Elucidation of the Genetic Diversity within Some In Situ Shea Germplasm in Ghana
Abstract
:1. Introduction
2. Materials and Methods
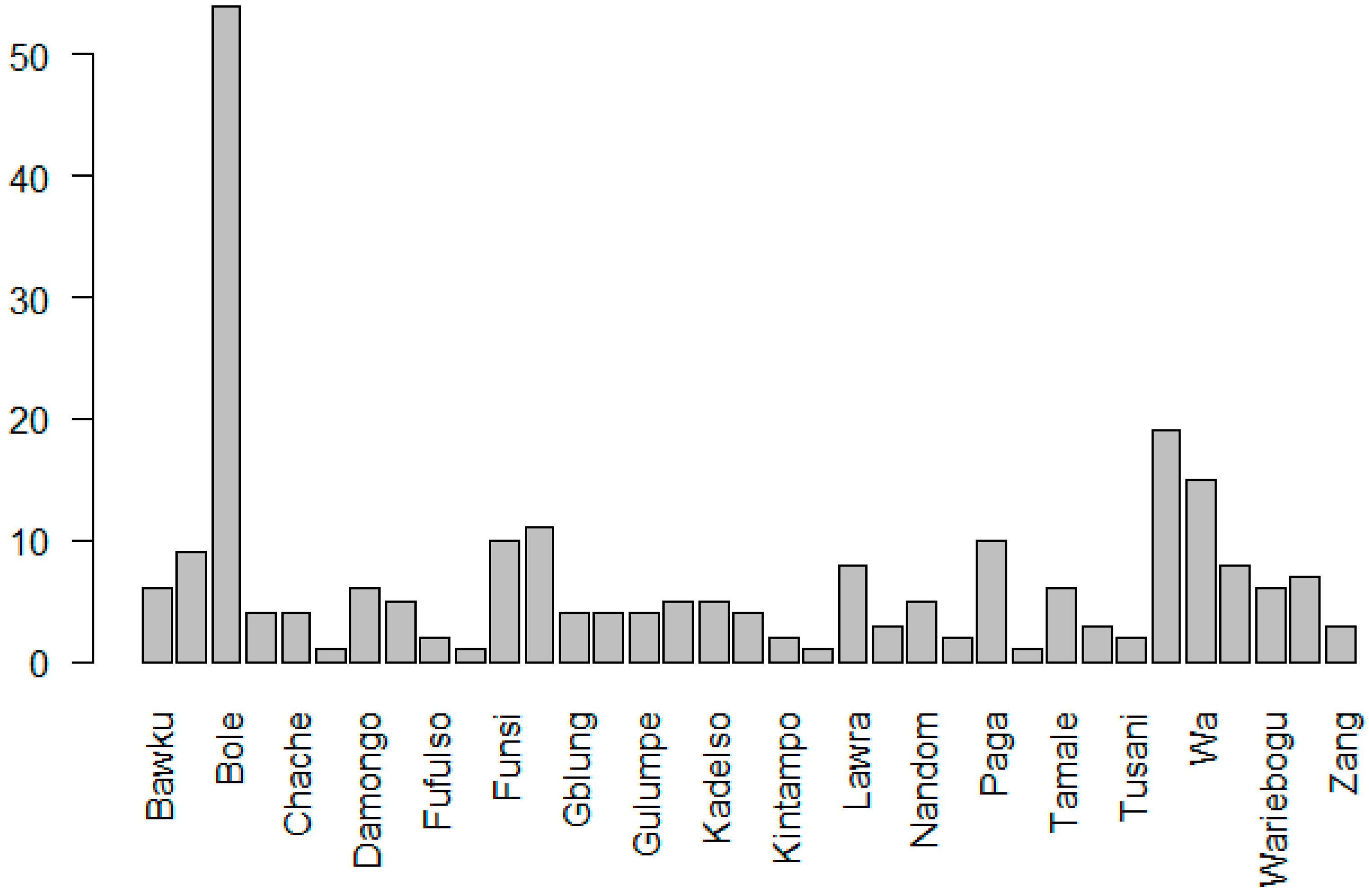
2.1. Evaluated Genotypes and Study Area
2.2. Data Collection
2.2.1. Sampling of Leaves for Genotyping-by-Sequencing
2.2.2. Sequencing
2.3. Data Analysis
3. Results
3.1. Morphological
3.2. Multivariate Analysis
3.2.1. Correlation among Traits
3.2.2. Genotype-Trait Association
3.3. Single Nucleotide Polymorphism Analysis
3.4. Diversity and Genetic Relationship
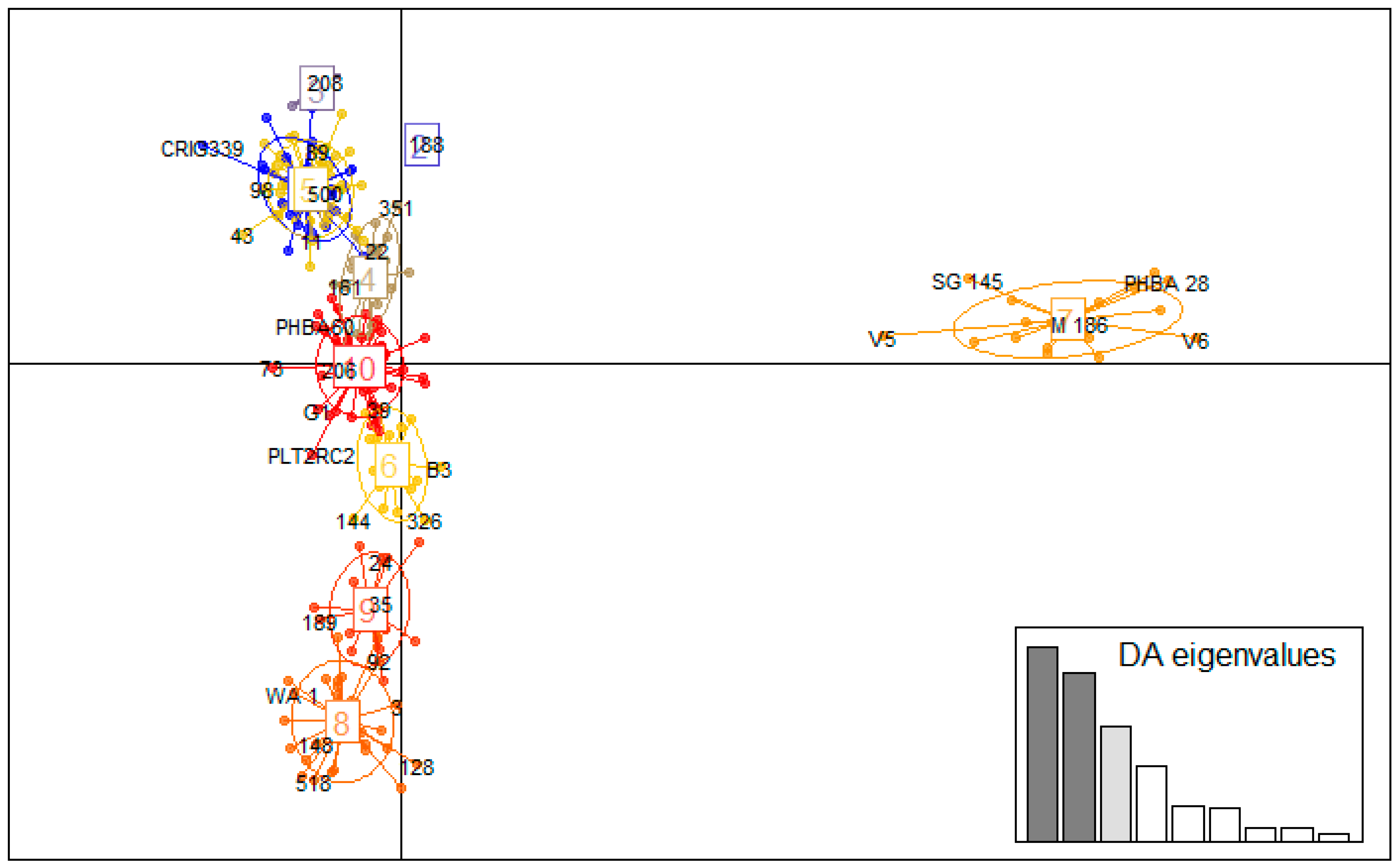
3.5. Multivariate Analysis
Genetic Clustering of Germplasms Using DAPC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maranz, S.; Wiesman, Z. Influence of climate on the tocopherol content of shea butter. J. Agric. Food Chem. 2004, 52, 2934–2937. [Google Scholar] [CrossRef] [PubMed]
- Lovett, P.N. The shea butter Industry expanding in West Africa. Inform 2005, 16, 273–275. [Google Scholar]
- Abbiw, D.K. Useful Plants of Ghana: West African Uses of Wild and Cultivated Plants; Intermediate Technology Publications: London, UK; The Royal Botanic Gardens, Kew: London, UK, 1990; pp. 66–67. [Google Scholar]
- Yidana, J.A. Pollination in Shea Trees; Ghana Annual Report 1992/93; Cocoa Research Institute: Bole, Ghana, 1994. [Google Scholar]
- Stout, J.C.; Nombre, I.; de Bruijn, B.; Delaney, A.; Doke, D.A.; Gyimah, T.; Kamano, F.; Kelly, R.; Lovett, P.; Marshall, E.; et al. Insect pollination improves yield of shea (Vitellaria paradoxa subsp. paradoxa) in the agroforestry parklands of West Africa. J. Pollinat. Ecol. 2018, 22, 11–20. [Google Scholar] [CrossRef]
- Jaeger, P. Note provisoire sur le role des chauves-souris dans la dissemination des fruits du karite (Butyrospermum parkii Kot.). Bull. Inst. Fr. Afr. Noire Ser. A Sci. Nat. 1962, 24, 113–115. [Google Scholar]
- Miège, J. Nombres chromosomiques et répartition géographique de quelques plantes tropicales et équatoriales. Rev. Cytol. Biol. Végétales Bot. 1954, 15, 312–348. [Google Scholar]
- Johnson, M. Cytology. In The Genera of Sapotaceae; Pennington, T.D., Ed.; Royal Botanic Gardens, Kew: London, UK, 1991; pp. 15–22. [Google Scholar]
- Gwali, S.; Vaillant, A.; Nakabonge, G.; Okullo, J.B.; Eilu, G.; Muchugi, A.; Bouvet, J.-M. Genetic diversity in shea tree (Vitellaria paradoxa subspecies nilotica) ethno-varieties in Uganda assessed with microsatellite markers. For. Trees Livelihoods 2014, 24, 163–175. [Google Scholar] [CrossRef]
- Allal, F.; Sanou, H.; Millet, L. Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity 2011, 107, 174–186. [Google Scholar] [CrossRef]
- Bouvet, J.M.; Fontaine, C.; Sanou, H.; Cardi, C. An analysis of the pattern of genetic variation in Vitellaria paradoxa using RAPD markers. Agrofor. Syst. 2004, 60, 61–69. [Google Scholar] [CrossRef]
- Lovett, P.N.; Haq, N. Diversity of the Sheanut tree (Vitellaria paradoxa C.F. Gaertn.) in Ghana. Genet. Resour. Crop. Evol. 2000, 47, 293–304. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Huq, M.A.; Akter, S.; Jung, Y.J.; Nou, I.S.; Cho, Y.G.; Kang, K.K. Identification of functional SNPs in genes and their effects on plant phenotypes. J. Plant Biotechnol. 2016, 43, 1–11. [Google Scholar] [CrossRef]
- Bacilieri, R.; Ducousso, A.; Kremer, A. Comparison of morphological characters and molecular markers for the analysis of hybridization in sessile and pedunculate oak. Sciences 1996, 53, 79–91. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H. Diversity arrays technology: A generic genome profiling technology on open platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1, new tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Djekota, C.; Diouf, D.; Sane, S.; Mbaye, M.S.; Noba, K. Morphological characterization of shea tree (Vitellaria paradoxa subsp. paradoxa) populations in the region of Mandoul in Chad. Int. J. Biodivers. Conserv. 2014, 6, 184–193. [Google Scholar] [CrossRef]
- Gwali, S.; Nakabonge, G.; Okullo, J.B.L.; Eilu, G.; Nyeko, P.; Vuzi, P. Morphological variation among shea tree (Vitellaria paradoxa subsp. nilotica) ‘ethnovarieties’ in Uganda. Genet. Resour. Crop. Evol. 2012, 59, 1883–1898. [Google Scholar] [CrossRef]
- Sanou, H.; Picard, N.; Lovett, P.N.; Dembélé, M.; Korbo, A.; Diarisso, D.; Bouvet, J.M. Phenotypic variation of agromorphological traits of the shea tree, Vitellaria paradoxa C.F. Gaertn., in Mali. Genetic Resour. Crop Evol. 2006, 53, 145–161. [Google Scholar] [CrossRef]
- Xiong, H.; Shi, A.; Mou, B.; Qin, J.; Motes, D.; Lu, W. Genetic Diversity and Population Structure of Cowpea (Vigna unguiculata L. Walp). PLoS ONE 2016, 11, e0160941. [Google Scholar] [CrossRef]
- Teklehaimanot, Z. Exploiting the potential of indigenous agroforestry trees: Parkia biglobosa and Vitellaria paradoxa in sub-Saharan Africa. New Vistas Agrofor. 2004, 1, 207–220. [Google Scholar]





| Variable | Minimum | Maximum | Mean | Std. Deviation |
|---|---|---|---|---|
| Blade length (cm) | 12.28 | 20.68 | 14.89 | 1.83 |
| Blade width (cm) | 4.17 | 9.21 | 5.10 | 0.93 |
| Total Length (cm) | 18.63 | 30.85 | 22.33 | 2.70 |
| Petiole length (cm) | 5.53 | 10.20 | 7.41 | 1.16 |
| BTPR (cm) | 1.41 | 2.69 | 2.07 | 0.27 |
| Latitude | Longitude | Blade Length | Petiole Length | Breadth | Total Length | |
|---|---|---|---|---|---|---|
| longitude | −0.11 | |||||
| Blade length | −0.47 ** | 0.34 | ||||
| Petiole length | −0.82 *** | −0.01 | 0.57 *** | |||
| Breadth | −0.45 ** | 0.09 | 0.27 | 0.49 ** | ||
| Total Length | −0.64 *** | 0.27 | 0.87 *** | 0.78 *** | 0.41 * | |
| Blade To Petiole ratio | 0.55 *** | 0.39 * | 0.21 | −0.67 *** | −0.33 | −0.14 |
| Leaf Parameters | Correlation | Cos2 | Contribution | |||
|---|---|---|---|---|---|---|
| Dim.1 | Dim.2 | Dim.1 | Dim.2 | Dim.1 | Dim.2 | |
| Blade length | 0.758 | 0.631 | 0.575 | 0.398 | 20.055 | 28.606 |
| Petiole length | 0.933 | −0.252 | 0.871 | 0.063 | 30.392 | 4.560 |
| Breadth | 0.632 | −0.261 | 0.399 | 0.068 | 13.923 | 4.915 |
| Total leaf length | 0.914 | 0.324 | 0.836 | 0.105 | 29.168 | 7.547 |
| Blade to petiole ratio | −0.430 | 0.869 | 0.185 | 0.756 | 6.461 | 54.372 |
| Ho | Hs | Ht | Dst | Htp | Dstp | Fst | Fstp | Fis | Dest |
|---|---|---|---|---|---|---|---|---|---|
| 0.0341 | 0.0406 | 0.0407 | 0.0001 | 0.0407 | 0.0001 | 0.0022 | 0.0023 | 0.1616 | 0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anyomi, W.E.; Barnor, M.T.; Eleblu, J.S.Y.; Danquah, A.; Avicor, S.W.; Ofori, K.; Hale, I.; Padi, F.K.; Danquah, E.Y. Elucidation of the Genetic Diversity within Some In Situ Shea Germplasm in Ghana. Agronomy 2023, 13, 2256. https://doi.org/10.3390/agronomy13092256
Anyomi WE, Barnor MT, Eleblu JSY, Danquah A, Avicor SW, Ofori K, Hale I, Padi FK, Danquah EY. Elucidation of the Genetic Diversity within Some In Situ Shea Germplasm in Ghana. Agronomy. 2023; 13(9):2256. https://doi.org/10.3390/agronomy13092256
Chicago/Turabian StyleAnyomi, Wisdom Edem, Michael Teye Barnor, John Saviour Yaw Eleblu, Agyemang Danquah, Silas Wintuma Avicor, Kwadwo Ofori, Iago Hale, Francis Kwame Padi, and Eric Yirenkyi Danquah. 2023. "Elucidation of the Genetic Diversity within Some In Situ Shea Germplasm in Ghana" Agronomy 13, no. 9: 2256. https://doi.org/10.3390/agronomy13092256









