Comprehensive Morphometric and Biochemical Characterization of Seven Basil (Ocimum basilicum L.) Genotypes: Focus on Light Use Efficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Biological Materials
2.2. Experimental Design and Growing Practices
2.3. Plant Morphometric Characterization
2.4. Light Use Efficiency (LUE) Determination
2.5. Analyses of Volatile Components
2.6. Identification of Components of Essential Oils
2.7. Shading System Set-Up
2.8. Data Analysis
3. Results
3.1. Climatic Conditions
3.2. Morphometric Characterization of Harvested Plants at Different Stages
3.3. Plant Morphometric Characterization
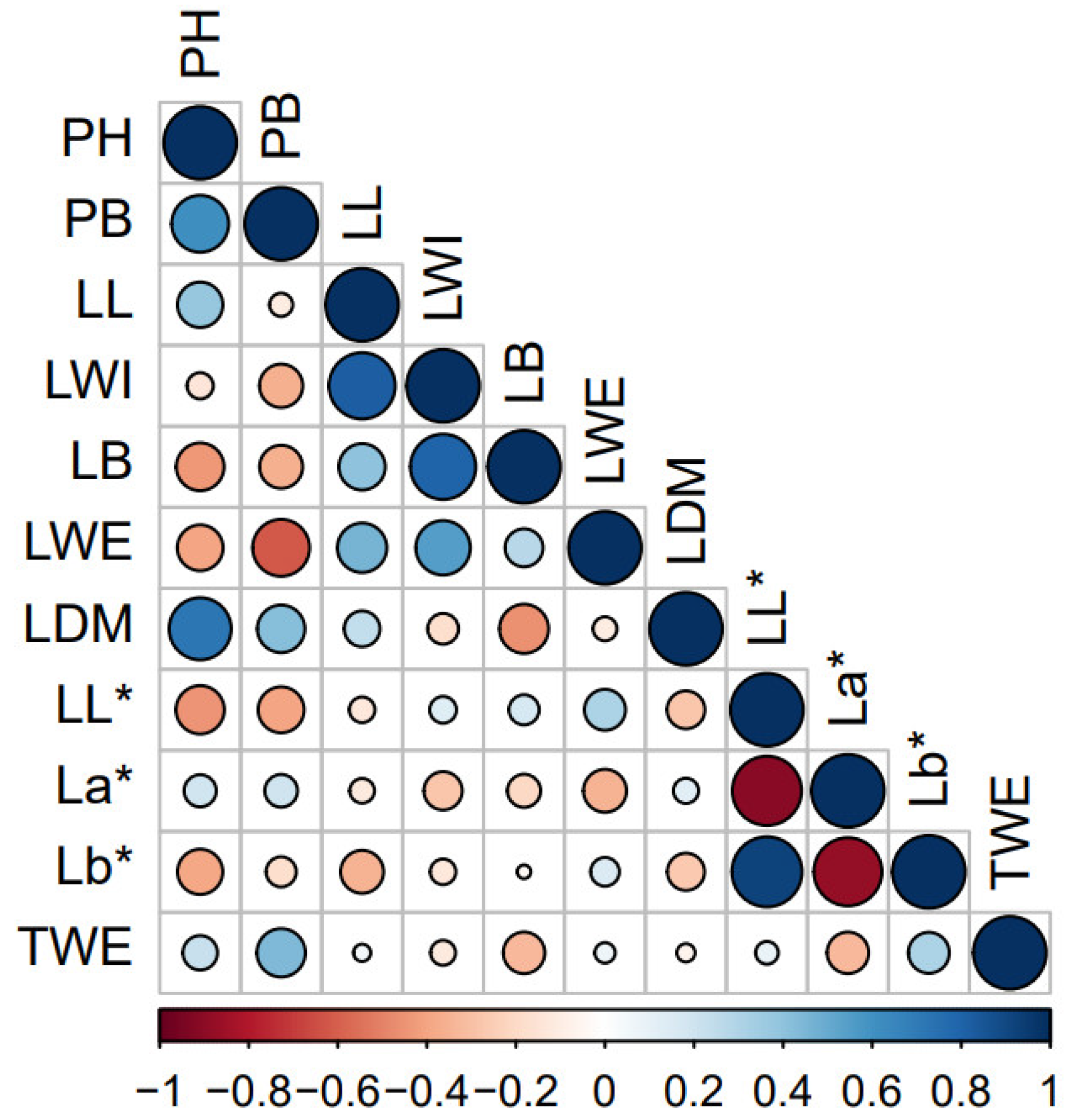
3.4. Pearson’s Correlation between Traits
3.5. Biochemical Characterization of the Volatile Compounds
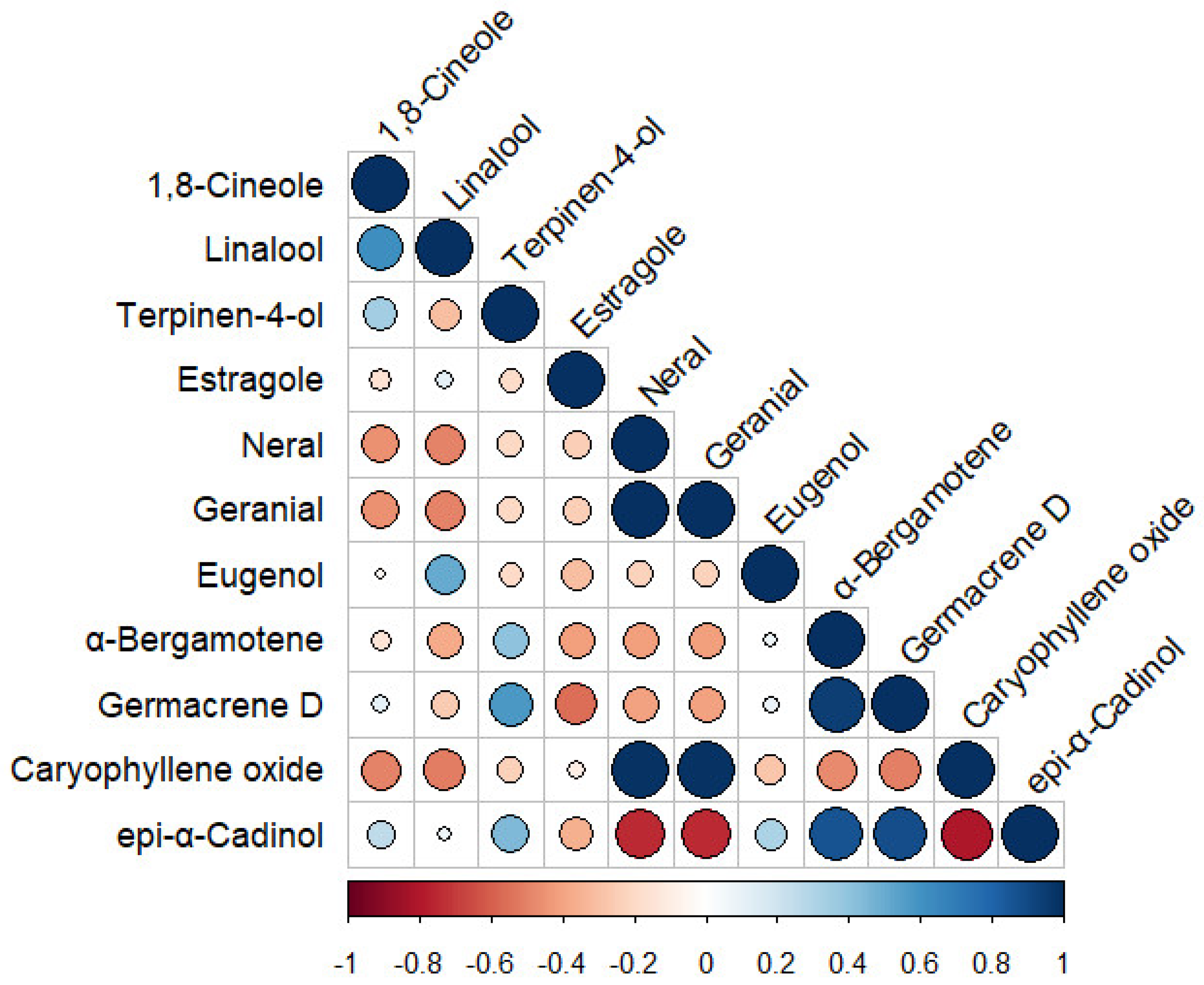
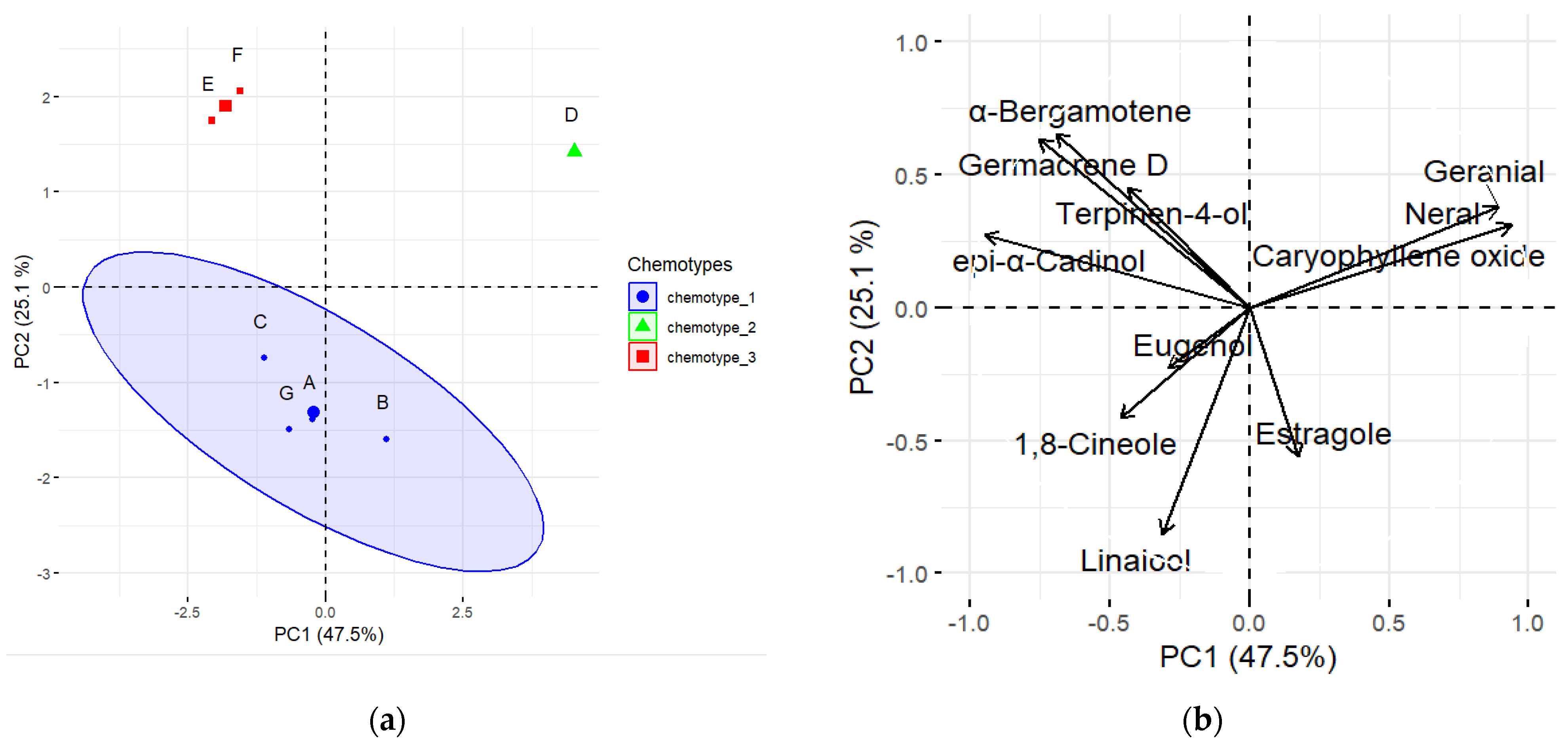
3.6. Pearson’s Correlation and Chemotypes Determination among the Aromatic Compounds
3.7. Variations in Harvesting Weight in Relation to the Solar Radiation Conditions
3.8. Light Use Efficiency (LUE) in Relation to the Solar Radiation Conditions
3.9. Variation in the Morphometric Traits in Relation to the Solar Radiation Conditions
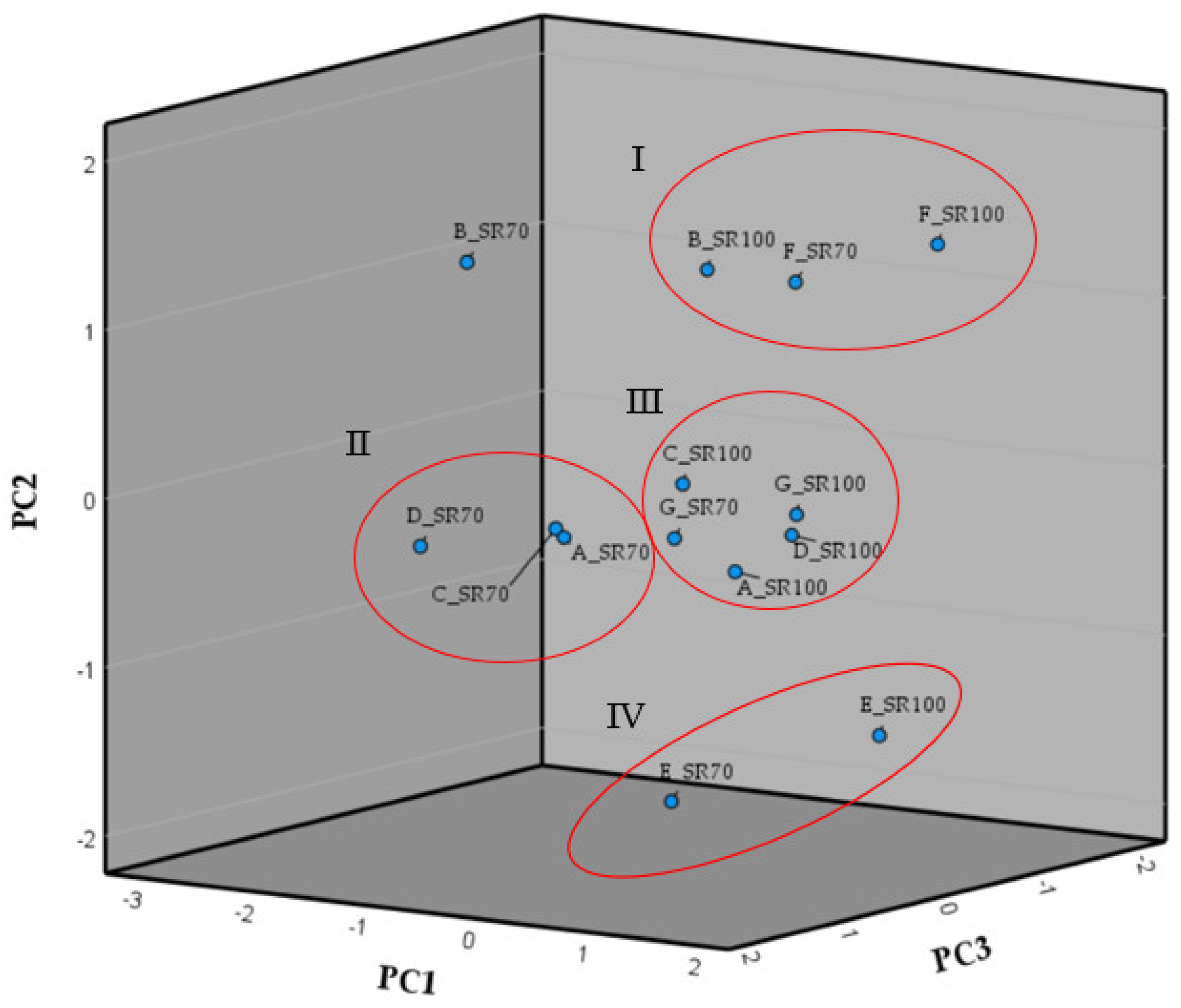
3.10. Principal Component Analysis of the Morphological Trait Variations as Result of the Different Solar Radiation Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yaldiz, G.; Camlica, M. Agro-morphological and Phenotypic Variability of Sweet Basil Genotypes for Breeding Purposes. Crop Sci. 2021, 61, 621–642. [Google Scholar] [CrossRef]
- Purushothaman, B.; Srinivasan, R.P.; Suganthi, P.; Ranganathan, B.; Gimbun, J.; Shanmugam1, K. A Comprehensive Review on Ocimum basilicum. J. Nat. Remedies 2018, 18, 71–85. [Google Scholar] [CrossRef]
- Dudai, N.; Nitzan, N.; Gonda, I. Ocimum basilicum L. (Basil). In Medicinal, Aromatic and Stimulant Plants; Springer: Berlin/Heidelberg, Germany, 2020; pp. 377–405. [Google Scholar]
- Rubab, S.; Bahadur, S.; Hanif, U.; Durrani, A.I.; Sadiqa, A.; Shafique, S.; Zafar, U.; Shuaib, M.; Urooj, Z.; Nizamani, M.M.; et al. Phytochemical and Antimicrobial Investigation of Methanolic Extract/Fraction of Ocimum basilicum L. Biocatal. Agric. Biotechnol. 2021, 31, 101894. [Google Scholar] [CrossRef]
- Filip, S. Basil (Ocimum basilicum L.) a Source of Valuable Phytonutrients. Int. J. Clin. Nutr. Diet. 2017, 3, 118. [Google Scholar] [CrossRef]
- Almalki, D. Renoprotective Effect of Ocimum basilicum (Basil) Against Diabetes-Induced Renal Affection in Albino Rats. Mater. Socio Med. 2019, 31, 236. [Google Scholar] [CrossRef]
- Pratama, Y.; Ulfah, T.; Bintoro, V.P. Effect of Basil (Ocimum americanum L.) Proportion on Physical and Organoleptical Properties of Basil Cracker. J. Appl. Food Technol. 2018, 5, 1–5. [Google Scholar] [CrossRef]
- Shahrajabian, M.H.; Sun, W.; Cheng, Q. Chemical Components and Pharmacological Benefits of Basil (Ocimum basilicum): A Review. Int. J. Food Prop. 2020, 23, 1961–1970. [Google Scholar] [CrossRef]
- Zhan, Y.; An, X.; Wang, S.; Sun, M.; Zhou, H. Basil Polysaccharides: A Review on Extraction, Bioactivities and Pharmacological Applications. Bioorg. Med. Chem. 2020, 28, 115179. [Google Scholar] [CrossRef]
- Yaldız, G.; Çamlıca, M.; Özen, F.; Eratalar, S.A. Effect of Poultry Manure on Yield and Nutrient Composition of Sweet Basil (Ocimum basilicum L.). Commun. Soil Sci. Plant Anal. 2019, 50, 838–852. [Google Scholar] [CrossRef]
- Ciriello, M.; Pannico, A.; El-Nakhel, C.; Formisano, L.; Cristofano, F.; Duri, L.G.; Pizzolongo, F.; Romano, R.; De Pascale, S.; Colla, G.; et al. Sweet Basil Functional Quality as Shaped by Genotype and Macronutrient Concentration Reciprocal Action. Plants 2020, 9, 1786. [Google Scholar] [CrossRef]
- Du, P.; Yuan, H.; Chen, Y.; Zhou, H.; Zhang, Y.; Huang, M.; Jiangfang, Y.; Su, R.; Chen, Q.; Lai, J.; et al. Identification of Key Aromatic Compounds in Basil (Ocimum L.) Using Sensory Evaluation, Metabolomics and Volatilomics Analysis. Metabolites 2023, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- D’Imperio, M.; Renna, M.; Cardinali, A.; Buttaro, D.; Santamaria, P.; Serio, F. Silicon Biofortification of Leafy Vegetables and Its Bioaccessibility in the Edible Parts. J. Sci. Food Agric. 2016, 96, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Raiciu, A.D.; Livadariu, O.; Șerbanica, C.-P. The Assesment of the Influence Induced by LED-s Irradiation on Basil Sprouts (Ocimum basilicum L.). Rom. Biotechnol. Lett. 2020, 25, 1334–1339. [Google Scholar] [CrossRef]
- Puccinelli, M.; Malorgio, F.; Pintimalli, L.; Rosellini, I.; Pezzarossa, B. Biofortification of Lettuce and Basil Seedlings to Produce Selenium Enriched Leafy Vegetables. Horticulturae 2022, 8, 801. [Google Scholar] [CrossRef]
- Stetsenko, L.A.; Pashkovsky, P.P.; Voloshin, R.A.; Kreskavski, V.D.; Kuznetsov, V.V.; Allakhverdiev, S.I. Role of Anthocyanin and Carotenoids in the Adaptation of the Photosynthetic Apparatus of Purple- and Green-Leaved Cultivars of Sweet Basil (Ocimum basilicum) to High-Intensity Light. Photosynthetica 2020, 58, 890–901. [Google Scholar] [CrossRef]
- Barbi, S.; Barbieri, F.; Taurino, C.; Bertacchini, A.; Montorsi, M. Quantitative Calculation of the Most Efficient LED Light Combinations at Specific Growth Stages for Basil Indoor Horticulture: Modeling through Design of Experiments. Appl. Sci. 2023, 13, 2004. [Google Scholar] [CrossRef]
- Milić Komić, S.; Živanović, B.; Dumanović, J.; Kolarž, P.; Sedlarević Zorić, A.; Morina, F.; Vidović, M.; Veljović Jovanović, S. Differential Antioxidant Response to Supplemental UV-B Irradiation and Sunlight in Three Basil Varieties. Int. J. Mol. Sci. 2023, 24, 15350. [Google Scholar] [CrossRef]
- Kotilainen, T.; Robson, T.M.; Hernández, R. Light Quality Characterization under Climate Screens and Shade Nets for Controlled-Environment Agriculture. PLoS ONE 2018, 13, e0199628. [Google Scholar] [CrossRef]
- Pennisi, G.; Pistillo, A.; Orsini, F.; Cellini, A.; Spinelli, F.; Nicola, S.; Fernandez, J.A.; Crepaldi, A.; Gianquinto, G.; Marcelis, L.F.M. Optimal Light Intensity for Sustainable Water and Energy Use in Indoor Cultivation of Lettuce and Basil under Red and Blue LEDs. Sci. Hortic. 2020, 272, 109508. [Google Scholar] [CrossRef]
- Lim, S.; Kim, J. Light Quality Affects Water Use of Sweet Basil by Changing Its Stomatal Development. Agronomy 2021, 11, 303. [Google Scholar] [CrossRef]
- Tabbert, J.M.; Riewe, D.; Schulz, H.; Krähmer, A. Facing Energy Limitations—Approaches to Increase Basil (Ocimum basilicum L.) Growth and Quality by Different Increasing Light Intensities Emitted by a Broadband LED Light Spectrum (400–780 Nm). Front. Plant Sci. 2022, 13, 1055352. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Martínez, J.F.; Beltran, V.; Martínez, N.L. Decision Support Systems for Agriculture 4.0: Survey and Challenges. Comput. Electron. Agric. 2020, 170, 105256. [Google Scholar] [CrossRef]
- Hossain, A.; Krupnik, T.J.; Timsina, J.; Mahboob, M.G.; Chaki, A.K.; Farooq, M.; Bhatt, R.; Fahad, S.; Hasanuzzaman, M. Agricultural Land Degradation: Processes and Problems Undermining Future Food Security. In Environment, Climate, Plant and Vegetation Growth; Springer International Publishing: Cham, Switzerland, 2020; pp. 17–61. [Google Scholar]
- Argento, S.; Treccarichi, S.; Arena, D.; Rizzo, G.F.; Branca, F. Exploitation of a Grafting Technique for Improving the Water Use Efficiency of Eggplant (Solanum melongena L.) Grown in a Cold Greenhouse in Mediterranean Climatic Conditions. Agronomy 2023, 13, 2705. [Google Scholar] [CrossRef]
- Ragusa, L.; Picchi, V.; Tribulato, A.; Cavallaro, C.; Lo Scalzo, R.; Branca, F. The Effect of the Germination Temperature on the Phytochemical Content of Broccoli and Rocket Sprouts. Int. J. Food Sci. Nutr. 2017, 68, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Tschora, H.; Cherubini, F. Co-Benefits and Trade-Offs of Agroforestry for Climate Change Mitigation and Other Sustainability Goals in West Africa. Glob. Ecol. Conserv. 2020, 22, e00919. [Google Scholar] [CrossRef]
- Haggar, J.; Casanoves, F.; Cerda, R.; Cerretelli, S.; Gonzalez-Mollinedo, S.; Lanza, G.; Lopez, E.; Leiva, B.; Ospina, A. Shade and Agronomic Intensification in Coffee Agroforestry Systems: Trade-Off or Synergy? Front. Sustain. Food Syst. 2021, 5, 645958. [Google Scholar] [CrossRef]
- Caudill, S.A.; DeClerck, F.J.A.; Husband, T.P. Connecting Sustainable Agriculture and Wildlife Conservation: Does Shade Coffee Provide Habitat for Mammals? Agric. Ecosyst. Environ. 2015, 199, 85–93. [Google Scholar] [CrossRef]
- Xie, X.; Cheng, H.; Hou, C.; Ren, M. Integration of Light and Auxin Signaling in Shade Plants: From Mechanisms to Opportunities in Urban Agriculture. Int. J. Mol. Sci. 2022, 23, 3422. [Google Scholar] [CrossRef]
- Toscano, S.; Ferrante, A.; Branca, F.; Romano, D. Enhancing the Quality of Two Species of Baby Leaves Sprayed with Moringa Leaf Extract as Biostimulant. Agronomy 2021, 11, 1399. [Google Scholar] [CrossRef]
- Picchi, V.; Lo Scalzo, R.; Tava, A.; Doria, F.; Argento, S.; Toscano, S.; Treccarichi, S.; Branca, F. Phytochemical Characterization and In Vitro Antioxidant Properties of Four Brassica Wild Species from Italy. Molecules 2020, 25, 3495. [Google Scholar] [CrossRef]
- Specht, K.; Siebert, R.; Hartmann, I.; Freisinger, U.B.; Sawicka, M.; Werner, A.; Thomaier, S.; Henckel, D.; Walk, H.; Dierich, A. Urban Agriculture of the Future: An Overview of Sustainability Aspects of Food Production in and on Buildings. Agric. Hum. Values 2014, 31, 33–51. [Google Scholar] [CrossRef]
- Ramin Shamshiri, R.; Kalantari, F.; Ting, K.C.; Thorp, K.R.; Hameed, I.A.; Weltzien, C.; Ahmad, D.; Mojgan Shad, Z. Advances in Greenhouse Automation and Controlled Environment Agriculture: A Transition to Plant Factories and Urban Agriculture. Int. J. Agric. Biol. Eng. 2018, 11, 1–22. [Google Scholar] [CrossRef]
- Walters, S.; Stoelzle Midden, K. Sustainability of Urban Agriculture: Vegetable Production on Green Roofs. Agriculture 2018, 8, 168. [Google Scholar] [CrossRef]
- Guenet, B.; Gabrielle, B.; Chenu, C.; Arrouays, D.; Balesdent, J.; Bernoux, M.; Bruni, E.; Caliman, J.; Cardinael, R.; Chen, S.; et al. Can N2O Emissions Offset the Benefits from Soil Organic Carbon Storage? Glob. Chang. Biol. 2021, 27, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Blaser, W.J.; Oppong, J.; Hart, S.P.; Landolt, J.; Yeboah, E.; Six, J. Climate-Smart Sustainable Agriculture in Low-to-Intermediate Shade Agroforests. Nat. Sustain. 2018, 1, 234–239. [Google Scholar] [CrossRef]
- Drolet, G.G.; Middleton, E.M.; Huemmrich, K.F.; Hall, F.G.; Amiro, B.D.; Barr, A.G.; Black, T.A.; McCaughey, J.H.; Margolis, H.A. Regional Mapping of Gross Light-Use Efficiency Using MODIS Spectral Indices. Remote Sens. Environ. 2008, 112, 3064–3078. [Google Scholar] [CrossRef]
- Yuan, W.; Cai, W.; Xia, J.; Chen, J.; Liu, S.; Dong, W.; Merbold, L.; Law, B.; Arain, A.; Beringer, J.; et al. Global Comparison of Light Use Efficiency Models for Simulating Terrestrial Vegetation Gross Primary Production Based on the LaThuile Database. Agric. For. Meteorol. 2014, 192–193, 108–120. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gamon, J.A. The Need for a Common Basis for Defining Light-Use Efficiency: Implications for Productivity Estimation. Remote Sens. Environ. 2015, 156, 196–201. [Google Scholar] [CrossRef]
- Pei, Y.; Dong, J.; Zhang, Y.; Yuan, W.; Doughty, R.; Yang, J.; Zhou, D.; Zhang, L.; Xiao, X. Evolution of Light Use Efficiency Models: Improvement, Uncertainties, and Implications. Agric. For. Meteorol. 2022, 317, 108905. [Google Scholar] [CrossRef]
- Godefroot, M.; Sandra, P.; Verzele, M. New Method for Quantitative Essential Oil Analysis. J. Chromatogr. A 1981, 203, 325–335. [Google Scholar] [CrossRef]
- Schulz, H.; Schrader, B.; Quilitzsch, R.; Pfeffer, S.; Krüger, H. Rapid Classification of Basil Chemotypes by Various Vibrational Spectroscopy Methods. J. Agric. Food Chem. 2003, 51, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Barut, M.; Nadeem, M.A.; Akgür, Ö.; Tansi, L.S.; Aasim, M.; Altaf, M.T.; Baloch, F.S. Medicinal and Aromatic Plants in the Omics Era: Application of Plant Breeding Andbiotechnology for Plant Secondary Metabolite Production. Turk. J. Agric. For. 2022, 46, 182–203. [Google Scholar] [CrossRef]
- Zheljazkov, V.D.; Cantrell, C.L.; Tekwani, B.; Khan, S.I. Content, Composition, and Bioactivity of the Essential Oils of Three Basil Genotypes as a Function of Harvesting. J. Agric. Food Chem. 2008, 56, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Carlo, N.; Silvia, S.; Stefano, B.; Paolo, S. Influence of Cut Number on Qualitative Traits in Different Cultivars of Sweet Basil. Ind. Crops Prod. 2013, 44, 465–472. [Google Scholar] [CrossRef]
- Ciriello, M.; Formisano, L.; El-Nakhel, C.; Corrado, G.; Pannico, A.; De Pascale, S.; Rouphael, Y. Morpho-Physiological Responses and Secondary Metabolites Modulation by Preharvest Factors of Three Hydroponically Grown Genovese Basil Cultivars. Front. Plant Sci. 2021, 12, 671026. [Google Scholar] [CrossRef] [PubMed]
- Gavrić, T.; Jurković, J.; Gadžo, D.; Čengić, L.; Sijahović, E.; Bašić, F. Fertilizer Effect on Some Basil Bioactive Compounds and Yield. Ciênc. Agrotecnol. 2021, 45. [Google Scholar] [CrossRef]
- Corrado, G.; Chiaiese, P.; Lucini, L.; Miras-Moreno, B.; Colla, G.; Rouphael, Y. Successive Harvests Affect Yield, Quality and Metabolic Profile of Sweet Basil (Ocimum basilicum L.). Agronomy 2020, 10, 830. [Google Scholar] [CrossRef]
- Zhao, Z.; Moghadasian, M.H. Chemistry, Natural Sources, Dietary Intake and Pharmacokinetic Properties of Ferulic Acid: A Review. Food Chem. 2008, 109, 691–702. [Google Scholar] [CrossRef]
- Thorn, A.M.; Orians, C.M. Partial Defoliation and Hydraulic Integration in Ocimum Basilicum (Lamiaceae): Testing a Model for Sectored Xylem Flow Using 15 N Labeling. Am. J. Bot. 2011, 98, 1816–1824. [Google Scholar] [CrossRef]
- Iqbal, N.; Masood, A.; Khan, N.A. Analyzing the Significance of Defoliation in Growth, Photosynthetic Compensation and Source-Sink Relations. Photosynthetica 2012, 50, 161–170. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Hou, X.; Zhao, W.; Xu, G.; Li, Z. Effects of Leaf Zeatin and Zeatin Riboside Induced by Different Clipping Heights on the Regrowth Capacity of Ryegrass. Ecol. Res. 2014, 29, 167–180. [Google Scholar] [CrossRef]
- Skalák, J.; Vercruyssen, L.; Claeys, H.; Hradilová, J.; Černý, M.; Novák, O.; Plačková, L.; Saiz-Fernández, I.; Skaláková, P.; Coppens, F.; et al. Multifaceted Activity of Cytokinin in Leaf Development Shapes Its Size and Structure in Arabidopsis. Plant J. 2019, 97, 805–824. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sharma, S.; Pathania, V. Effect of Shading and Plant Density on Growth, Yield and Oil Composition of Clary Sage (Salvia sclarea L.) in North Western Himalaya. J. Essent. Oil Res. 2013, 25, 23–32. [Google Scholar] [CrossRef]
- Kesumawati, E.; Apriyatna, D.; Rahmawati, M. The Effect of Shading Levels and Varieties on the Growth and Yield of Chili Plants (Capsicum annuum L.). IOP Conf. Ser. Earth Environ. Sci. 2020, 425, 012080. [Google Scholar] [CrossRef]
- Stagnari, F.; Di Mattia, C.; Galieni, A.; Santarelli, V.; D’Egidio, S.; Pagnani, G.; Pisante, M. Light Quantity and Quality Supplies Sharply Affect Growth, Morphological, Physiological and Quality Traits of Basil. Ind. Crops Prod. 2018, 122, 277–289. [Google Scholar] [CrossRef]
- Solis-Toapanta, E.; Gómez, C. Growth and Photosynthetic Capacity of Basil Grown for Indoor Gardening under Constant or Increasing Daily Light Integrals. Horttechnology 2019, 29, 880–888. [Google Scholar] [CrossRef]
- Yu, K.M.J.; McKinley, B.; Rooney, W.L.; Mullet, J.E. High Planting Density Induces the Expression of GA3-Oxidase in Leaves and GA Mediated Stem Elongation in Bioenergy Sorghum. Sci. Rep. 2021, 11, 46. [Google Scholar] [CrossRef]
- Ballaré, C.L.; Pierik, R. The Shade-avoidance Syndrome: Multiple Signals and Ecological Consequences. Plant Cell Environ. 2017, 40, 2530–2543. [Google Scholar] [CrossRef]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple Pathways Regulate Shoot Branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef]
- Castronuovo, D.; Russo, D.; Libonati, R.; Faraone, I.; Candido, V.; Picuno, P.; Andrade, P.; Valentao, P.; Milella, L. Influence of Shading Treatment on Yield, Morphological Traits and Phenolic Profile of Sweet Basil (Ocimum basilicum L.). Sci. Hortic. 2019, 254, 91–98. [Google Scholar] [CrossRef]
- Jakovljević, D.; Momčilović, J.; Bojović, B.; Stanković, M. The Short-Term Metabolic Modulation of Basil (Ocimum basilicum L. Cv. ‘Genovese’) after Exposure to Cold or Heat. Plants 2021, 10, 590. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, R.; Bhandari, K.; Nayyar, H. Temperature Stress and Redox Homeostasis in Agricultural Crops. Front. Environ. Sci. 2015, 3, 11. [Google Scholar] [CrossRef]
- Radulović, N.S.; Blagojević, P.D.; Miltojević, A.B. α-Linalool—A Marker Compound of Forged/Synthetic Sweet Basil (Ocimum basilicum L.) Essential Oils. J. Sci. Food Agric. 2013, 93, 3292–3303. [Google Scholar] [CrossRef] [PubMed]
- Amadei, G.; Ross, B.M. Quantification of Character-impacting Compounds in Ocimum basilicum and “Pesto Alla Genovese” with Selected Ion Flow Tube Mass Spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; De Luca, L.; Aiello, A.; Pagano, R.; Di Pierro, P.; Pizzolongo, F.; Masi, P. Basil (Ocimum basilicum L.) Leaves as a Source of Bioactive Compounds. Foods 2022, 11, 3212. [Google Scholar] [CrossRef] [PubMed]
- Singh-Sangwan, N.; Sangwan, R.; Luthra, R.; Thakur, R. Geraniol Dehydrogenase: A Determinant of Essential Oil Quality in Lemongrass1. Planta Med. 1993, 59, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Iijima, Y.; Gang, D.R.; Fridman, E.; Lewinsohn, E.; Pichersky, E. Characterization of Geraniol Synthase from the Peltate Glands of Sweet Basil. Plant Physiol. 2004, 134, 370–379. [Google Scholar] [CrossRef]
- Majdi, C.; Pereira, C.; Dias, M.I.; Calhelha, R.C.; Alves, M.J.; Rhourri-Frih, B.; Charrouf, Z.; Barros, L.; Amaral, J.S.; Ferreira, I.C.F.R. Phytochemical Characterization and Bioactive Properties of Cinnamon Basil (Ocimum basilicum Cv. ‘Cinnamon’) and Lemon Basil (Ocimum × Citriodorum). Antioxidants 2020, 9, 369. [Google Scholar] [CrossRef]
- Hamad, A.; Djalil, A.D.; Dewi, D.Y.S.; Hartanti, D. Development of Lemon Basil Essential Oil as a Natural Chicken Meat Preservative. IOP Conf. Ser. Earth Environ. Sci. 2021, 803, 012028. [Google Scholar] [CrossRef]
- Ngamprasertsith, S.; Menwa, J.; Sawangkeaw, R. Caryophyllene Oxide Extraction from Lemon Basil (Ocimum citriodorum Vis.) Straw by Hydrodistillation and Supercritical CO2. J. Supercrit. Fluids 2018, 138, 1–6. [Google Scholar] [CrossRef]
- Wesołowska, A.; Jadczak, D.; Wesołowska, A. Mineral Elements and Chemical Composition Essential Oil from Leaves and Flowers of Selected Lemon-Scented Ocimum Species. J. Elem. 2023, 28, 123–143. [Google Scholar] [CrossRef]



| Accession | Code | Common Name | Origin |
|---|---|---|---|
| UNICT 2106 | A | A foglia larga | Catania |
| UNICT 2111 | B | Violetto | Iran |
| UNICT 2102 | C | Genovese | Hortus sementi |
| UNICT 2630 | D | Citrodora | Romania |
| UNICT 2112 | E | Nano | Catania |
| UNICT 2125 | F | Foglia di lattuga | La Rosa |
| UNICT 2094 | G | Genovese Gigante | Royal Sluis |
| Code | Trait |
|---|---|
| PH | Plant height (cm) |
| PB | Number of plant branches (n) |
| LL | Leaf lamina length (cm) |
| LWI | Leaf lamina width (cm) |
| LB | Leaf blistering (absent: 0; present: 1) |
| LWE | Leaf weight percentage (%) |
| LDM | Leaf dry matter percentage (%) |
| LL* | Leaf chromatic parameter L* |
| La* | Leaf chromatic parameter a* |
| Lb* | Leaf chromatic parameter b* |
| TWE | Harvesting weight of the total fresh biomass (g) |
| DAT | A | B | C | D | E | F | G | Mean |
|---|---|---|---|---|---|---|---|---|
| 35 | 88.00 | 69.00 | 93.70 | 56.80 | 54.00 | 54.60 | 61.00 | 68.16 |
| 56 | 225.53 | 327.33 | 134.37 | 185.60 | 272.33 | 170.90 | 86.23 | 200.33 |
| 84 | 211.57 | 0.00 | 218.57 | 317.20 | 0.00 | 19.50 | 0.00 | 109.55 |
| 119 | 97.63 | 28.30 | 157.57 | 267.67 | 289.30 | 159.17 | 95.30 | 156.42 |
| TWE | 622.73 | 424.63 | 604.20 | 827.27 | 615.63 | 404.17 | 242.53 | 534.45 |
| Trait | A | B | C | D | E | F | G | Mean |
|---|---|---|---|---|---|---|---|---|
| PH | 36.80 | 42.18 | 44.08 | 39.23 | 22.94 | 19.12 | 40.83 | 35.03 |
| PB | 10.56 | 9.35 | 8.63 | 5.03 | 9.97 | 8.10 | 8.54 | 8.59 |
| LL | 6.82 | 6.60 | 7.00 | 7.78 | 2.25 | 8.60 | 5.81 | 6.41 |
| LWI | 4.33 | 3.13 | 3.94 | 4.17 | 1.04 | 7.83 | 3.99 | 4.06 |
| LB | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.33 | 0.19 |
| LWE | 54.09 | 49.59 | 60.03 | 58.38 | 44.22 | 56.78 | 35.38 | 51.21 |
| LDM | 11.90 | 12.90 | 13.90 | 9.90 | 9.70 | 9.40 | 12.90 | 11.51 |
| LL* | 48.79 | 39.62 | 45.25 | 47.76 | 48.39 | 46.14 | 48.49 | 46.35 |
| La* | −11.88 | −0.55 | −10.58 | −11.77 | −9.53 | −9.91 | −11.59 | −9.40 |
| Lb* | 19.28 | 3.12 | 15.96 | 18.08 | 22.60 | 12.98 | 18.69 | 15.81 |
| Molecule | A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|---|
| 1,8-Cineole | 5.0 | 2.2 | 3.3 | t * | 7.2 | 0.2 | 11.2 |
| Linalool | 31.4 | 15.0 | 30.8 | 0.7 | 6.7 | 2.1 | 33.5 |
| Terpinen-4-ol | 0.1 | 0.2 | 0.1 | - | 4.4 | 0.2 | 0.2 |
| Estragole | 26.2 | 56.9 | 0.1 | 0.7 | 3.0 | 1.2 | 1.3 |
| Neral | - | t | 0.1 | 23.7 | - | - | - |
| Geranial | - | t | 0.1 | 31.2 | - | - | - |
| Eugenol | 1.5 | t | 7.2 | 0.3 | 0.5 | 0.8 | 0.8 |
| α-Bergamotene | 8.6 | 3.1 | 13.6 | 1.9 | 23.0 | 33.6 | 5.8 |
| Germacrene D | 2.5 | 0.2 | 3.0 | 0.9 | 5.6 | 5.7 | 2.2 |
| Caryophyllene Oxide | 0.1 | 1.5 | 0.1 | 8.6 | 0.1 | 0.1 | 0.1 |
| epi-α-Cadinol | 6.2 | 4.6 | 10.0 | 0.6 | 11.5 | 12.2 | 7.6 |
| SR70 | SR100 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAT | A | B | C | D | E | F | G | Mean | A | B | C | D | E | F | G | Mean |
| 35 | 147.70 | 183.70 | 156.82 | 201.00 | 167.30 | 151.00 | 85.50 | 156.15a | 88.00 | 69.00 | 93.70 | 56.80 | 54.00 | 54.60 | 61.00 | 68.16b |
| 56 | 269.30 | 186.13 | 220.73 | 370.83 | 523.17 | 235.67 | 92.47 | 271.19a | 225.53 | 327.33 | 134.37 | 185.60 | 272.33 | 170.90 | 86.23 | 200.33b |
| 84 | 228.13 | 166.77 | 165.27 | 191.47 | 0.00 | 139.10 | 174.70 | 152.20a | 211.57 | 0.00 | 218.57 | 317.20 | 0.00 | 19.50 | 0.00 | 109.55b |
| 119 | 148.83 | 50.47 | 223.60 | 140.40 | 344.70 | 225.17 | 95.43 | 175.51a | 97.63 | 28.30 | 157.57 | 267.67 | 289.30 | 159.17 | 95.30 | 156.42ab |
| TWE | 793.97 | 587.07 | 766.42 | 903.70 | 1035.17 | 750.93 | 448.10 | 755.05a | 622.73 | 424.63 | 604.20 | 827.27 | 615.63 | 404.17 | 242.53 | 534.45b |
| Means for each genotype | ||||||||||||||||
| A | B | C | D | E | F | G | ||||||||||
| 35 | 117.85 | 126.35 | 125.26 | 128.90 | 110.65 | 102.80 | 73.25 | |||||||||
| 56 | 247.42 | 256.73 | 177.55 | 278.22 | 397.75 | 203.28 | 89.35 | |||||||||
| 84 | 219.85 | 83.38 | 191.92 | 254.33 | 0.00 | 79.30 | 87.35 | |||||||||
| 119 | 123.23 | 39.38 | 190.58 | 204.03 | 317.00 | 192.17 | 95.37 | |||||||||
| TWE | 708.35b | 505.85de | 685.31c | 865.48a | 825.40ab | 577.55d | 345.32f | |||||||||
| Significance of the differences using ANOVA Student–Newman–Keuls | ||||||||||||||||
| SR | GE | HT | SR × GE | SR × HT | GE × HT | SR × GE × HT | ||||||||||
| TWE | *** | *** | *** | *** | *** | *** | *** | |||||||||
| SR70 | SR100 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | Mean | A | B | C | D | E | F | G | Mean | |
| LUE | 40.60 | 30.02 | 39.19 | 46.21 | 52.93 | 38.40 | 22.91 | 38.61a | 22.29 | 15.20 | 21.63 | 29.61 | 22.04 | 14.47 | 8.68 | 19.13b |
| Mean for each genotype | ||||||||||||||||
| A | B | C | D | E | F | G | ||||||||||
| LUE | 31.45b | 22.61d | 30.41b | 37.91a | 37.49a | 26.43c | 15.80e | |||||||||
| Significance of the differences using ANOVA | ||||||||||||||||
| SR | *** | |||||||||||||||
| GE | *** | |||||||||||||||
| SR × GE | *** | |||||||||||||||
| SR70 | SR100 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAT | A | B | C | D | E | F | G | Mean | A | B | C | D | E | F | G | Mean |
| PH | 47.97 | 71.13 | 59.33 | 78.87 | 33.68 | 34.43 | 53.25 | 54.09a | 36.80 | 42.18 | 44.08 | 39.23 | 22.94 | 19.12 | 40.83 | 35.03b |
| PB | 12.56 | 16.46 | 12.73 | 15.78 | 16.29 | 9.06 | 11.42 | 13.47a | 10.56 | 9.35 | 8.63 | 5.03 | 9.97 | 8.10 | 8.54 | 8.59b |
| LL | 8.47 | 7.49 | 8.04 | 9.62 | 2.86 | 10.38 | 6.20 | 7.58a | 6.82 | 6.60 | 7.00 | 7.78 | 2.25 | 8.60 | 5.81 | 6.41a |
| LWI | 4.59 | 3.02 | 3.93 | 4.76 | 1.30 | 8.64 | 3.21 | 4.21a | 4.33 | 3.13 | 3.94 | 4.17 | 1.04 | 7.83 | 3.99 | 4.06a |
| LB | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.33 | 0.19a | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.33 | 0.19a |
| LWE | 63.30 | 30.76 | 53.19 | 36.82 | 34.12 | 61.71 | 40.56 | 45.78b | 54.09 | 49.59 | 60.03 | 58.38 | 44.22 | 56.78 | 35.38 | 51.21a |
| LDM | 15.27 | 13.60 | 13.60 | 13.60 | 10.71 | 10.32 | 13.70 | 12.97a | 11.90 | 12.90 | 13.90 | 9.90 | 9.70 | 9.40 | 12.90 | 11.51a |
| LL* | 47.50 | 35.41 | 44.95 | 46.15 | 45.91 | 46.46 | 47.61 | 44.86a | 48.79 | 39.62 | 45.25 | 47.76 | 48.39 | 46.14 | 48.49 | 46.35a |
| La* | −12.44 | 0.88 | −12.47 | −12.29 | −11.20 | −12.85 | −11.96 | −10.3a | −11.88 | −0.55 | −10.58 | −11.77 | −9.53 | −9.91 | −11.59 | −9.40a |
| Mean for each genotype | ||||||||||||||||
| A | B | C | D | E | F | G | ||||||||||
| PH | 42.39b | 56.65a | 51.71ab | 59.05a | 28.31c | 26.78c | 47.04ab | |||||||||
| PB | 11.56a | 12.90a | 10.68a | 10.40a | 13.13a | 8.58a | 9.98a | |||||||||
| LL | 7.64ab | 7.04ab | 7.52ab | 8.70ab | 2.56c | 9.49a | 6.01b | |||||||||
| LWI | 4.46b | 3.08c | 3.93bc | 4.47b | 1.17d | 8.24a | 3.60c | |||||||||
| LB | 0.00b | 0.00b | 0.00b | 0.00b | 0.00b | 1.00a | 0.33b | |||||||||
| LWE | 58.70a | 40.18bc | 56.61a | 47.60b | 39.17bc | 59.25a | 37.97bc | |||||||||
| LDM | 13.59a | 13.25a | 13.75a | 11.75b | 10.21bc | 9.86c | 13.30a | |||||||||
| LL* | 48.15a | 37.52b | 45.10a | 46.96a | 47.15a | 46.30a | 48.05a | |||||||||
| La* | −12.16b | 0.16a | −11.53b | −12.03b | −10.37b | −11.38b | −11.78b | |||||||||
| Lb* | 18.67ab | 1.76d | 16.05bc | 17.02bc | 22.00a | 13.97c | 17.87b | |||||||||
| Significance of the differences using ANOVA Student−Newman−Keuls | ||||||||||||||||
| PH | PB | LL | LWI | LB | LWE | LDM | LL* | La* | Lb* | |||||||
| SR | *** | *** | n.s. | n.s. | n.s. | * | n.s. | n.s. | * | n.s. | ||||||
| GE | *** | n.s. | *** | *** | *** | *** | *** | *** | *** | *** | ||||||
| SR × GE | ** | n.s. | n.s. | *** | n.s. | * | n.s. | n.s. | n.s. | n.s. | ||||||
| Trait | PC1 | PC2 | PC3 |
|---|---|---|---|
| PH | −0.714 | 0.132 | 0.603 |
| PB | −0.703 | −0.234 | 0.364 |
| LL | 0.141 | 0.774 | 0.596 |
| LWI | 0.553 | 0.748 | 0.288 |
| LB | 0.598 | 0.566 | −0.137 |
| LWE | 0.644 | 0.333 | 0.221 |
| LDM | −0.570 | 0.085 | 0.506 |
| LL* | 0.790 | −0.484 | 0.218 |
| La* | −0.716 | 0.382 | −0.535 |
| Lb* | 0.626 | −0.731 | 0.227 |
| TWE | −0.044 | −0.393 | 0.595 |
| Variance (%) | 35.996 | 24.898 | 18.152 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Branca, F.; Treccarichi, S.; Ruberto, G.; Renda, A.; Argento, S. Comprehensive Morphometric and Biochemical Characterization of Seven Basil (Ocimum basilicum L.) Genotypes: Focus on Light Use Efficiency. Agronomy 2024, 14, 224. https://doi.org/10.3390/agronomy14010224
Branca F, Treccarichi S, Ruberto G, Renda A, Argento S. Comprehensive Morphometric and Biochemical Characterization of Seven Basil (Ocimum basilicum L.) Genotypes: Focus on Light Use Efficiency. Agronomy. 2024; 14(1):224. https://doi.org/10.3390/agronomy14010224
Chicago/Turabian StyleBranca, Ferdinando, Simone Treccarichi, Giuseppe Ruberto, Agatino Renda, and Sergio Argento. 2024. "Comprehensive Morphometric and Biochemical Characterization of Seven Basil (Ocimum basilicum L.) Genotypes: Focus on Light Use Efficiency" Agronomy 14, no. 1: 224. https://doi.org/10.3390/agronomy14010224
APA StyleBranca, F., Treccarichi, S., Ruberto, G., Renda, A., & Argento, S. (2024). Comprehensive Morphometric and Biochemical Characterization of Seven Basil (Ocimum basilicum L.) Genotypes: Focus on Light Use Efficiency. Agronomy, 14(1), 224. https://doi.org/10.3390/agronomy14010224








