 | 68.3 ± 12.7 | 31.7 ± 12.7 | 0 ± 0 |
| PD3 NPA 0.01 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 NPA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 NPA 0.5 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 NPA 1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PO1 IBA 0 | 10 ± 7.7 | 89.3 ± 9 | 0 ± 0 |
| PO1 IBA 0.1 | 19.4 ± 14.9 | 63.6 ± 13.2 | 16.3 ± 12.3 |
| PO1 NPA 0.01 | 55 ± 7.5 | 36.2 ± 14.4 | 8.8 ± 8.4 |
| PO1 NPA 0.1 | 93.2 ± 10.3 | 6.8 ± 10.3 | 0 ± 0 |
| PO1 NPA 0.5 | n.d. | n.d. | n.d. |
| PO1 NPA 1 | n.d. | n.d. | n.d. |
| PO2 IBA 0 | 0 ± 0 | 11.2 ± 9.9 | 90.2 ± 10 |
| PO2 IBA 0.1 | 51.8 ± 14.9 | 45.1 ± 17.2 | 3.1 ± 5.1 |
| PO2 NPA 0.01 | 64.7 ± 14.3 | 35.3 ± 14.3 | 0 ± 0 |
| PO2 NPA 0.1 | 91.1 ± 10.2 | 8.9 ± 10.2 | 0 ± 0 |
| PO2 NPA 0.5 | n.d. | n.d. | n.d. |
| PO2 NPA 1 | n.d. | n.d. | n.d. |
| PA IBA 0 | 0 ± 0 | 13.4 ± 7.9 | 86.6 ± 7.9 |
| PA IBA 0.1 | 22.2 ± 10 | 56.8 ± 13.7 | 21 ± 10 |
| PA NPA 0.01 | 81.4 ± 11.1 | 18.6 ± 11.1 | 0 ± 0 |
| PA NPA 0.1 | 93.4 ± 8.5 | 6.6 ± 8.5 | 0 ± 0 |
| PA NPA 0.5 | n.d. | n.d. | n.d. |
| PA NPA 1 | n.d. | n.d. | n.d. |
| Size Classification of Lateral Shoots | |||
|---|---|---|---|
| <5 mm | 5~10 mm | >10 mm | |
| Mean [%] ± SD | Mean [%] ± SD | Mean [%] ± SD | |
| WT IBA 0 | 74 ± 16.2 | 26 ± 16.2 | 0 ± 0 |
| WT IBA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT PCIB 0.01 | 60 ± 12.8 | 29.5 ± 13.2 | 10.6 ± 7.6 |
| WT PCIB 0.1 | 37.9 ± 17.4 | 44.4 ± 13.7 | 17.7 ± 10.5 |
| WT PCIB 0.5 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT PCIB 1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 IBA 0 | 62 ± 10.3 | 38 ± 10.3 | 0 ± 0 |
| PD3 IBA 0.1 | 68.3 ± 12.7 | 31.7 ± 12.7 | 0 ± 0 |
| PD3 PCIB 0.01 | 42.2 ± 13.3 | 40.2 ± 11.7 | 17.7 ± 8.5 |
| PD3 PCIB 0.1 | 66.8 ± 14.1 | 33.2 ± 14.1 | 0 ± 0 |
| PD3 PCIB 0.5 | 91.6 ± 9.4 | 8.4 ± 9.4 | 0 ± 0 |
| PD3 PCIB 1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PO1 IBA 0 | 10 ± 7.7 | 89.3 ± 9 | 0 ± 0 |
| PO1 IBA 0.1 | 19.4 ± 14.9 | 63.6 ± 13.2 | 16.3 ± 12.3 |
| PO1 PCIB 0.01 | 26.5 ± 12 | 48.9 ± 11 | 24.7 ± 17.7 |
| PO1 PCIB 0.1 | 73.6 ± 15.5 | 26.4 ± 15.5 | 0 ± 0 |
| PO1 PCIB 0.5 | n.d. | n.d. | n.d. |
| PO1 PCIB 1 | n.d. | n.d. | n.d. |
| PO2 IBA 0 | 0 ± 0 | 11.2 ± 9.9 | 90.2 ± 10 |
| PO2 IBA 0.1 | 51.8 ± 14.9 | 45.1 ± 17.2 | 3.1 ± 5.1 |
| PO2 PCIB 0.01 | 27.1 ± 13.2 | 59 ± 14.2 | 13.9 ± 10.9 |
| PO2 PCIB 0.1 | 70.3 ± 10.8 | 29.7 ± 10.8 | 0 ± 0 |
| PO2 PCIB 0.5 | n.d. | n.d. | n.d. |
| PO2 PCIB 1 | n.d. | n.d. | n.d. |
| PA IBA 0 | 0 ± 0 | 13.4 ± 7.9 | 86.6 ± 7.9 |
| PA IBA 0.1 | 22.2 ± 10 | 56.8 ± 13.7 | 21 ± 10 |
| PA PCIB 0.01 | 23.3 ± 12.9 | 53.6 ± 11 | 23.2 ± 16.6 |
| PA PCIB 0.1 | 68.8 ± 13.4 | 31.3 ± 13.4 | 0 ± 0 |
| PA PCIB 0.5 | n.d. | n.d. | n.d. |
| PA PCIB 1 | n.d. | n.d. | n.d. |
| Size Classification of Lateral Shoots | |||
|---|---|---|---|
| <5 mm | 5~10 mm | >10 mm | |
| Mean [%] ± SD | Mean [%] ± SD | Mean [%] ± SD | |
| WT IBA 0 | 74 ± 16.2 | 26 ± 16.2 | 0 ± 0 |
| WT IBA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT TIBA 0.01 | 91.9 ± 8.8 | 8.1 ± 8.8 | 0 ± 0 |
| WT TIBA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT TIBA 1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| WT TIBA 2 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 IBA 0 | 62 ± 10.3 | 38 ± 10.3 | 0 ± 0 |
| PD3 IBA 0.1 | 68.3 ± 12.7 | 31.7 ± 12.7 | 0 ± 0 |
| PD3 TIBA 0.01 | 71.9 ± 16.1 | 28.1 ± 16.1 | 0 ± 0 |
| PD3 TIBA 0.1 | 55.7 ± 10.3 | 30.7 ± 12.4 | 13.7 ± 9.5 |
| PD3 TIBA 1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PD3 TIBA 2 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PO1 IBA 0 | 10 ± 7.7 | 89.3 ± 9 | 0 ± 0 |
| PO1 IBA 0.1 | 19.4 ± 14.9 | 63.6 ± 13.2 | 16.3 ± 12.3 |
| PO1 TIBA 0.01 | 65.6 ± 12.5 | 30.3 ± 12.5 | 14.3 ± 41.6 |
| PO1 TIBA 0.1 | 86.2 ± 12 | 13.8 ± 12 | 0 ± 0 |
| PO1 TIBA 1 | n.d. | n.d. | n.d. |
| PO1 TIBA 2 | n.d. | n.d. | n.d. |
| PO2 IBA 0 | 0 ± 0 | 11.2 ± 9.9 | 90.2 ± 10 |
| PO2 IBA 0.1 | 51.8 ± 14.9 | 45.1 ± 17.2 | 3.1 ± 5.1 |
| PO2 TIBA 0.01 | 74.7 ± 13.4 | 25.3 ± 13.4 | 0 ± 0 |
| PO2 TIBA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PO2 TIBA 1 | n.d. | n.d. | n.d. |
| PO2 TIBA 2 | n.d. | n.d. | n.d. |
| PA IBA 0 | 0 ± 0 | 13.4 ± 7.9 | 86.6 ± 7.9 |
| PA IBA 0.1 | 22.2 ± 10 | 56.8 ± 13.7 | 21 ± 10 |
| PA TIBA 0.01 | 75.9 ± 11.4 | 24.1 ± 11.4 | 0 ± 0 |
| PA TIBA 0.1 | 100 ± 0 | 0 ± 0 | 0 ± 0 |
| PA TIBA1 | n.d. | n.d. | n.d. |
| PA TIBA 2 | n.d. | n.d. | n.d. |
| Fresh Weight of Growth Cluster Per Neo-Formed Lateral Shoot (mg/Shoot) | |||||
|---|---|---|---|---|---|
| Mean ± SE | Mean ± SE | Mean ± SE | |||
| WT IBA 0 | 94.1 ± 1.9 b | WT IBA 0 | 94.1 ± 1.9 b | WT IBA 0 | 94.1 ± 1.9 b |
| WT IBA 0.1 | 106 ± 3.2 a | WT IBA 0.1 | 106 ± 3.2 a | WT IBA 0.1 | 106 ± 3.2 a |
| WT NPA 0.01 | 58.7 ± 1.7 g | WT PCIB 0.01 | 76.8 ± 1.7 cd | WT TIBA 0.01 | 76.6 ± 2.3 def |
| WT NPA 0.1 | 64.9 ± 1.2 ef | WT PCIB 0.1 | 70.8 ± 1.1 efg | WT TIBA 0.1 | 79.9 ± 2.5 de |
| WT NPA 0.5 | 41.4 ± 0.7 i | WT PCIB 0.5 | 59.1 ± 1 j | WT TIBA 1 | 54.2 ± 1.1 kl |
| WT NPA 1 | 45.7 ± 1.4 hi | WT PCIB 1 | 48.9 ± 0.6 k | WT TIBA 2 | 48.1 ± 0.9 mn |
| PD3 IBA 0 | 79.7 ± 1.9 c | PD3 IBA 0 | 79.7 ± 1.9 c | PD3 IBA 0 | 79.7 ± 1.9 d |
| PD3 IBA 0.1 | 93.7 ± 2.3 b | PD3 IBA 0.1 | 93.7 ± 2.3 b | PD3 IBA 0.1 | 93.7 ± 2.3 b |
| PD3 NPA 0.01 | 63.5 ± 0.9 f | PD3 PCIB 0.01 | 75.1 ± 1.5 cde | PD3 TIBA 0.01 | 85.8 ± 2 c |
| PD3 NPA 0.1 | 67.4 ± 1.4 def | PD3 PCIB 0.1 | 67 ± 1.3 gh | PD3 TIBA 0.1 | 77.3 ± 1.1 de |
| PD3 NPA 0.5 | 47.5 ± 0.7 h | PD3 PCIB 0.5 | 50.1 ± 1 k | PD3 TIBA 1 | 46 ± 1.1 n |
| PD3 NPA 1 | 47.9 ± 0.9 h | PD3 PCIB 1 | 46.8 ± 0.9 k | PD3 TIBA 2 | 46.2 ± 1.1 n |
| PO1 IBA 0 | 79.7 ± 2.1 c | PO1 IBA 0 | 79.7 ± 2.1 c | PO1 IBA 0 | 79.7 ± 2.1 d |
| PO1 IBA 0.1 | 58.3 ± 1.3 g | PO1 IBA 0.1 | 58.3 ± 1.3 j | PO1 IBA 0.1 | 58.3 ± 1.3 jk |
| PO1 NPA 0.01 | 78.6 ± 1.6 c | PO1 PCIB 0.01 | 73.8 ± 1.7 de | PO1 TIBA 0.01 | 74.7 ± 1.7 ef |
| PO1 NPA 0.1 | 55.1 ± 0.6 g | PO1 PCIB 0.1 | 70.9 ± 1.7 efg | PO1 TIBA 0.1 | 60.9 ± 0.8 ij |
| PO1 NPA 0.5 | n.d. | PO1 PCIB 0.5 | n.d. | PO1 TIBA 1 | n.d. |
| PO1 NPA 1 | n.d. | PO1 PCIB 1 | n.d. | PO1 TIBA 2 | n.d. |
| PO2 IBA 0 | 77.7 ± 1.6 c | PO2 IBA 0 | 77.7 ± 1.6 cd | PO2 IBA 0 | 77.7 ± 1.6 de |
| PO2 IBA 0.1 | 65.2 ± 1.8 ef | PO2 IBA 0.1 | 65.2 ± 1.8 hi | PO2 IBA 0.1 | 65.2 ± 1.8 hi |
| PO2 NPA 0.01 | 70.8 ± 1.5 d | PO2 PCIB 0.01 | 74.3 ± 2.1 de | PO2 TIBA 0.01 | 63.8 ± 1.8 hi |
| PO2 NPA 0.1 | 46.8 ± 0.7 h | PO2 PCIB 0.1 | 66.6 ± 1.5 gh | PO2 TIBA 0.1 | 54.2 ± 1 kl |
| PO2 NPA 0.5 | n.d. | PO2 PCIB 0.5 | n.d. | PO2 TIBA 1 | n.d. |
| PO2 NPA 1 | n.d. | PO2 PCIB 1 | n.d. | PO2 TIBA 2 | n.d. |
| PA IBA 0 | 76.9 ± 1.7 c | PA IBA 0 | 76.9 ± 1.7 cd | PA IBA 0 | 76.9 ± 1.7 d |
| PA IBA 0.1 | 68 ± 1.6 de | PA IBA 0.1 | 68 ± 1.6 fgh | PA IBA 0.1 | 68 ± 1.6 gh |
| PA NPA 0.01 | 64.5 ± 1.1 ef | PA PCIB 0.01 | 72 ± 2 ef | PA TIBA 0.01 | 71.9 ± 2 fg |
| PA NPA 0.1 | 45.5 ± 0.8 hi | PA PCIB 0.1 | 61.4 ± 0.9 ij | PA TIBA 0.1 | 52.3 ± 0.9 lm |
| PA NPA 0.5 | n.d. | PA PCIB 0.5 | n.d. | PA TIBA1 | n.d. |
| PA NPA 1 | n.d. | PA PCIB 1 | n.d. | PA TIBA 2 | n.d. |
| Genotypes Comparisons | t | p |
|---|---|---|
| Colt-PO1 vs. Colt-PO2 | −3.294 | 0.0140 |
| Colt-PO1 vs. Colt-PA | −8.335 | <0.0001 |
| Colt-PO1 vs. Colt-PD3 | −25.213 | <0.0001 |
| Colt-PO1 vs. Colt-wt | −40.297 | <0.0001 |
| Colt-PO2 vs. Colt-PA | −1.615 | 0.0728 |
| Colt-PO2 vs. Colt-PD3 | −15.076 | <0.0001 |
| Colt-PO2 vs. Colt-wt | −32.704 | <0.0001 |
| Colt-PA vs. Colt-PD3 | −14.607 | <0.0001 |
| Colt-PA vs. Colt-wt | −32.960 | <0.0001 |
| Colt-PD3 vs. Colt-wt | −220.98 | <0.0001 |
| IBA0 | IBA 0.1 | NPA0.01 | NPA0.1 | NPA0.5 | NPA1 | PCIB0.01 | PCIB0.1 | PCIB0.5 | PCIB1 | TIBA0.01 | TIBA0.1 | TIBA1 | TIBA2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IBA0 | - | |||||||||||||
| IBA0.1 | t: −8.52 p < 0.001 | - | ||||||||||||
| NPA0.01 | t: −54.52 p < 0.001 | t: −39.60 p < 0.001 | - | |||||||||||
| NPA0.1 | t: −74.44 p < 0.001 | −62.77 p < 0.001 | −23.84 p < 0.001 | - | ||||||||||
| NPA0.5 | t: −53.00 p < 0.001 | t: −44.88 p < 0.001 | t: −30.11 p < 0.001 | t: −12.45 p < 0.001 | - | |||||||||
| NPA1 | t: −56.98 p < 0.001 | t: −48.11 p < 0.001 | t: −35.14 p < 0.001 | t: −16.48 p < 0.001 | t: −8.82 p = 0.001 | - | ||||||||
| PCIB0.01 | t: −23.20 p < 0.001 | t: −16.34 p < 0.001 | t: −39.33 p < 0.001 | t: −67.29 p < 0.001 | t: −52.36 p < 0.001 | t: −55.90 p < 0.001 | - | |||||||
| PCIB0.1 | t: −44.78 p < 0.001 | t: −33.02 p < 0.001 | t: −12.55 p < 0.001 | t: −32.60 p < 0.001 | t: −24.51 p < 0.001 | t: −27.86 p < 0.001 | t: −32.64 p < 0.001 | - | ||||||
| PCIB0.5 | T: −49.60 p < 0.001 | T: −36.78 p < 0.001 | T: −17.03 p < 0.001 | T: −2.07 p = 0.046 | T: −13.01 p < 0.001 | T: −20.65 p < 0.001 | T: −44.81 p < 0.001 | T: −19.98 p < 0.001 | - | |||||
| PCIB1 | t: −60.13 p < 0.001 | t: −52.08 p < 0.001 | t: −42.36 p < 0.001 | t: −23.93 p < 0.001 | t: −25.67 p < 0.001 | t: −16.07 p < 0.001 | t: −57.42 p < 0.001 | t: −38.33 p < 0.001 | t: −24.91 p < 0.001 | - | ||||
| TIBA0.01 | t: −41.44 p < 0.001 | t: −26.45 p < 0.001 | t: −8.32 p < 0.001 | t: −42.99 p < 0.001 | t: −40.07 p < 0.001 | t: −44.64 p < 0.001 | t: −22.96 p < 0.001 | t: −21.30 p < 0.001 | t: −26.65 p < 0.001 | t: −46.89 p < 0.001 | - | |||
| TIBA0.1 | t: −56.71 p < 0.001 | t: −46.41 p < 0.001 | t: −8.01 p < 0.001 | t: −5.14 p = 0.002 | t: −12.02 p < 0.001 | t: −15.88 p < 0.001 | t: −47.31 p < 0.001 | t: −14.70 p < 0.001 | t: −2.68 p = 0.025 | t: −22.27 p < 0.001 | t: −20.65 p < 0.001 | - | ||
| TIBA1 | t: −53.21 p < 0.001 | t: −42.43 p < 0.001 | t: −26.99 p < 0.001 | t: −6.82 p = 0.001 | t: −7.28 p = 0.001 | t: −4.18 p = 0.004 | t: −51.77 p < 0.001 | t: −24.05 p < 0.001 | t: −13.09 p < 0.001 | t: −16.73 p < 0.001 | t: −38.10 p < 0.001 | t: −8.80 p < 0.001 | - | |
| TIBA2 | t: −58.10 p < 0.001 | t: −47.62 p < 0.001 | t: −36.95 p < 0.001 | t: −18.32 p < 0.001 | t: −20.22 p < 0.001 | t: −9.72 p < 0.001 | t: −56.89 p < 0.001 | t: −32.58 p < 0.001 | t: −22.70 p < 0.001 | t: −4.81 p = 0.003 | t: −44.41 p < 0.001 | t: −15.62 p < 0.001 | t: −11.69 p < 0.001 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacona, C.; Medori, G.; Thomas, B.; Roncasaglia, R.; Dradi, G.; Radicetti, E.; Mancinelli, R.; Muleo, R.; Forgione, I. Apical Dominance and Branching in Plantlets of Colt Cherry Lines Expressing Different Light and Auxin Signalling Sensitivities. Agronomy 2023, 13, 2018. https://doi.org/10.3390/agronomy13082018
Iacona C, Medori G, Thomas B, Roncasaglia R, Dradi G, Radicetti E, Mancinelli R, Muleo R, Forgione I. Apical Dominance and Branching in Plantlets of Colt Cherry Lines Expressing Different Light and Auxin Signalling Sensitivities. Agronomy. 2023; 13(8):2018. https://doi.org/10.3390/agronomy13082018
Chicago/Turabian StyleIacona, Calogero, Gabriele Medori, Brian Thomas, Romano Roncasaglia, Giuliano Dradi, Emanuele Radicetti, Roberto Mancinelli, Rosario Muleo, and Ivano Forgione. 2023. "Apical Dominance and Branching in Plantlets of Colt Cherry Lines Expressing Different Light and Auxin Signalling Sensitivities" Agronomy 13, no. 8: 2018. https://doi.org/10.3390/agronomy13082018







