Transcriptome Analyses Reveal the Mechanism of Changes in the Sugar Constituents of Jujube Fruits under Saline–Alkali Stress
Abstract
:1. Introduction
2. Results
2.1. Effect of Saline–Alkali Stress on Jujube Fruit Development
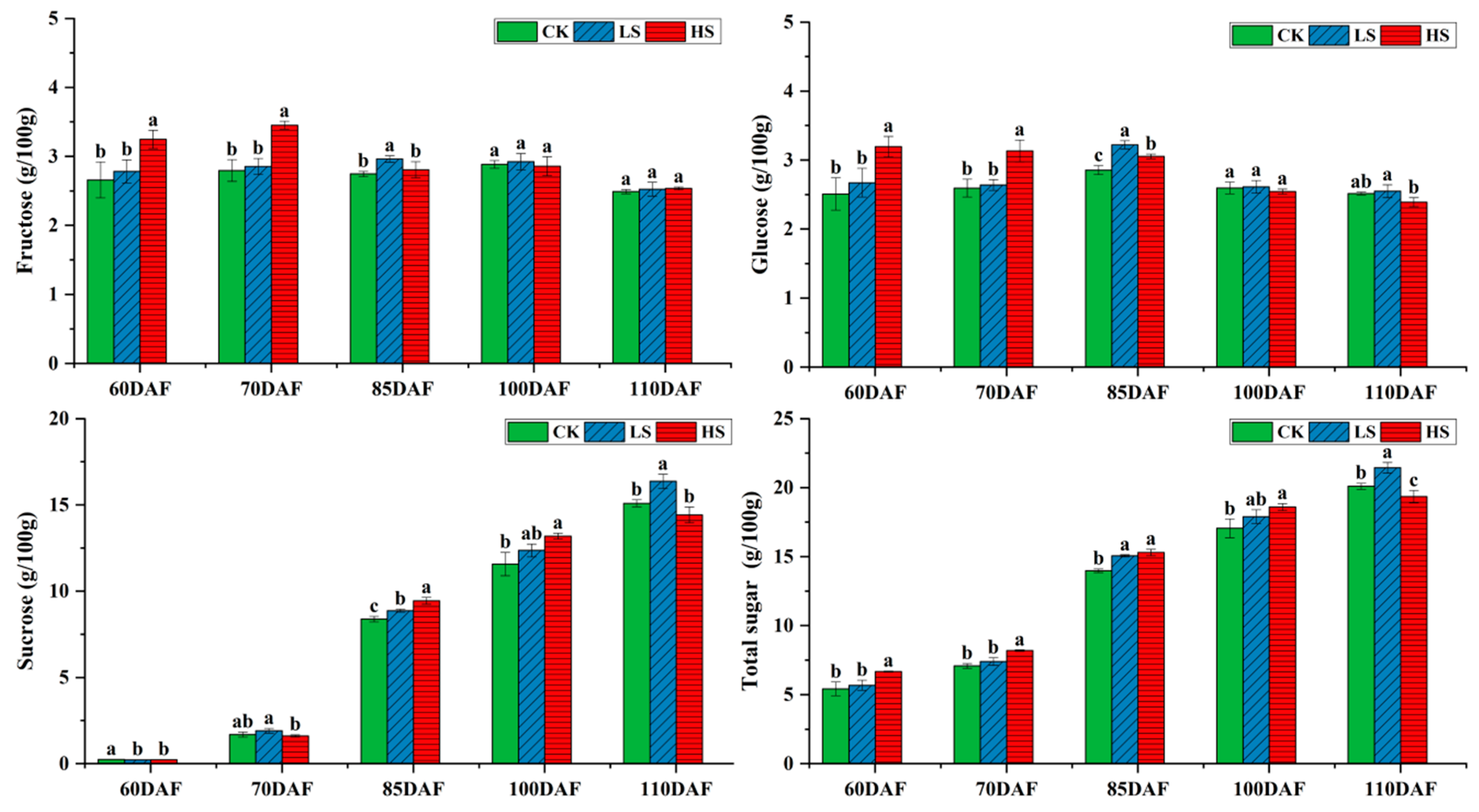
2.2. Changes in Sugar Constituents of Jujube Fruits at Different Developmental Stages under Saline–Alkali Stress
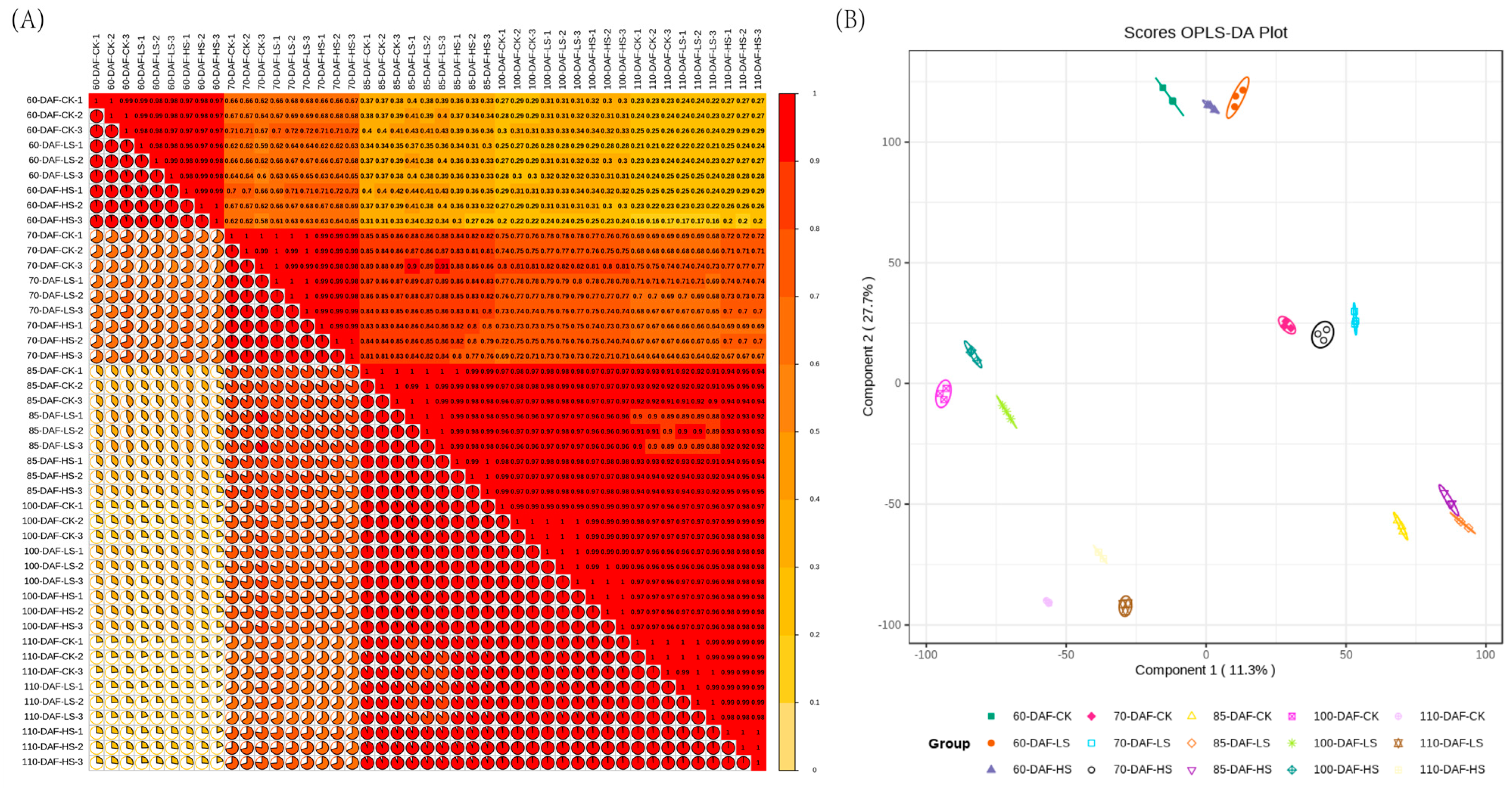
2.3. Transcriptomic Sequencing Analysis
2.4. Screening and Analysis of DEGs in Jujube Fruits under Saline–Alkali Stress
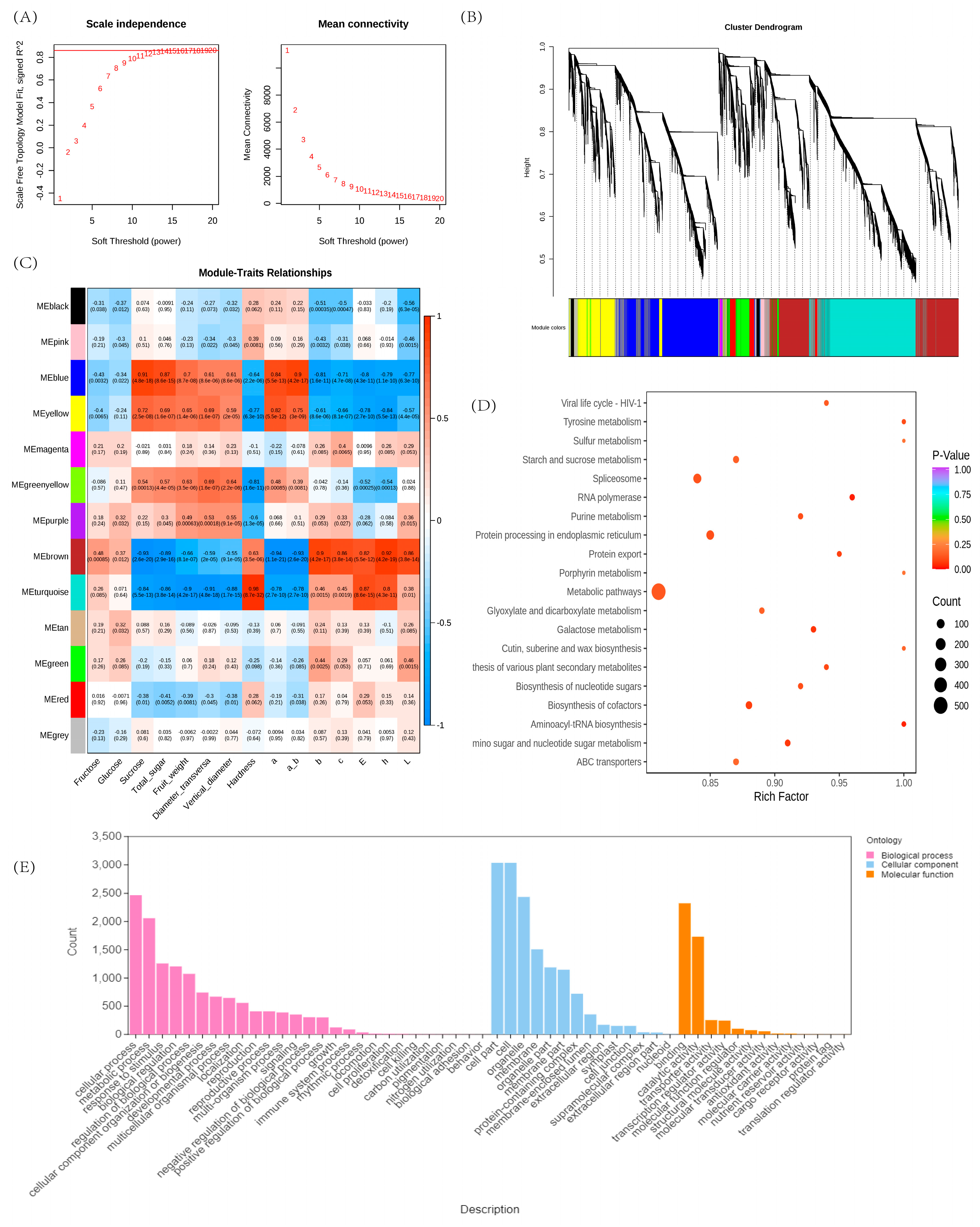
2.5. Co-Expression Network Analysis of Weighted Genes
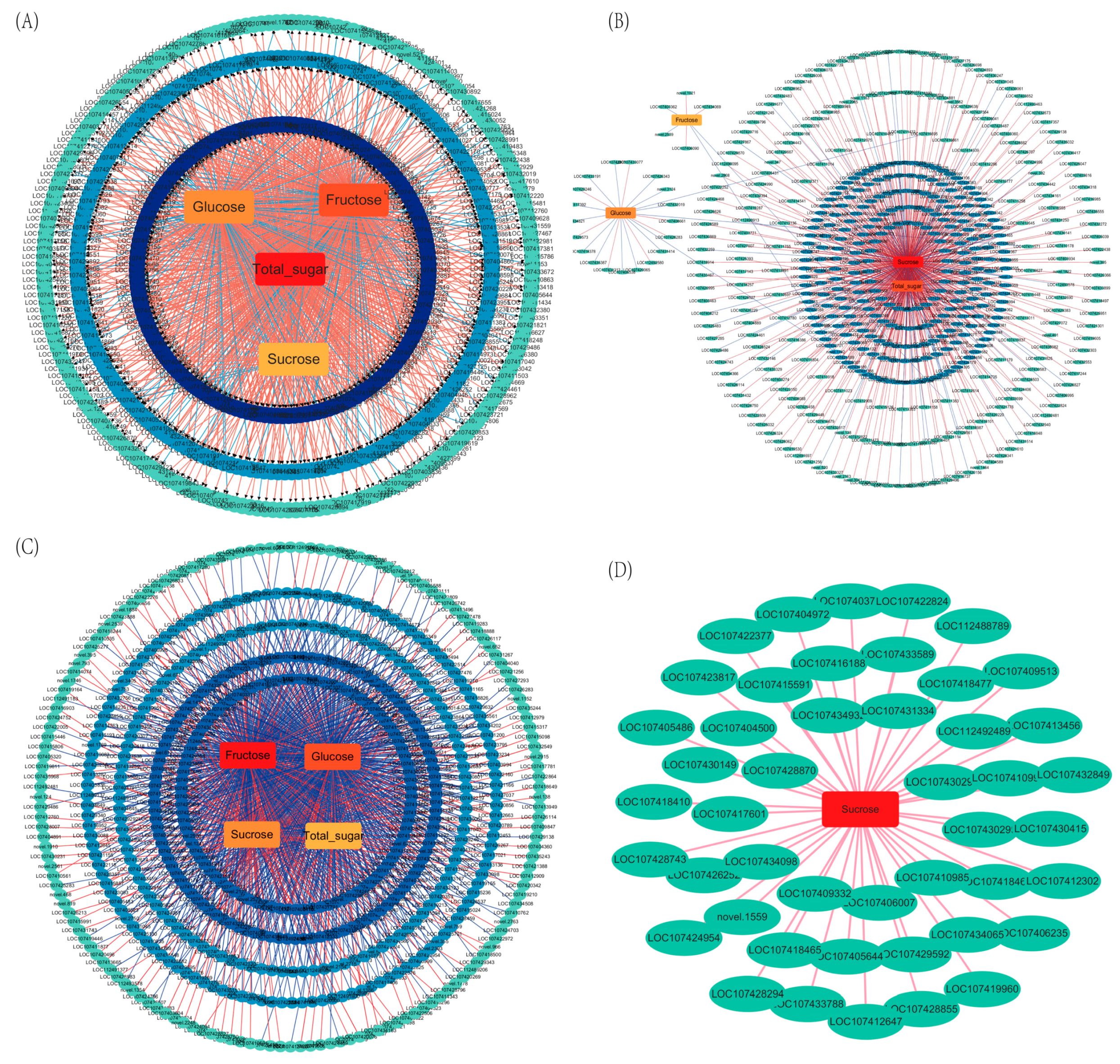
2.6. Co-Expression Network Analysis of DEGs and Sugar Components
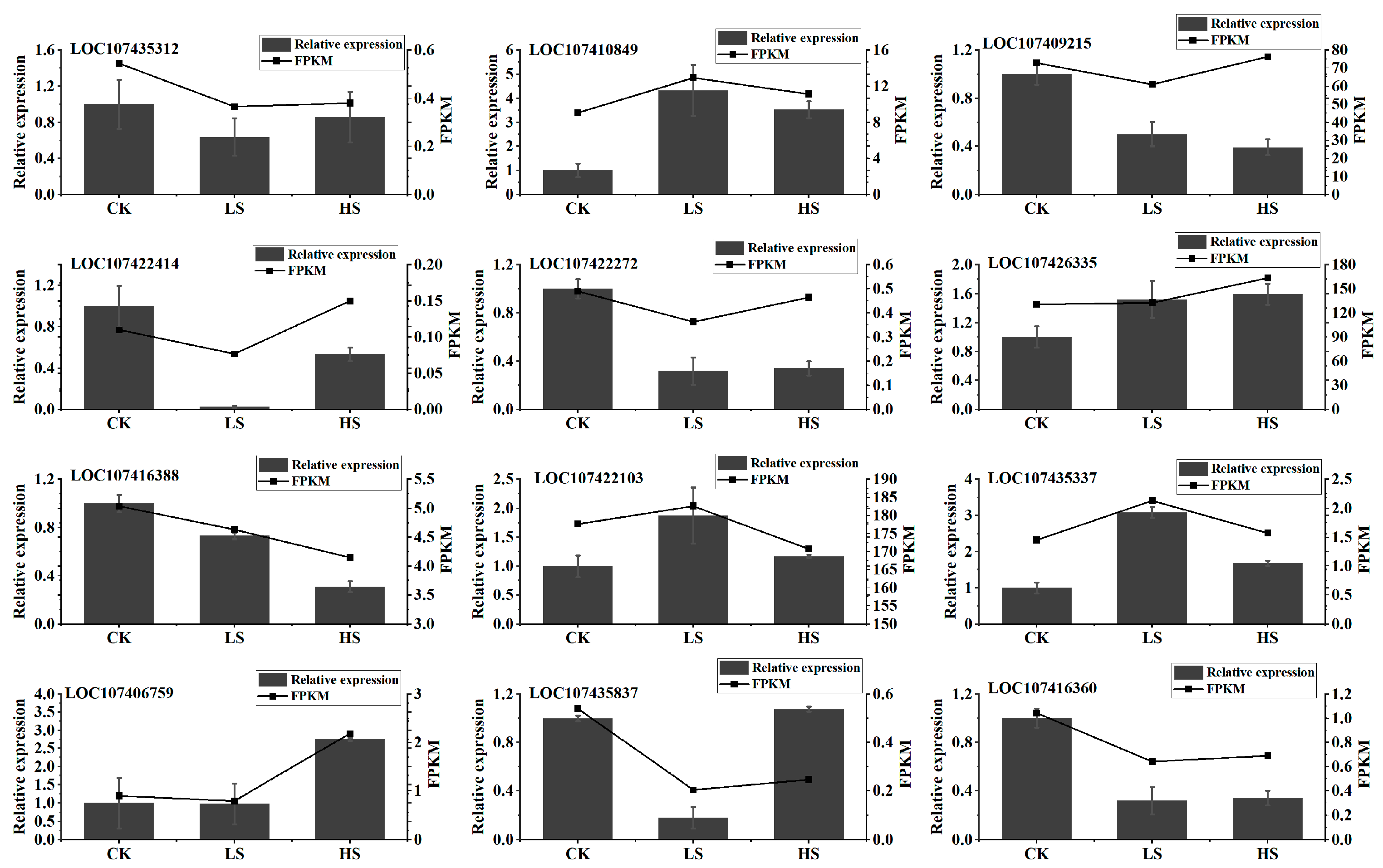
2.7. Quantitative Reverse Transcription-Polymerase Chain Reaction Validation
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Sample Collection
4.3. Determination of Sugar Composition
4.4. Transcriptomic Sequencing
4.5. Quantitative Real-Time PCR (qRT-PCR)
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Raw Reads | Clean Reads | Clean Base (G) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| 60-DAF-CK-1 | 46896952 | 44790862 | 6.72 | 97.76 | 93.71 | 43.39 |
| 60-DAF-CK-2 | 46268966 | 44318064 | 6.65 | 97.68 | 93.52 | 43.37 |
| 60-DAF-CK-3 | 46738576 | 44178678 | 6.63 | 97.74 | 93.64 | 42.99 |
| 60-DAF-LS-1 | 48469392 | 46220260 | 6.93 | 97.76 | 93.78 | 43.12 |
| 60-DAF-LS-2 | 47989768 | 45883648 | 6.88 | 97.49 | 93.09 | 43.98 |
| 60-DAF-LS-3 | 44300870 | 42534000 | 6.38 | 97.79 | 93.76 | 43.99 |
| 60-DAF-HS-1 | 46304818 | 44249608 | 6.64 | 97.73 | 93.59 | 43.87 |
| 60-DAF-HS-2 | 45167064 | 43123706 | 6.47 | 97.81 | 93.81 | 43.90 |
| 60-DAF-HS-3 | 46913320 | 44776204 | 6.72 | 97.67 | 93.47 | 43.85 |
| 70-DAF-CK-1 | 45702590 | 43270396 | 6.49 | 97.84 | 93.89 | 44.03 |
| 70-DAF-CK-2 | 45251794 | 42978476 | 6.45 | 97.82 | 93.86 | 44.06 |
| 70-DAF-CK-3 | 47918850 | 45174426 | 6.78 | 97.89 | 93.95 | 44.00 |
| 70-DAF-LS-1 | 46171330 | 43471364 | 6.52 | 97.69 | 93.50 | 44.10 |
| 70-DAF-LS-2 | 47294346 | 44766250 | 6.71 | 97.84 | 93.90 | 44.01 |
| 70-DAF-LS-3 | 47601724 | 44852966 | 6.73 | 97.87 | 93.95 | 43.72 |
| 70-DAF-HS-1 | 47649234 | 43937814 | 6.59 | 97.52 | 93.11 | 43.56 |
| 70-DAF-HS-2 | 48466654 | 46189112 | 6.93 | 97.71 | 93.59 | 44.31 |
| 70-DAF-HS-3 | 47033076 | 44819574 | 6.72 | 97.83 | 93.88 | 44.25 |
| 85-DAF-CK-1 | 45329698 | 43119728 | 6.47 | 97.76 | 93.66 | 44.00 |
| 85-DAF-CK-2 | 45602142 | 43519408 | 6.53 | 97.72 | 93.58 | 44.28 |
| 85-DAF-CK-3 | 46332666 | 43766548 | 6.56 | 97.92 | 94.07 | 44.28 |
| 85-DAF-LS-1 | 46455296 | 43700858 | 6.56 | 97.60 | 93.31 | 44.02 |
| 85-DAF-LS-2 | 46716526 | 44282652 | 6.64 | 97.92 | 94.06 | 44.32 |
| 85-DAF-LS-3 | 45544790 | 43441038 | 6.52 | 97.79 | 93.74 | 43.98 |
| 85-DAF-HS-1 | 48330934 | 45424220 | 6.81 | 97.86 | 93.91 | 43.93 |
| 85-DAF-HS-2 | 46370896 | 42795930 | 6.42 | 97.81 | 93.77 | 43.63 |
| 85-DAF-HS-3 | 48670574 | 44679440 | 6.70 | 97.72 | 93.58 | 43.74 |
| 100-DAF-CK-1 | 46304964 | 43829080 | 6.57 | 97.69 | 93.61 | 43.49 |
| 100-DAF-CK-2 | 53693900 | 51642144 | 7.75 | 97.68 | 93.48 | 44.02 |
| 100-DAF-CK-3 | 46537854 | 44558040 | 6.68 | 97.91 | 94.05 | 44.03 |
| 100-DAF-LS-1 | 46128808 | 43973312 | 6.60 | 97.65 | 93.52 | 44.12 |
| 100-DAF-LS-2 | 47574422 | 45121984 | 6.77 | 97.71 | 93.54 | 44.15 |
| 100-DAF-LS-3 | 45359914 | 43115012 | 6.47 | 97.71 | 93.55 | 43.72 |
| 100-DAF-HS-1 | 44515002 | 42489486 | 6.37 | 97.65 | 93.40 | 43.50 |
| 100-DAF-HS-2 | 43910128 | 42084182 | 6.31 | 97.9 | 93.99 | 44.03 |
| 100-DAF-HS-3 | 47919786 | 45738762 | 6.86 | 97.91 | 94.02 | 43.57 |
| 110-DAF-CK-1 | 45991250 | 44276892 | 6.64 | 97.64 | 93.40 | 43.47 |
| 110-DAF-CK-2 | 46090012 | 44023642 | 6.60 | 97.83 | 93.88 | 43.10 |
| 110-DAF-CK-3 | 44091626 | 42254784 | 6.34 | 97.65 | 93.39 | 43.28 |
| 110-DAF-LS-1 | 44423566 | 41550134 | 6.23 | 97.78 | 93.73 | 42.96 |
| 110-DAF-LS-2 | 47358900 | 44506524 | 6.68 | 97.68 | 93.59 | 44.03 |
| 110-DAF-LS-3 | 44703024 | 41101524 | 6.17 | 97.55 | 93.29 | 44.05 |
| 110-DAF-HS-1 | 47172650 | 44336140 | 6.65 | 97.78 | 93.67 | 44.25 |
| 110-DAF-HS-2 | 47821584 | 44808270 | 6.72 | 97.80 | 93.78 | 44.18 |
| 110-DAF-HS-3 | 48162024 | 45115442 | 6.77 | 97.92 | 94.05 | 44.15 |
| Sample | Total Reads | Reads Mapped | Unique Mapped | Multi Mapped | Read1 Mapped | Read2 Mapped |
|---|---|---|---|---|---|---|
| 60-DAF-CK-1 | 44790862 | 89.94% | 77.31% | 2.64% | 38.75% | 38.56% |
| 60-DAF-CK-2 | 44318064 | 90.18% | 77.68% | 2.51% | 38.95% | 38.73% |
| 60-DAF-CK-3 | 44178678 | 89.89% | 77.36% | 2.53% | 38.78% | 38.58% |
| 60-DAF-HS-1 | 44249608 | 89.85% | 77.51% | 2.35% | 38.87% | 38.63% |
| 60-DAF-HS-2 | 43123706 | 89.91% | 77.50% | 2.41% | 38.82% | 38.67% |
| 60-DAF-HS-3 | 44776204 | 89.72% | 77.29% | 2.43% | 38.77% | 38.52% |
| 60-DAF-LS-1 | 46220260 | 90.21% | 77.63% | 2.58% | 38.85% | 38.78% |
| 60-DAF-LS-2 | 45883648 | 89.31% | 77.15% | 2.15% | 38.74% | 38.41% |
| 60-DAF-LS-3 | 42534000 | 89.97% | 77.69% | 2.28% | 38.92% | 38.77% |
| 70-DAF-CK-1 | 43270396 | 89.74% | 77.64% | 2.10% | 38.90% | 38.74% |
| 70-DAF-CK-2 | 42978476 | 89.48% | 77.39% | 2.09% | 38.78% | 38.61% |
| 70-DAF-CK-3 | 45174426 | 89.77% | 77.76% | 2.01% | 38.95% | 38.81% |
| 70-DAF-HS-1 | 43937814 | 89.04% | 76.96% | 2.08% | 38.68% | 38.28% |
| 70-DAF-HS-2 | 46189112 | 89.69% | 77.71% | 1.98% | 38.97% | 38.74% |
| 70-DAF-HS-3 | 44819574 | 89.71% | 77.61% | 2.10% | 38.88% | 38.72% |
| 70-DAF-LS-1 | 43471364 | 89.39% | 77.29% | 2.10% | 38.73% | 38.56% |
| 70-DAF-LS-2 | 44766250 | 89.83% | 77.59% | 2.24% | 38.86% | 38.73% |
| 70-DAF-LS-3 | 44852966 | 89.67% | 77.50% | 2.18% | 38.82% | 38.68% |
| 85-DAF-CK-1 | 43119728 | 89.74% | 78.12% | 1.63% | 39.14% | 38.97% |
| 85-DAF-CK-2 | 43519408 | 89.76% | 78.23% | 1.53% | 39.24% | 38.99% |
| 85-DAF-CK-3 | 43766548 | 89.60% | 77.90% | 1.69% | 39.02% | 38.88% |
| 85-DAF-HS-1 | 45424220 | 89.49% | 77.56% | 1.93% | 38.85% | 38.71% |
| 85-DAF-HS-2 | 42795930 | 88.71% | 77.04% | 1.67% | 38.61% | 38.43% |
| 85-DAF-HS-3 | 44679440 | 88.85% | 77.12% | 1.73% | 38.67% | 38.45% |
| 85-DAF-LS-1 | 43700858 | 89.34% | 77.86% | 1.49% | 39.08% | 38.78% |
| 85-DAF-LS-2 | 44282652 | 89.85% | 78.17% | 1.68% | 39.13% | 39.04% |
| 85-DAF-LS-3 | 43441038 | 89.81% | 78.18% | 1.63% | 39.17% | 39.01% |
| 100-DAF-CK-1 | 43829080 | 89.65% | 77.95% | 1.70% | 39.04% | 38.91% |
| 100-DAF-CK-2 | 51642144 | 89.92% | 78.39% | 1.53% | 39.34% | 39.05% |
| 100-DAF-CK-3 | 44558040 | 90.17% | 78.49% | 1.68% | 39.30% | 39.19% |
| 100-DAF-HS-1 | 42489486 | 89.96% | 78.25% | 1.72% | 39.25% | 39.00% |
| 100-DAF-HS-2 | 42084182 | 90.34% | 78.42% | 1.92% | 39.30% | 39.12% |
| 100-DAF-HS-3 | 45738762 | 90.26% | 78.41% | 1.84% | 39.27% | 39.14% |
| 100-DAF-LS-1 | 43973312 | 89.56% | 77.90% | 1.66% | 39.01% | 38.89% |
| 100-DAF-LS-2 | 45121984 | 89.92% | 78.25% | 1.67% | 39.25% | 39.00% |
| 100-DAF-LS-3 | 43115012 | 89.84% | 78.22% | 1.63% | 39.23% | 38.99% |
| 110-DAF-CK-1 | 44276892 | 89.83% | 77.89% | 1.94% | 39.07% | 38.82% |
| 110-DAF-CK-2 | 44023642 | 89.84% | 77.82% | 2.02% | 38.97% | 38.85% |
| 110-DAF-CK-3 | 42254784 | 89.79% | 78.02% | 1.77% | 39.12% | 38.90% |
| 110-DAF-HS-1 | 44336140 | 89.49% | 77.74% | 1.75% | 38.97% | 38.77% |
| 110-DAF-HS-2 | 44808270 | 89.38% | 77.57% | 1.82% | 38.87% | 38.70% |
| 110-DAF-HS-3 | 45115442 | 89.49% | 77.73% | 1.77% | 38.93% | 38.80% |
| 110-DAF-LS-1 | 41550134 | 89.44% | 77.44% | 2.00% | 38.80% | 38.65% |
| 110-DAF-LS-2 | 44506524 | 88.95% | 77.24% | 1.71% | 38.68% | 38.57% |
| 110-DAF-LS-3 | 41101524 | 87.87% | 76.37% | 1.50% | 38.26% | 38.11% |
| Gene ID | Primer Sequence (5′–3′) | Primer Sequence (3′–5′) |
|---|---|---|
| LOC107406759 | CCTTCCTCAAGAGTATCGGT | AGAGTTTCATCCAAACGCTC |
| LOC107409215 | GCATGGTTATTATGCGATTCTT | AATGCTTTGACCAATGTCGT |
| LOC107410849 | TACTTGCATGAAGCCTTGAA | CCGGATCCATCTTCTGTAAC |
| LOC107416388 | GGGGTTTTCTACTTCACCAC | ATTCCTTTGATTCAGGCACC |
| LOC107422103 | TGAGCATATGGGATAAGGCA | TGTCATGCCACAATGACAAT |
| LOC107422414 | CATGCCTCAGCAAATACTGT | TCTTGGGTTCTGTGCCTTAT |
| LOC107426335 | GATCGATGAGGTGGACAATG | CTTCCTGTGCGGATCTTATG |
| LOC107427369 | TACGATTCCTTAGGTGGCAT | TCTAATGGTATTTACTTGATAGCG |
| LOC107430742 | AGGCTAAACCCATATCGGAA | CTTCGTACGGAAAATCGACA |
| LOC107435312 | CCGAATTGCTTGGAGAGAAA | AGATACTGGGAGTGAGAAGC |
| LOC107435837 | CTGCTGCAAACTCCAAGTAT | ATTTTCTAAACCAACGCCGA |
| LOC112492534 | GCGTACTCACAGAAACTTGT | CCAGCACTATATCTGCCTCT |
References
- Liu, M.J.; Wang, J.R.; Wang, L.L.; Liu, P.; Zhao, J. The historical and current research progress on jujube—A superfruit for the future. Hortic. Res. 2020, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.H.; Wu, C.S.; Wang, M. The jujube (Ziziphus jujuba Mill.) fruit: A review of current knowledge of fruit composition and health benefits. J. Agric. Food Chem. 2013, 61, 3351–3363. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.H.; Wu, C.S.; Yu, J.G.; Wang, M.; Ma, Y.J.; Li, C.L. Textural characteristic, antioxidant activity, sugar, organic acid, and phenolic profiles of 10 promising jujube (Ziziphus jujuba Mill.) selections. J. Food Sci. 2012, 77, C1218–C1225. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.J.; Xiong, J.R.; Yang, X.L.; Kadambot, H.M.; Du, T.S. Carbon footprint analysis of sweet sorghum–based bioethanol production in the potential saline—Alkali land of northwest China. J. Clean. Prod. 2022, 349, 131476. [Google Scholar] [CrossRef]
- Sagervanshi, A.; Geilfus, C.M.; Kaiser, H.; Mühling, K.H. Alkali salt stress causes fast leaf apoplastic alkalinization together with shifts in ion and metabolite composition and transcription of key genes during the early adaptive response of Vicia faba L. Plant Sci. 2022, 319, 111253. [Google Scholar] [CrossRef]
- Yan, K.; Cui, J.X.; Zhi, Y.B.; Su, H.Y.; Yu, S.Y.; Zhou, S.W. Deciphering salt tolerance in tetraploid honeysuckle (Lonicera japonica Thunb.) from ion homeostasis, water balance and antioxidant defense. Plant Physiol. Biochem. 2023, 195, 266–274. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Li, S.H.; Tang, T.; Liu, Y.D.; Muhammad, M.T.; Wang, C.; Meng, Z.Q.; Niu, J.X.; Yang, W.W.; Ma, J.J.; et al. Comparison of morphological, physiological, and related–gene expression responses to saline–alkali stress in eight apple rootstock genotypes. Sci. Hortic. 2022, 306, 111455. [Google Scholar] [CrossRef]
- Effler, R.S.; Goyer, R.A. Baldcypress and water tupelo sapling response to multiple stress agents and reforestation implications for Louisiana swamps. For. Ecol. Manag. 2006, 226, 330–340. [Google Scholar] [CrossRef]
- Zushi, K.; Higashijima, M. Correlation network analysis identified the key interactions of texture profiles with the sensory, physical, and organoleptic properties of cherry tomato cultivars grown under salt stress. Sci. Hortic. 2022, 293, 110754. [Google Scholar] [CrossRef]
- Wang, P.J.; Xu, Z.J.; Zhang, Y.; Ma, Y.B.; Yang, J.Y.; Zhou, F.; Gao, Y.; Li, G.B.; Hu, X.H. Over-expression of spermidine synthase 2 (SlSPDS2) in tomato plants improves saline–alkali stress tolerance by increasing endogenous polyamines content to regulate antioxidant enzyme system and ionic homeostasis. Plant Physiol. Biochem. 2022, 192, 172–185. [Google Scholar] [CrossRef]
- Shi, X.L.; Zhou, Y.F.; Zhao, X.H.; Guo, P.; Ren, J.Y.; Zhang, H.; Dong, Q.Q.; Zhang, Z.; Yu, H.Q.; Wan, S.B. Soil metagenome and metabolome of peanut intercropped with sorghum reveal a prominent role of carbohydrate metabolism in salt–stress response. Environ. Exp. Bot. 2023, 209, 105274. [Google Scholar] [CrossRef]
- Li, C.H.; Wan, Y.F.; Shang, X.L.; Fang, S.Z. Integration of transcriptomic and metabolomic analysis unveils the response mechanism of sugar metabolism in Cyclocarya paliurus seedlings subjected to PEG–induced drought stress. Plant Physiol. Biochem. 2023, 201, 107856. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.Q.; Li, N.; Zhang, D.P.; Meinhardt, L.; Cao, B.; Li, Y.J.; Song, L.H. Elevated temperature and drought stress significantly affect fruit quality and activity of anthocyanin–related enzymes in jujube (Ziziphus jujuba Mill. cv. ‘Lingwuchangzao’). PLoS ONE 2020, 15, e0241491. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.M.; Liu, Z.Y.; Chang, X.Y.; Zhang, Z.J.; Shi, W.C.; Zhang, Z.R.; Zhao, B.L. Analysis of the Alternative Splicing Events of Exogenousδ–Aminolevulinic Acid under NaCl Stress in Wild Jujube Seedlings. Forests 2022, 12, 2076. [Google Scholar] [CrossRef]
- Zhang, Z.; Xing, Y.; Ramakrishnan, M.; Chen, C.; Xie, F.; Hua, Q.; Chen, J.; Zhang, R.; Zhao, J.; Hu, G.; et al. Transcriptomics–based identification and characterization of genes related to sugar metabolism in ‘Hongshuijing’ pitaya. Hortic. Plant J. 2022, 8, 450–460. [Google Scholar] [CrossRef]
- Hao, Q.; Yang, L.; Fan, D.; Zeng, B.; Jin, J. The transcriptomic response to heat stress of a jujube (Ziziphus jujuba Mill.) cultivar is featured with changed expression of long noncoding RNAs. PLoS ONE 2021, 16, e0249663. [Google Scholar] [CrossRef]
- Zhao, J.; Li, S.; Xu, Y.; Ahmad, N.; Kuang, B.; Feng, M.; Wei, N.; Yang, X. The subgenome Saccharum spontaneum contributes to sugar accumulation in sugarcane as revealed by full–length transcriptomic analysis. J. Adv. Res. 2023. [Google Scholar] [CrossRef]
- Chen, X.; Chen, R.H.; Wang, Y.F.; Wu, C.Y.; Huang, J. Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress. Genes 2019, 10, 360. [Google Scholar] [CrossRef]
- Wu, Z.; Li, H.; Wang, Y.D.; Yang, D.J.; Tan, H.J.; Zhan, Y.; Luo, Y.; Chen, G. Optimization extraction, structural features and antitumor activity of polysaccharides from Z. jujuba cv. Ruoqiangzao seeds. Int. J. Biol. Macromol. 2019, 135, 1151–1161. [Google Scholar] [CrossRef]
- Zhang, C.M.; Bian, Y.; Hou, S.H.; Li, X.G. Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta 2018, 248, 1187–1199. [Google Scholar] [CrossRef]
- Wang, L.N.; Chen, C.; He, R.J.; Cyren, M.R.; Mao, Q.Y.; Sun, P.L. Tree age and maturity stage affect reducing sugars, organic acids and minerals in Ziziphus jujuba Mill. cv. Huping fruits. J. Food Compos. Anal. 2023, 115, 105007. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, L.H.; Li, M.Z.; Deng, X.L.; Ji, R.H. Predicting apple sugar content based on spectral characteristics of apple tree leaf in different phenological phases. Comput. Electron. Agric. 2015, 112, 20–27. [Google Scholar] [CrossRef]
- Liu, X.M.; Hu, C.X.; Liu, X.D.; Riaz, M.; Liu, Y.; Dong, Z.H.; Tan, Q.L.; Sun, X.S.; Wu, S.W.; Tan, Z.H. Effect of magnesium application on the fruit coloration and sugar accumulation of navel orange (Citrus sinensis Osb.). Sci. Hortic. 2022, 304, 111282. [Google Scholar] [CrossRef]
- Wu, B.H.; Quilot, B.; Génard, M.; Kervella, J.; Li, S.H. Changes in sugar and organic acid concentrations during fruit maturation in peaches, P. davidiana and hybrids as analyzed by principal component analysis. Sci. Hortic. 2005, 103, 429–439. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Luo, Z.; Wang, L.; Liu, M.J. Genome-wide identification and expression profile analysis of SWEET genes in Chinese jujube. PeerJ 2023, 11, e14704. [Google Scholar] [CrossRef] [PubMed]
- Qing, H.G.; Wu, C.S.; Wang, M. Effect of Drying of Jujubes (Ziziphus jujuba Mill.) on the Contents of Sugars, Organic Acids, α-Tocopherol, β-Carotene, and Phenolic. J. Agric. Food Chem. 2012, 60, 9642–9648. [Google Scholar]
- Guo, S.; Duan, J.A.; Qian, D.W.; Tang, Y.P.; Wu, D.W.; Su, S.L.; Wang, H.Q.; Zhao, Y.N. Content variations of triterpenic acid, nucleoside, nucleobase, and sugar in jujube (Ziziphus jujuba) fruit during ripening. Food Chem. 2015, 167, 468–474. [Google Scholar] [CrossRef]
- Yamada, H.; Takimoto, S.; Toshinari, D.; Kataoka, K.; Habu, T. Effect of Seawater Irrigation on Water Relations and Fruit Quality in Potted Citrus Trees. Hortic. J. 2015, 84, 195–201. [Google Scholar] [CrossRef]
- Debouba, M.; Gouia, H.; Suzuki, A.; Ghorbel, M.H. NaCl stress effects on enzymes involved in nitrogen assimilation pathway in tomato “Lycopersicon esculentum” seedlings. J. Plant Physiol. 2006, 163, 1247–1258. [Google Scholar] [CrossRef]
- Galli, V.; Messias, R.S.; Perin, E.C.; Borowski, J.M.; Bamberg, A.L.; Rombaldi, C.V. Mild salt stress improves strawberry fruit quality. LWT 2016, 2016, 693–699. [Google Scholar] [CrossRef]
- Akrami, M.; Arzani, A. Inheritance of fruit yield and quality in melon (Cucumis melo L.) grown under feld salinity stress. Sci. Rep. 2019, 9, 7249. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.X.; Sun, H.Q.; Wang, Y.F.; Wang, X.; Guo, Y.P. Effects of water stress on quality and sugar metabolism in ‘Gala’ apple fruit. Hortic. Plant J. 2023, 9, 60–72. [Google Scholar] [CrossRef]
- Yu, L.; Shao, X.F.; Wei, Y.Y.; Xu, F.; Wang, H.F. Sucrose degradation is regulated by 1–methycyclopropene treatment and is related to chilling tolerance in two peach cultivars. Postharvest Biol. Technol. 2017, 124, 25–34. [Google Scholar] [CrossRef]
- Kheir, A.; Ali, E.F.; He, Z.; Ali, O.A.M.; Feike, T. Recycling of sugar crop disposal to boost the adaptation of canola (Brassica napus L.) to abiotic stress through different climate zones. J. Environ. Manag. 2021, 281, 111881. [Google Scholar] [CrossRef] [PubMed]
- Kosar, F.; Akram, N.A.; Sadiq, M.; Al-Qurainy, F.; Ashraf, M. Trehalose: A Key Organic Osmolyte Effectively Involved in Plant Abiotic Stress Tolerance. J. Plant Growth Regul. 2019, 38, 606–618. [Google Scholar] [CrossRef]
- Tan, J.; Wang, X.J.; Wang, L.; Zhou, X.Q.; Liu, C.L.; Ge, J.L.; Bian, L.; Chen, S.Q. Transcriptomic responses to air exposure stress in coelomocytes of the sea cucumber, Apostichopus japonicus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 42, 100963. [Google Scholar] [CrossRef]
- Liang, Y.H.; Wang, S.S.; Zhao, C.H.; Ma, X.W.; Zhao, Y.Y.; Shao, J.; Li, Y.B.; Li, H.L.; Song, H.W.; Ma, H.; et al. Transcriptional regulation of bark freezing tolerance in apple (Malus domestica Borkh.). Hortic. Res. 2020, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Dai, M.S.; Cai, D.Y.; Shi, Z.B. Proteome and transcriptome profile analysis reveals regulatory and stress–responsive networks in the russet fruit skin of sand pear. Hortic. Res. 2020, 7, 16. [Google Scholar] [CrossRef]
- Jia, X.M.; Zhu, Y.F.; Hu, Y.; Zhang, R.; Cheng, L.; Zhu, Z.L.; Zhao, T.; Zhang, X.Y.; Wang, Y.X. Integrated physiologic, proteomic, and metabolomic analyses of Malus halliana adaptation to saline–alkali stress. Hortic. Res. 2019, 6, 91. [Google Scholar] [CrossRef]
- Deng, C.H.; Zhang, Z.B.; Yan, G.R.; Wang, F.; Zhao, L.J.; Liu, N.; Abudukeyoumu, A.; Li, Y.S.; Wang, W.; Shi, S.B. Salt–responsive transcriptome analysis of triticale reveals candidate genes involved in the key metabolic pathway in response to salt stress. Sci. Rep. 2020, 10, 20669. [Google Scholar] [CrossRef]
- Guo, H.J.; Huang, Z.J.; Li, M.Q.; Hou, Z.N. Growth, ionic homeostasis, and physiological responses of cotton under different salt and alkali stresses. Sci. Rep. 2020, 10, 21844. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, C.M.; Zhao, X.; Fei, Z.J.; Wan, K.K.; Zhang, Z.; Pang, X.M.; Yin, X.; Bai, Y.; Sun, X.Q.; et al. The Jujube Genome Provides Insights into Genome Evolution and the Domestication of Sweetness/Acidity Taste in Fruit Trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Zhao, P.C.; Cui, L.W.; Fang, J.G.; Jia, H.F. Over-expression of Gene FaASR Promotes Strawberry Fruit Coloring. Hortic. Plant J. 2015, 3, 147–154. [Google Scholar]
- Xu, W.J.; Wei, Y.Y.; Wang, X.X.; Han, P.P.; Chen, Y.; Xu, F.; Shao, X.F. Molecular cloning and expression analysis of hexokinase genes in peach fruit under postharvest disease stress. Postharvest Biol. Technol. 2021, 172, 111377. [Google Scholar] [CrossRef]
- Zhu, X.D.; Wang, M.Q.; Li, X.P.; Jiu, S.T.; Wang, C.; Fang, J.G. Genome–Wide Analysis of the Sucrose Synthase Gene Family in Grape (Vitis vinifera): Structure, Evolution, and Expression Profiles. Genes 2017, 8, 111. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, Y.F.; Yan, M.; Yu, J.; Zhou, X.F.; Bao, J.K.; Zhang, Q.Q.; Wu, C.Y. Effect of Saline–Alkali Stress on Sugar Metabolism of Jujube Fruit. Horticulturae 2022, 8, 474. [Google Scholar] [CrossRef]
- Kenneth, J.L.; Thomas, D.S. Analysis of Relative Gene Expression Data Using RealTime Quantitative PCR and the 2−∆∆Ct Method. Methods 2001, 4, 402–408. [Google Scholar]

| Group | Upregulated | Downregulated | DEGs |
|---|---|---|---|
| 60-DAF-CK_vs_LS | 176 | 94 | 270 |
| 70-DAF-CK_vs_LS | 37 | 63 | 100 |
| 85-DAF-CK_vs_LS | 22 | 11 | 33 |
| 100-DAF-CK_vs_LS | 33 | 31 | 64 |
| 110-DAF-CK_vs_LS | 34 | 34 | 68 |
| 60-DAF-CK_vs_HS | 368 | 244 | 612 |
| 70-DAF-CK_vs_HS | 99 | 60 | 159 |
| 85-DAF-CK_vs_HS | 207 | 176 | 383 |
| 100-DAF-CK_vs_HS | 206 | 197 | 403 |
| 110-DAF-CK_vs_HS | 305 | 434 | 739 |
| 60-DAF-LS_vs_HS | 69 | 65 | 134 |
| 70-DAF-LS_vs_HS | 73 | 126 | 199 |
| 85-DAF-LS_vs_HS | 128 | 230 | 358 |
| 100-DAF-LS_vs_HS | 170 | 220 | 390 |
| 110-DAF-LS_vs_HS | 282 | 262 | 544 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Feng, Y.; Yan, M.; Pu, X.; Lu, D.; Yuan, H.; Wu, C. Transcriptome Analyses Reveal the Mechanism of Changes in the Sugar Constituents of Jujube Fruits under Saline–Alkali Stress. Agronomy 2023, 13, 2243. https://doi.org/10.3390/agronomy13092243
Wang Y, Feng Y, Yan M, Pu X, Lu D, Yuan H, Wu C. Transcriptome Analyses Reveal the Mechanism of Changes in the Sugar Constituents of Jujube Fruits under Saline–Alkali Stress. Agronomy. 2023; 13(9):2243. https://doi.org/10.3390/agronomy13092243
Chicago/Turabian StyleWang, Yan, Yifeng Feng, Min Yan, Xiaoqiu Pu, Dengyang Lu, Hengzhou Yuan, and Cuiyun Wu. 2023. "Transcriptome Analyses Reveal the Mechanism of Changes in the Sugar Constituents of Jujube Fruits under Saline–Alkali Stress" Agronomy 13, no. 9: 2243. https://doi.org/10.3390/agronomy13092243
APA StyleWang, Y., Feng, Y., Yan, M., Pu, X., Lu, D., Yuan, H., & Wu, C. (2023). Transcriptome Analyses Reveal the Mechanism of Changes in the Sugar Constituents of Jujube Fruits under Saline–Alkali Stress. Agronomy, 13(9), 2243. https://doi.org/10.3390/agronomy13092243





