A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering
Abstract
1. Introduction
2. Materials and Methods
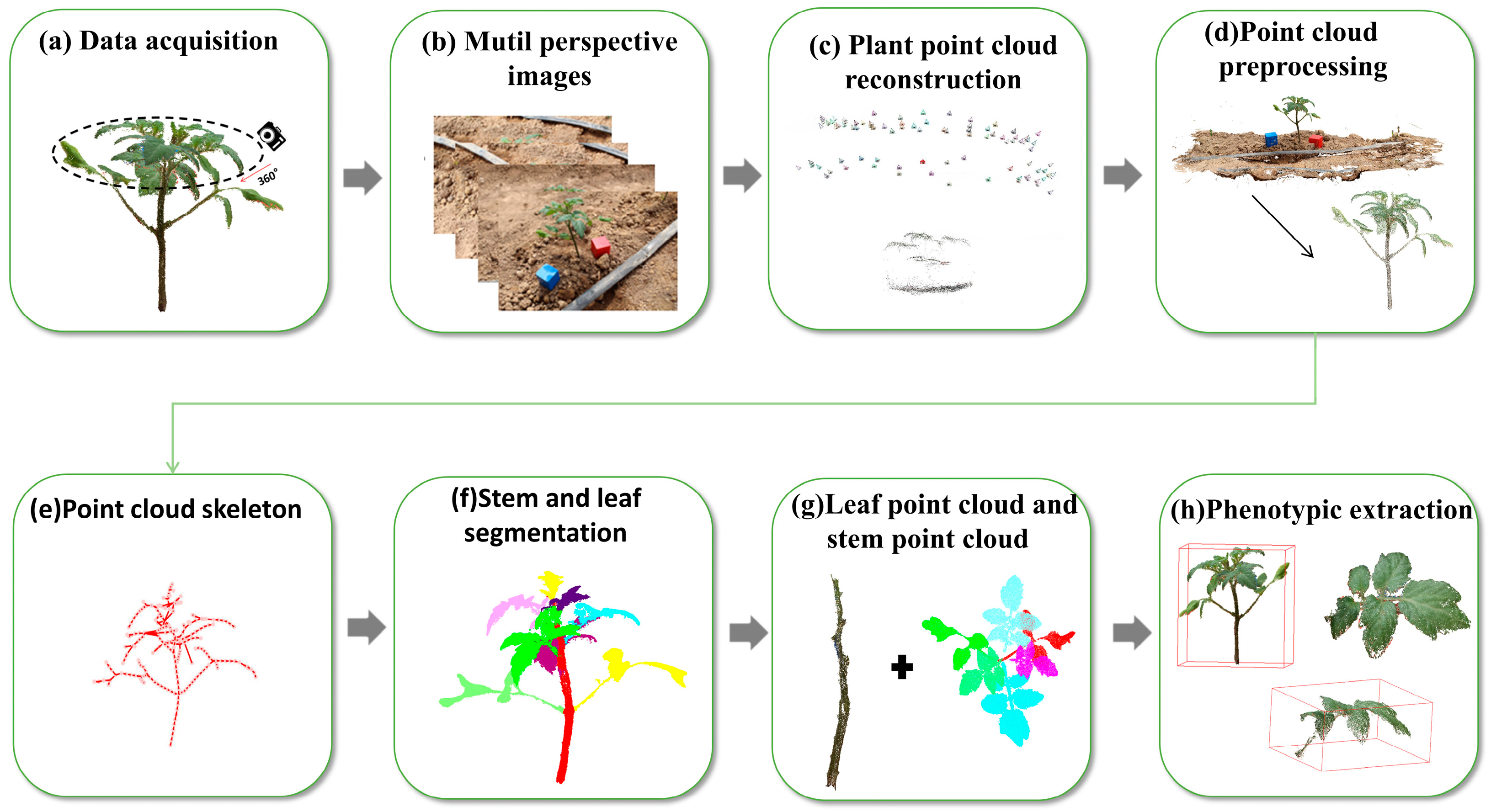
2.1. The Process of the Experiment
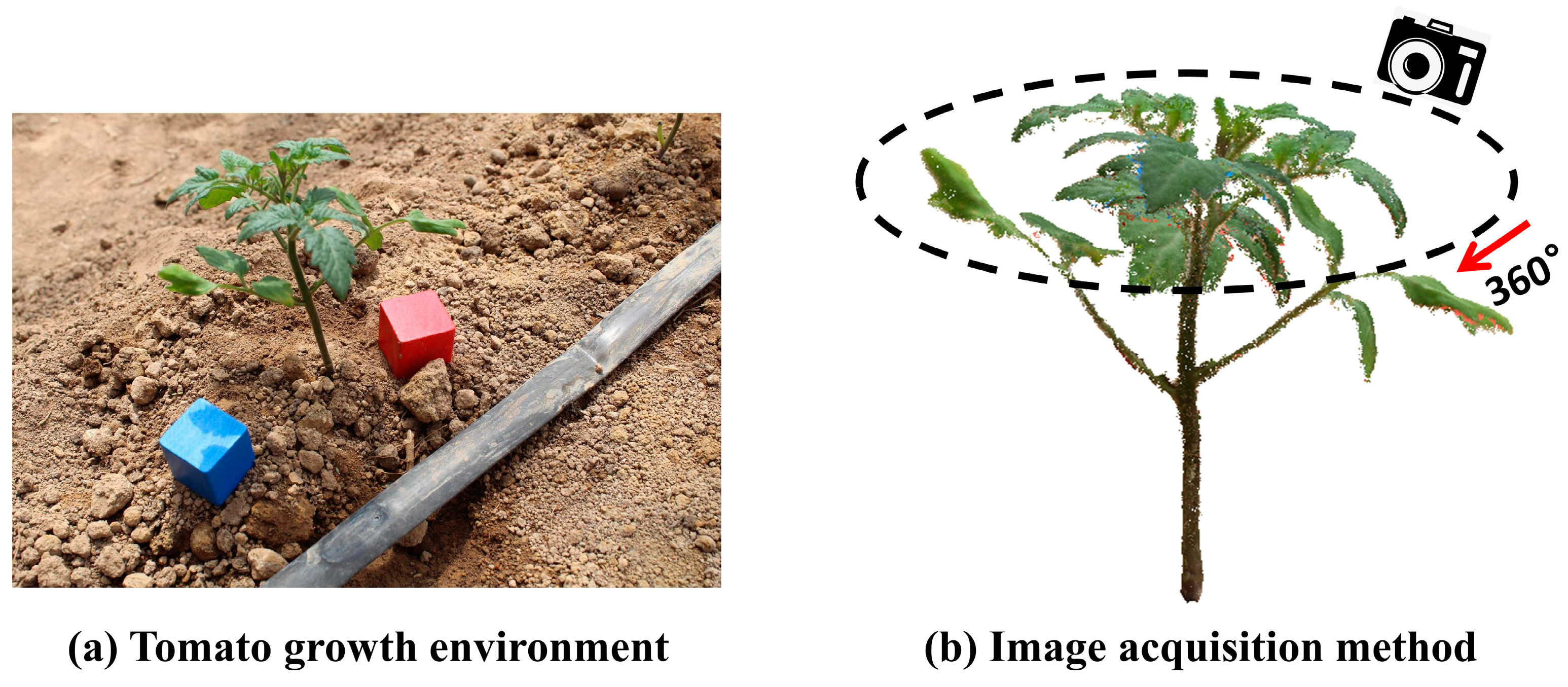
2.2. Image Data Acquisition
2.3. Point Cloud Reconstruction and Preprocessing
2.4. Stem and Leaf Segmentation Methods
2.4.1. Skeleton Extraction
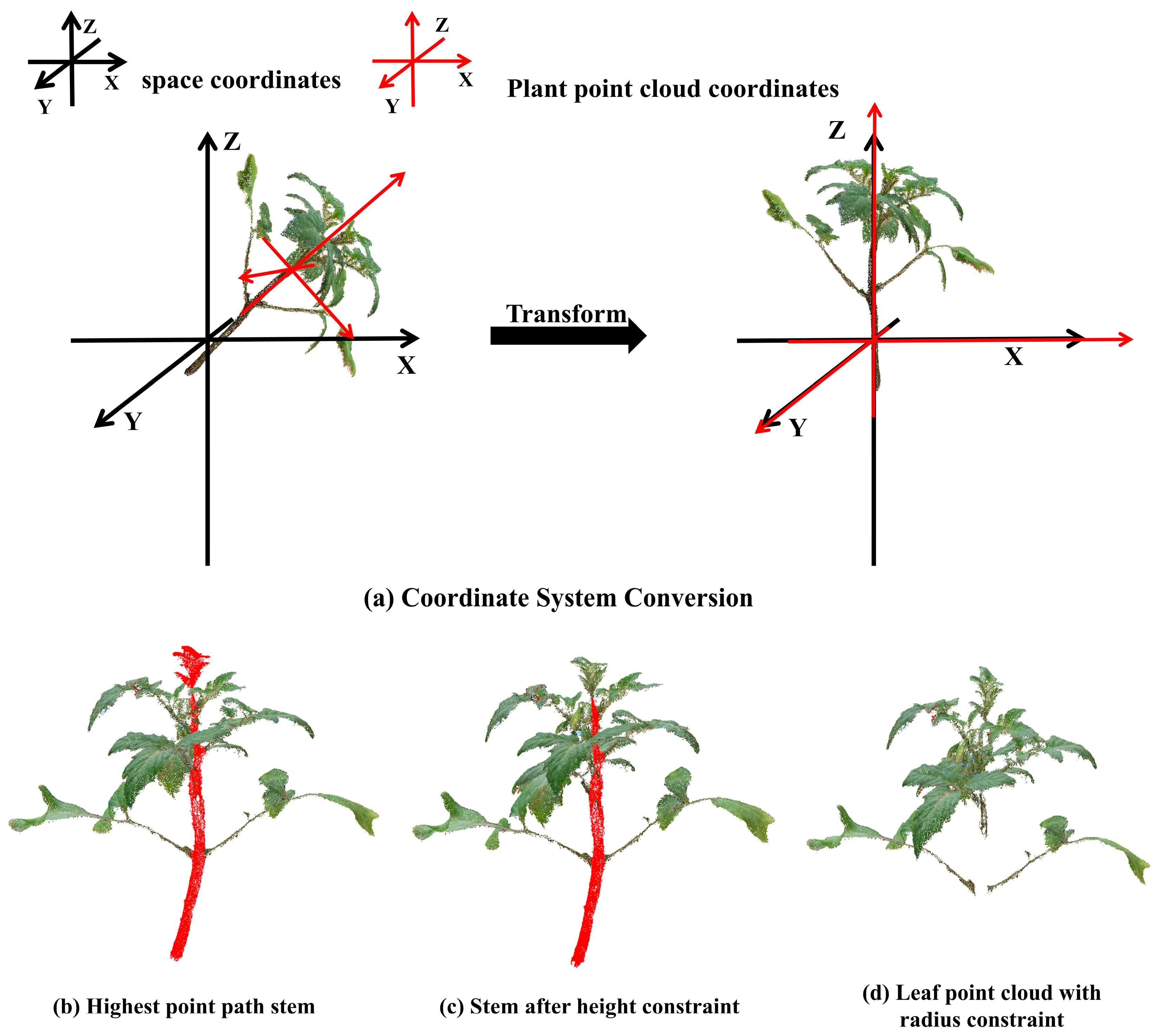
2.4.2. Skeleton Based Stem Extraction
Plant Coordinate System Correction
Finding the Path to the Highest Point of the Skeleton
Height Constraint
Radius Constraint
2.4.3. Supervoxel Clustering Based on Euclidean Distance
2.5. Phenotype Extraction Method for Tomato Plants
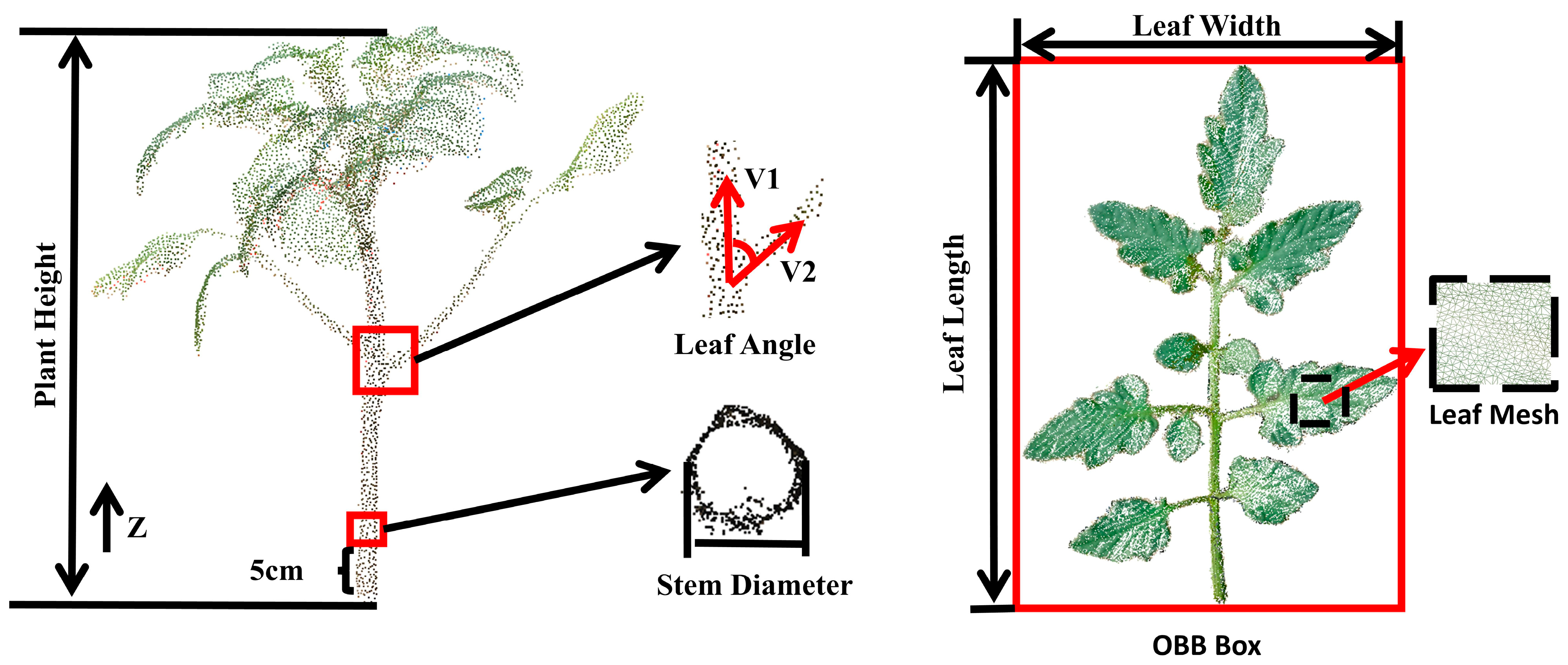
2.5.1. Phenotypic Parameter Extraction Method Based on Point Cloud
2.5.2. Phenotypic Parameter Real Value Acquisition
- Measurement of real data on plant height: using a tape measure, the distance from the ground part to the highest point of plant growth was measured as the plant height real value;
- Measurement of real data stem diameter: the stem at 5 cm above ground was uniformly selected, vernier calipers were used to measure the longitudinal and transverse distances of the stem using the crossover method, and, finally, the average of the two distances as the real value of the stem diameter was taken;
- Measurement of real data leaf angle: the angle between the leaf ventral surface and plant growth direction was measured with a protractor as the true value of the leaf angle;
- Measurement of real data leaf length: a tape measure was used to measure the distance from the petiole to the tip of the leaf as the true value of the leaf length;
- Measurement of real data leaf width: a tape measure was used to measure the distance of the maximum width of the middle part of the leaf as the true value of the leaf width;
- Measurement of real data leaf area: destructive sampling of the leaf was performed by placing the leaf on a black curtain containing a calibration block, using image processing techniques to extract the contours of the leaf and the calibration block, respectively, solving for their respective pixel points, and performing a pixel conversion to obtain the true leaf area of the leaf (Figure 8).
2.6. Evaluation of Stem and Leaf Segmentation and Phenotype Extraction
3. Results
3.1. Effectiveness and Evaluation of Different Stages of Stem and Leaf Division
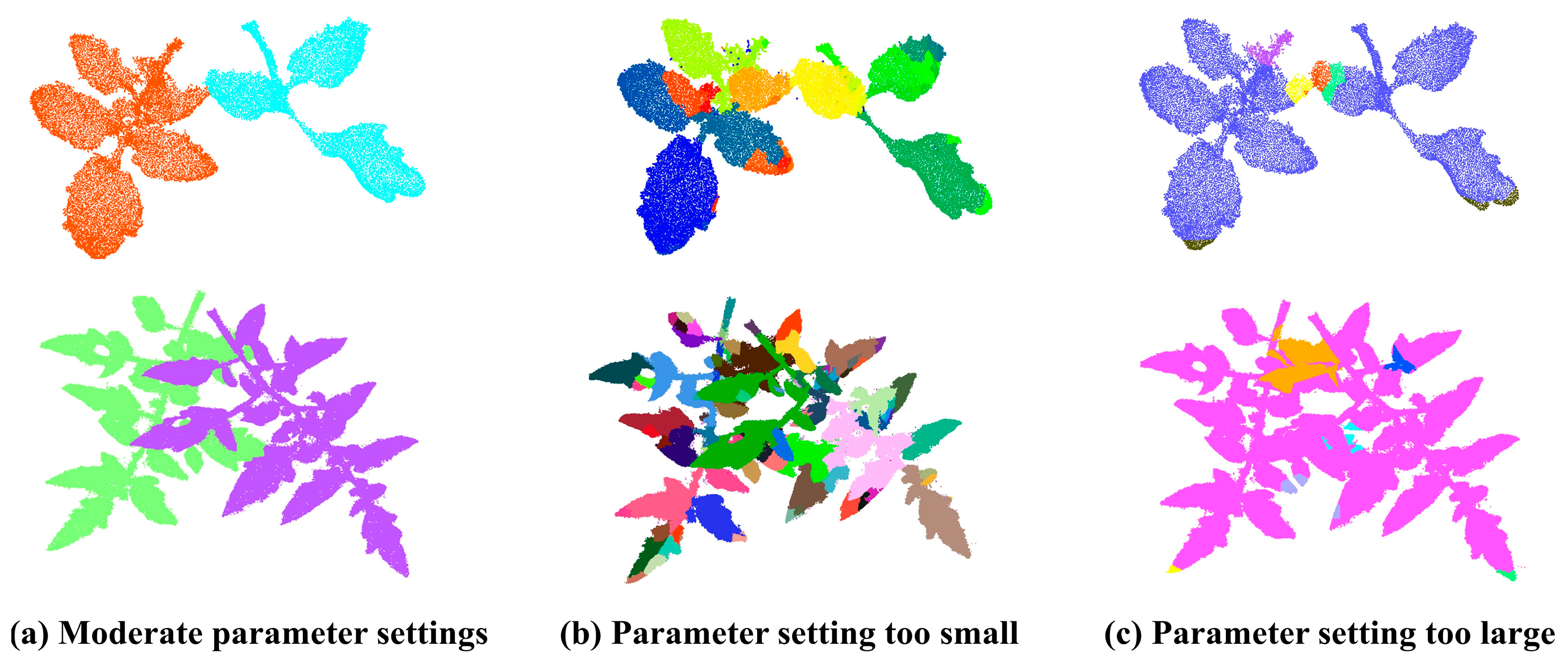
3.1.1. Effect of Algorithm Parameters on Blade Segmentation Accuracy
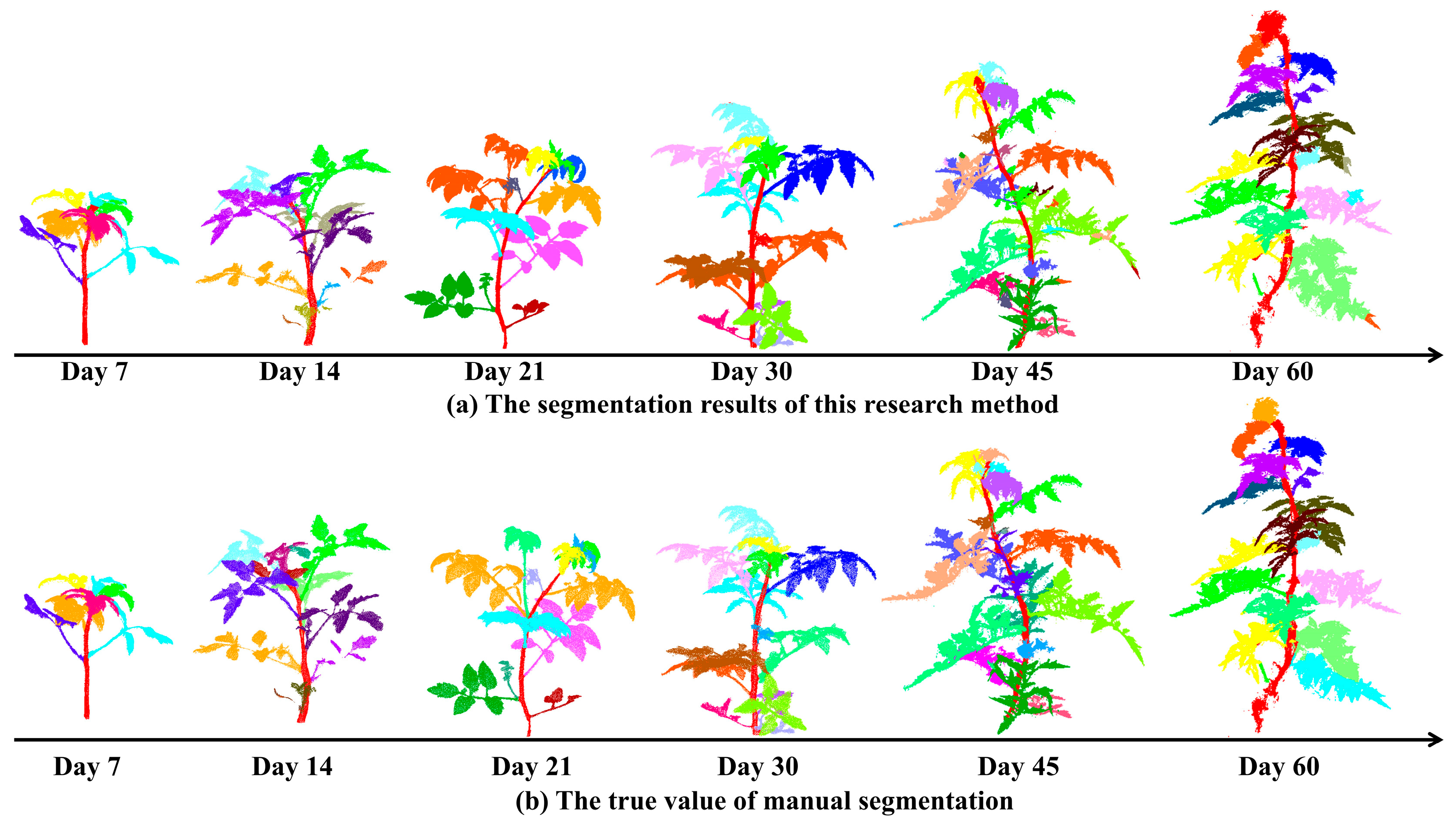
3.1.2. Stem and Leaf Segmentation Effect
- (1)
- As the plant grows, side shoots grow between the leaves and the stem, and our algorithm does not effectively segment them from the leaves, causing the side shoots and leaves to be grouped together, resulting in under-segmentation;
- (2)
- When there is a break between the leaves, our algorithm will split that leaf into multiple parts, obtaining many incomplete leaves, resulting in over-segmentation;
- (3)
- When there are mostly adhesions and occlusions between the blades, it is not possible to perform a complete segmentation by adjusting the parameters of the supervoxel, and, due to the large differences in the normal vectors between such leaves, it is unavoidable that the segmentation of a leaf into multiple parts occurs, resulting in an increase in the final number of leaves.
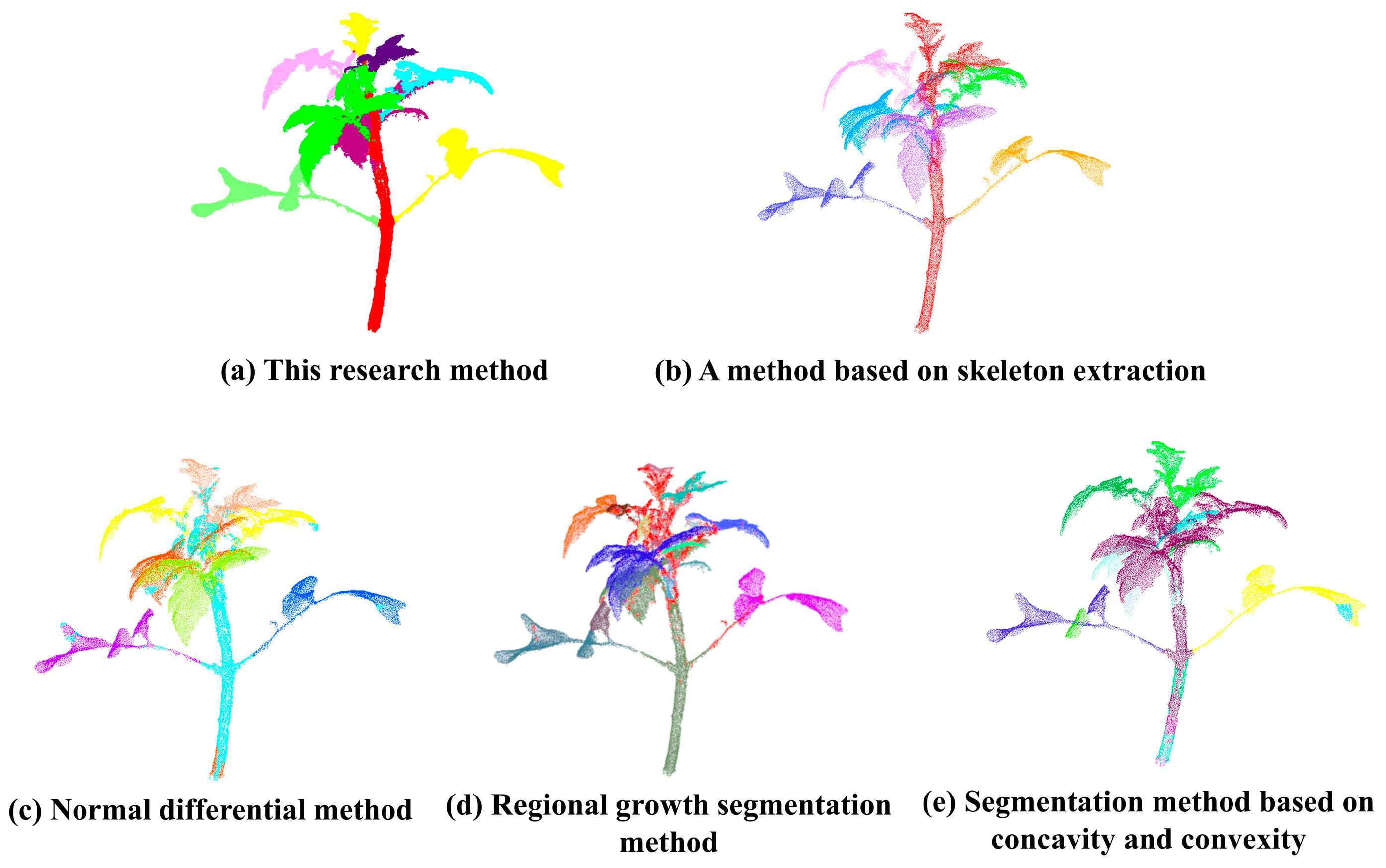
3.1.3. Comparison of Different Segmentation Methods
3.2. The Present Study on the Effectiveness of Stem and Leaf Division Methods for Division in Different Crops
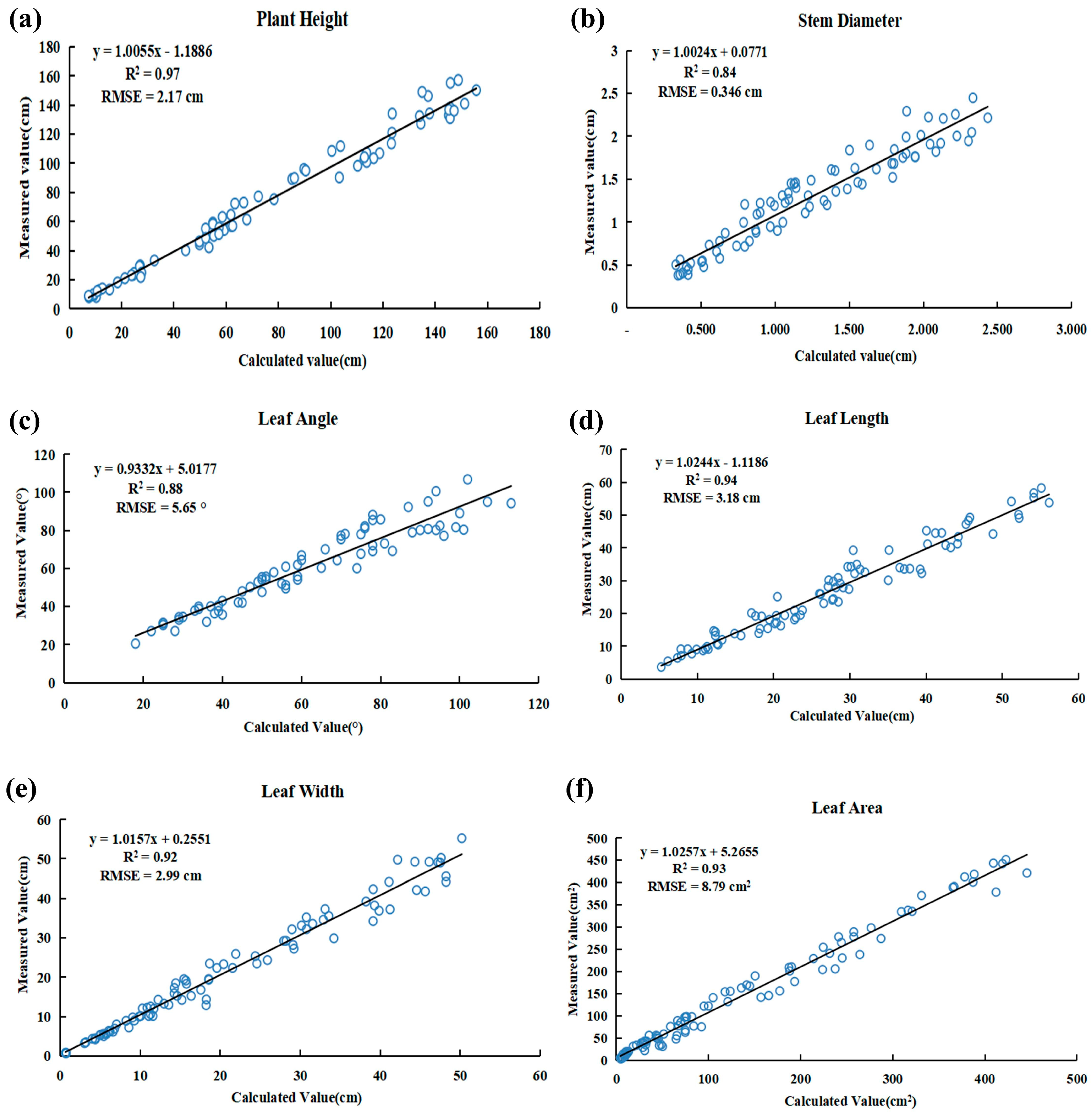
3.3. Phenotypic Parameter Measurement Results
3.4. Algorithm Efficiency Evaluation
4. Discussion
4.1. Comparison of Three-Dimensional Weighting Methods
4.2. Stem and Leaf Segmentation
4.3. Extraction of Phenotypic Parameters
4.4. Future Work
- (1)
- First, this study still acquires image data manually, which cannot be fully automated, and we will consider the combination of multiple cameras and mobile devices to realize the automatic acquisition of image data in a later stage;
- (2)
- Second, for the segmentation failure caused by mutual organ occlusion, the algorithm used will be subsequently improved to further increase the segmentation accuracy;
- (3)
- Third, the missing leaves seriously affected the phenotypic measurement results, and the point cloud complementation will be considered at a later stage to complement the mutilated leaves to further improve the measurement accuracy of the phenotypic parameters;
- (4)
- Finally, new methods will be further developed for phenotypic measurements of other organs of the tomato plant, such as fruits, roots, nodes and flowers, to provide a comprehensive picture of tomato growth dynamics. At the same time, according to the pattern of change in phenotypic parameters, there should be work with the water and fertilizer machine, to achieve automatic decision-making and irrigation.
5. Conclusions
- (1)
- Using the stem and leaf segmentation method proposed in this study to test tomato plants with different numbers of growth days, the average accuracy, average recall and average F1 score of stem and leaf segmentation were 0.88, 0.80 and 0.84, respectively, compared with the manual segmentation results; compared with the segmentation methods based on the skeleton, the normal differential difference method, the regional growth method and the segmentation method based on the concavity, the stem and leaf segmentation was successful. The average accuracy rate increased by 11, 24, 30 and 34 percentage points, respectively, indicating that the present research method can segment tomato organs more accurately. Meanwhile, segmentation tests were carried out on five other greenhouse crops, and the average accuracy rates were 0.98, 0.97, 0.97, 0.95 and 0.92, respectively, which had a certain degree of generalization, suggesting that the present research method can be effective in segmenting other crops as well. The method of this study has some reference value in the organ segmentation of multi-branched crops.
- (2)
- The correlation coefficients (R2) between the phenotype parameters measured using the proposed method and the true values obtained from manual measurements were 0.97 for plant height, 0.84 for stem diameter, 0.88 for leaf angle, 0.94 for leaf length, 0.92 for leaf width and 0.93 for leaf area. The relative errors (RMSE) were 2.17 cm for plant height, 0.346 cm for stem diameter, 5.65 degrees for leaf angle, 3.18 cm for leaf length, 2.99 cm for leaf width and 8.79 cm2 for leaf area. The algorithm’s measurement results exhibited a high correlation with the true values obtained from manual measurements, meeting the requirements for agricultural production.
- (3)
- The average elapsed time of the algorithm used in this study was 26.11 min, which can complete the point cloud reconstruction, stem and leaf segmentation, and phenotype extraction work faster, with high timeliness.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chitwood-Brown, J.; Vallad, G.E.; Lee, T.G.; Hutton, S.F. Breeding for Resistance to Fusarium Wilt of Tomato: A Review. Genes 2021, 12, 1673. [Google Scholar] [CrossRef] [PubMed]
- Roohanitaziani, R.; de Maagd, R.A.; Lammers, M.; Molthoff, J.; Meijer-Dekens, F.; van Kaauwen, M.P.; Bovy, A.G. Exploration of a resequenced tomato core collection forphenotypic and genotypic variation in plant growth and fruit quality traits. Genes 2020, 11, 1278. [Google Scholar] [CrossRef]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenoty** to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Huichun, Z.; Meng, Z.; Liming, B. Plant chlorophyll content estimation and visualization technology based on YOLO v5. J. Agric. Mach. 2022, 53, 313–321. [Google Scholar]
- Lu, C.P.; Liaw, J.J.; Wu, T.C.; Hung, T.F. Development of a mushroom growth measurement system applying deep learning for image recognition. Agronomy 2019, 9, 32. [Google Scholar] [CrossRef]
- Wang, D.; Li, C.; Song, H.; Xiong, H.; Liu, C.; He, D. Deep learning approach for apple edge detection to remotely monitor apple growth in orchards. IEEE Access 2020, 8, 26911–26925. [Google Scholar] [CrossRef]
- Lu, J.; Yubin, L.; Zhiyun, W. Optimization of Plant 3D Modeling ICP Point Cloud Registration. J. Agric. Eng. 2022, 38, 183–191. [Google Scholar]
- Yang, J.; Kai, T.; Weiguo, Z.; Xia, Y. Separation of stems and leaves of gramineous plants in mudflat wetlands from ground lidar point cloud data. China Laser 2022, 49, 122–130. [Google Scholar]
- Haibo, C.; Shengbo, L.; Lele, W.; Chang, Z. Automated 3D reconstruction and validation of single plant crops based on Kinect V3. J. Agric. Eng. 2022, 38, 215–223. [Google Scholar]
- Liu, X.; Wang, Y.; Kang, F.; Yue, Y.; Zheng, Y. Canopy parameter estimation of Citrus grandis var. Longanyou based on Lidar 3d point clouds. Remote Sens. 2021, 13, 1859. [Google Scholar] [CrossRef]
- Zhenghe, Y.; Chen, Y.; Huimin, Y.; Xhou, X. Extraction of geometric parameters of greenhouse tomato canopy based on LiDAR. Xinjiang Agric. Sci. 2021, 58, 1909. [Google Scholar]
- Jinqi, J.; Jingjing, H.; Jingxin, X. Acquisition of citrus canopy structure information based on mobile 3D LiDAR. J. Hunan Agric. Univ. 2022, 48, 109–113. [Google Scholar] [CrossRef]
- Lin, C.; Hu, F.; Peng, J.; Wang, J.; Zhai, R. Segmentation and stratification methods of field maize terrestrial LiDAR point cloud. Agriculture 2022, 12, 1450. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Wu, F.; Pang, S.; Gao, S.; Hu, T.; Guo, Q. Stem-Leaf Segmentation and Phenotypic Trait Extraction of Individual Maize Using Terrestrial LiDAR Data. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1336–1346. [Google Scholar] [CrossRef]
- Yuchao, L. Research on 3D Reconstruction and Phenotypic Measurement of Maize Plants at Seedling Stage Based on Multi Perspective Images; Hebei Agricultural University: Hebei, China, 2022. [Google Scholar]
- Johann, C.R.; Stefan, P.; Heiner, K. Accuracy Analysis of a Multi-View Stereo Approach for Phenotyping of Tomato Plants at the Organ Level. Sensors 2015, 15, 9651–9665. [Google Scholar]
- Shaochen, L.; Aiwu, Z.; Xizhen, Z.; Jang, Z.; Li, M. Method for extracting three-dimensional phenotypic information of maize seedlings at leaf scale. Prog. Laser Optoelectron. 2023, 60, 71–79. [Google Scholar]
- Chai, H.; Shao, K.; Yu, C.; Shao, J.; Wang, R.; Sui, Y.; Bai, K.; Liu, Y.; Ma, W. Extraction of Beet Root Phenotypic Parameters and Root Type Discrimination Based on 3D Point Cloud. J. Agric. Eng. 2020, 36, 181–188. [Google Scholar]
- Aobo, J.; Tianhao, D.; Yan, Z. Identification of tobacco plant type features in the field based on 3D point clouds and ensemble learning. J. Zhejiang Univ. 2022, 48, 393–402. [Google Scholar]
- Yitong, X.; Shuai, L.; Chenlian, H.; Liu, Q.; Li, F.; Zhang, W. Organ segmentation and phenotype analysis of soybean plants based on three-dimensional point clouds. China J. Agric. Sci. Technol. 2023, 25, 115–125. [Google Scholar] [CrossRef]
- Li, D.; Shi, G.; Kong, W.; Wang, S.; Chen, Y. A leaf segmentation and phenotypic feature extraction framework for multiview stereo plant point clouds. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 2321–2336. [Google Scholar] [CrossRef]
- Elnashef, B.; Filin, S.; Lati, R.N. Tensor-based classification and segmentation of three-dimensional point clouds for organ-level plant phenoty** and growth analysis. Comput. Electron. Agric. 2019, 156, 51–61. [Google Scholar] [CrossRef]
- Xu, Y. Research on Crop Phenotypic Parameter Acquisition and Dynamic Quantification Method Based on Multitemporal Point Cloud Data; Huazhong Agricultural University: Wuhan, China, 2019. [Google Scholar] [CrossRef]
- Qibing, Z.; Li, H.; Chen, Z. Recognition and counting method of potted kumquat fruits based on point cloud alignment. J. Agric. Mach. 2022, 53, 209–216. [Google Scholar]
- Li, D.; Shi, G.; Li, J.; Zhang, S.; Xiang, S.; Jin, S. PlantNet: A dual-function point cloud segmentation network for multiple plant species. ISPRS J. Photogramm. Remote Sens. 2022, 184, 243–263. [Google Scholar] [CrossRef]
- Guo, X.; Sun, Y.; Yang, H. FF-Net: Feature-Fusion-Based Network for Semantic Segmentation of 3D Plant Point Cloud. Plants 2023, 12, 1867. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, S.; Ren, H.; Yang, W.; Zhai, R. 3DPhenoMVS: A Low-Cost 3D Tomato Phenotyping Pipeline Using 3D Reconstruction Point Cloud Based on Multiview Images. Agronomy 2022, 12, 1865. [Google Scholar] [CrossRef]
- Cheng, P.; Shuai, L.; Yanlong, M. Tomato plant stem and leaf segmentation and phenotypic feature extraction based on 3D point cloud. J. Agric. Eng. 2022, 38, 187–194. [Google Scholar]
- Baiming, L.; Qian, W.; Jie, W.; Su, H. Optimization of crop 3D point cloud reconstruction strategy based on multi view automatic imaging system. J. Agric. Eng. 2023, 39, 8675. [Google Scholar]
- Xiuying, L.; Fengran, Z.; Huan, C.; Fan, B.; Li, H. 3D reconstruction and trait extraction of maize plants based on motion recovery structure. J. Agric. Mach. 2020, 51, 209–2019. [Google Scholar]
- Zhu, T.; Ma, X.; Guan, H.; Wu, X.; Wang, F. A method for detecting tomato canopies’ phenotypic traits based on improved skeleton extraction algorithm. Comput. Electron. Agric. 2023, 214, 108285. [Google Scholar] [CrossRef]
- Cao, J.; Tagliasacchi, A.; Olson, M.; Zhang, H.; Su, Z. Point cloud skeletons via Laplacian based contraction. In Proceedings of the SMI 2010, Shape Modeling International Conference, Aix en Provence, France, 21–23 June 2010; IEEE: Piscataway, NJ, USA, 2010; Volume 57, pp. 187–197. [Google Scholar]
- Wang, Y.; Chen, Y.; Zhang, X.; Gong, W. Research on measurement method of leaf length and width based on point cloud. Agriculture 2021, 11, 63. [Google Scholar] [CrossRef]
- Dijkstra, E.W. A note on two problems in connexion with graphs. Numer. Math. 1959, 1, 269–271. [Google Scholar] [CrossRef]
- Dongyu, R.; Xiaojuan, L.; Tao, L.; Tang, J. A three-dimensional reconstruction method for fruit tree branches based on Kinect v2 sensor. J. Agric. Mach. 2022, 53, 197–203. [Google Scholar]
- Wu, Q.; Liu, J.; Gao, C.; Wang, B.; Shen, G. Improved RANSAC point cloud spherical target detection and parameter estimation method based on principal curvature constraint. Sensors 2022, 22, 5850. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.S.; Han, Y.X. A Low-Cost 3D Phenotype Measurement Method of Leafy Vegetables Using Video Recordings from Smartphones. Sensors 2020, 20, 6068. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Tang, S.; Liao, J.; Li, X.; Wang, W.; Li, Y.; Guo, R. A supervoxel-based random forest method for robust and effective airborne LiDAR point cloud classification. Remote Sens. 2022, 14, 1516. [Google Scholar] [CrossRef]
- Zheng, L.; Wang, L.; Wang, M. Automatic 3D point cloud reconstruction of Leaf lettuce based on Kinect camera. Trans. Chin. Soc. Agric. Machinery 2001, 52, 159–168. [Google Scholar]
- Yibin, L.; Shenglian, L.; Tingting, Q.; Chen, M. Plant 3D point cloud segmentation. J. Appl. Sci. 2021, 39, 660–671. [Google Scholar]
- Xiao, S.; Liu, S.; Bi, K.; Du, M. A fast and accurate approach to the extraction of leaf midribs from point clouds. Remote Sens. Lett. 2020, 11, 255–264. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, X.; Guan, H.; Zhu, K.; Feng, J.; Yu, S. A method for calculating the leaf inclination of soybean canopy based on 3D point clouds. Int. J. Remote Sens. 2021, 42, 5719–5740. [Google Scholar] [CrossRef]
- Feldman, A.; Wang, H.; Fukano, Y.; Kato, Y.; Ninomiya, S.; Guo, W. EasyDCP: An affordable, high-throughput tool to measure plant phenotypic traits in 3D. Methods Ecol. Evol. 2021, 12, 1679–1686. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, J.; Zhang, M. Curve skeleton extraction via k-nearest-neighbors based contraction. Int. J. Appl. Math. Comput. Sci. 2020, 30, 123–132. [Google Scholar]






| Growth Days/d | |||
|---|---|---|---|
| 7 | 0.88 | 0.80 | 0.84 |
| 14 | 0.91 | 0.84 | 0.87 |
| 21 | 0.92 | 0.85 | 0.88 |
| 30 | 0.88 | 0.82 | 0.85 |
| 45 | 0.86 | 0.77 | 0.81 |
| 60 | 0.84 | 0.74 | 0.79 |
| Average | 0.88 | 0.80 | 0.84 |
| Methods | Average of Precision | Average of Recall Rates | Average of F1 |
|---|---|---|---|
| This research method | 0.88 | 0.80 | 0.84 |
| Segmentation method based on skeleton extraction | 0.77 | 0.63 | 0.76 |
| Normal differential method | 0.64 | 0.59 | 0.57 |
| Regional growth segmentation method | 0.58 | 0.53 | 0.59 |
| Segmentation method based on concavity and convexity | 0.54 | 0.57 | 0.62 |
| Crop Name | Average of Precision | Average of Recall Rates | -Score |
|---|---|---|---|
| Maize | 0.98 | 0.96 | 0.94 |
| Eggplant | 0.97 | 0.95 | 0.96 |
| Cucumber | 0.97 | 0.94 | 0.97 |
| Pepper | 0.95 | 0.93 | 0.92 |
| Squash | 0.92 | 0.89 | 0.92 |
| Processing Stage | Point Cloud Reconstruction/min | Stem and Leaf Segmentation/min | Phenotypic Extraction/min | Total Time/min |
|---|---|---|---|---|
| Minimum processing time | 7.37 | 3.26 | 1.72 | 12.35 |
| Maximum processing time | 26.44 | 7.26 | 3.59 | 37.29 |
| Average processing time | 17.89 | 5.25 | 2.97 | 26.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, Q.; Yang, J.; Ren, G.; Wang, W.; Zhang, W.; Li, F. A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering. Agronomy 2024, 14, 198. https://doi.org/10.3390/agronomy14010198
Wang Y, Liu Q, Yang J, Ren G, Wang W, Zhang W, Li F. A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering. Agronomy. 2024; 14(1):198. https://doi.org/10.3390/agronomy14010198
Chicago/Turabian StyleWang, Yaxin, Qi Liu, Jie Yang, Guihong Ren, Wenqi Wang, Wuping Zhang, and Fuzhong Li. 2024. "A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering" Agronomy 14, no. 1: 198. https://doi.org/10.3390/agronomy14010198
APA StyleWang, Y., Liu, Q., Yang, J., Ren, G., Wang, W., Zhang, W., & Li, F. (2024). A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering. Agronomy, 14(1), 198. https://doi.org/10.3390/agronomy14010198





