Genome-Wide Identification of the Genes of the Odorant-Binding Protein Family Reveal Their Role in the Olfactory Response of the Tomato Leaf Miner (Tuta absoluta) to a Repellent Plant
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Strains
2.2. Search and Identification of Odorant-Binding Protein Family Gene in T. absoluta
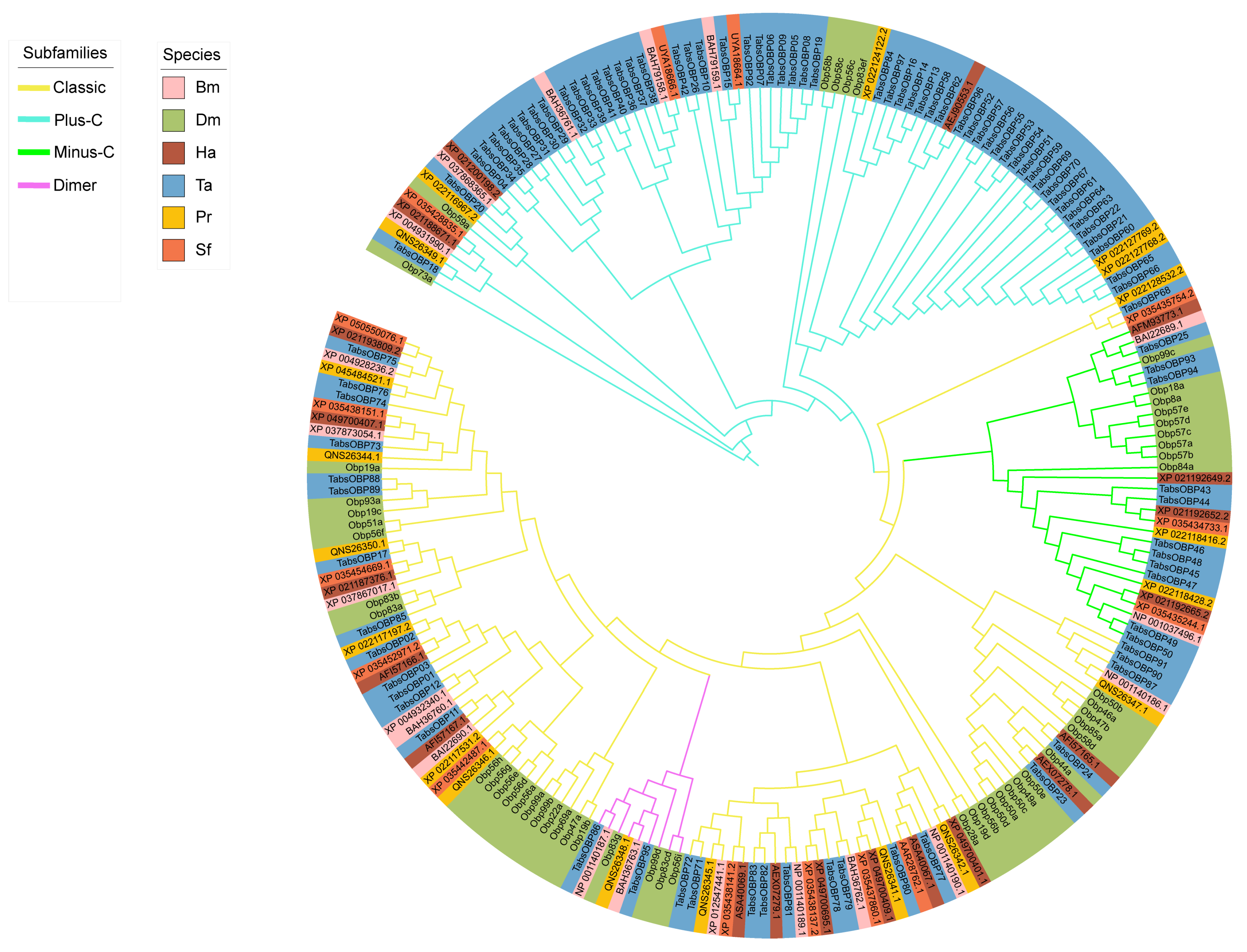
2.3. Phylogenetic Analysis of OBP Genes in T. absoluta
2.4. Developmental Stage-Specific Expression Profile of TaOBPs
2.5. Determination of the Taxis Behavior of T. absoluta to Plectranthus tomentosa
2.6. Induction Treatment of T. absoluta Larvae and Adults by Plant Volatiles
2.7. Total RNA Extraction and Reverse Transcription
2.8. Real-Time Quantitative PCR (RT-qPCR) Analyses of TaOBPs
2.9. Data Analysis
3. Results
3.1. Annotation and Identification of Genes from the Odorant-Binding Protein Family in T. absoluta
3.2. Phylogenetic Analysis and Chromosomal Location of OBP Genes in T. absoluta
3.3. Developmental Stage-Specific Expression Profile of TaOBPs
3.4. Behavioral Responses of T. absoluta to Plectranthus tomentosa
3.5. Transcriptional Responses of TaOBPs to Tomatoes and P. tomentosa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bruce, T.J.; Wadhams, L.J.; Woodcock, C.M. Insect host location: A volatile situation. Trends Plant Sci. 2005, 10, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Liu, Y.; Wang, G. Research progress on signal transduction mechanism of insect peripheral olfactory system. Sci. China Life Sci. 2016, 46, 573–583. [Google Scholar]
- Fan, J.; Francis, F.; Liu, Y.; Chen, J.L.; Cheng, D.F. An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet. Mol. Res. 2011, 10, 3056–3069. [Google Scholar] [CrossRef]
- Wang, G.; Guo, Y.; Wu, K. Advances in research on antennal odorant binding proteins of insects. Entomol. J. 2002, 45, 131–137. [Google Scholar]
- Schneider, D. Insect olfaction: Deciphering system for chemical messages. Science 1969, 163, 1031–1037. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Benton, R. Molecular mechanisms of olfactory detection in insects: Beyond receptors. Open Biol. 2020, 10, 200252. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Y.; Yang, K.; Liu, Y.; Xu, W.; Anderson, A.; Dong, S.L. Two general-odorant binding proteins in Spodoptera litura are differentially tuned to sex pheromones and plant odorants. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 180, 23–31. [Google Scholar] [CrossRef]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Issar, S.; Yadav, N.; Gaur, R. Role of volatile organic compounds in plant growth, communication and defense. Agrica 2021, 10, 111–119. [Google Scholar] [CrossRef]
- Beck, J.J.; Higbee, B.S.; Light, D.M.; Gee, W.S.; Merrill, G.B.; Hayashi, J.M. Hull split and damaged almond volatiles attract male and female navel orangeworm moths. J. Agric. Food Chem. 2012, 60, 8090–8096. [Google Scholar] [CrossRef] [PubMed]
- Nishida, R. Chemical ecology of insect–plant interactions: Ecological significance of plant secondary metabolites. Biosci. Biotechnol. Biochem. 2014, 78, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Parolin, P.; Bresch, C.; Desneux, N.; Brun, R.; Bout, A.; Boll, R.; Poncet, C. Secondary plants used in biological control: A review. Int. J. Pest Manag. 2012, 58, 91–100. [Google Scholar] [CrossRef]
- Holden, M.H.; Ellner, S.P.; Lee, D.H.; Nyrop, J.P.; Sanderson, J.P. Designing an effective trap cropping strategy: The effects of attraction, retention and plant spatial distribution. J. Appl. Ecol. 2012, 49, 715–722. [Google Scholar] [CrossRef]
- Lu, Y.-H.; Zheng, X.-S.; Lu, Z.-X. Application of vetiver grass Vetiveria zizanioides: Poaceae (L.) as a trap plant for rice stem borer Chilo suppressalis: Crambidae (Walker) in the paddy fields. J. Integr. Agric. 2019, 18, 797–804. [Google Scholar] [CrossRef]
- Moreau, T.L.; Warman, P.; Hoyle, J. An evaluation of companion planting and botanical extracts as alternative pest controls for the Colorado potato beetle. Biol. Agric. Hortic. 2006, 23, 351–370. [Google Scholar] [CrossRef]
- El-Shafie, H.A.F. Tuta absoluta (Meyrick)(Lepidoptera: Gelechiidae): An invasive insect pest threatening the world tomato production. In Invasive Species-Introduction Pathways, Economic Impact, Possible Management Options; IntechOpen: London, UK, 2020; pp. 1–16. [Google Scholar]
- Gharekhani, G.; Salek-Ebrahimi, H. Evaluating the damage of Tuta absoluta (Meyrick)(Lepidoptera: Gelechiidae) on some cultivars of tomato under greenhouse condition. Arch. Phytopathol. Plant Prot. 2014, 47, 429–436. [Google Scholar] [CrossRef]
- Saeidi, Z.; Raeesi, M. Integration of resistant variety and biological agent to control tomato leaf miner, Tuta absoluta (Meyrick), under greenhouse conditions. Bull. Entomol. Res. 2021, 111, 357–363. [Google Scholar] [CrossRef]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Catalán Ruescas, D.; Tabone, E.; Frandon, J. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Biondi, A.; Guedes, R.N.C.; Wan, F.-H.; Desneux, N. Ecology, worldwide spread, and management of the invasive South American tomato pinworm, Tuta absoluta: Past, present, and future. Annu. Rev. Entomol. 2018, 63, 239–258. [Google Scholar] [CrossRef]
- Gao, Y.; Lei, Z.; Reitz, S.R. Western flower thrips resistance to insecticides: Detection, mechanisms and management strategies. Pest Manag. Sci. 2012, 68, 1111–1121. [Google Scholar] [CrossRef]
- Soares, M.A.; Campos, M.R.; Passos, L.C.; Carvalho, G.A.; Haro, M.M.; Lavoir, A.-V.; Biondi, A.; Zappalà, L.; Desneux, N. Botanical insecticide and natural enemies: A potential combination for pest management against Tuta absoluta. J. Pest Sci. 2019, 92, 1433–1443. [Google Scholar] [CrossRef]
- Yarou, B.B.; Bawin, T.; Boullis, A.; Heukin, S.; Lognay, G.; Verheggen, F.J.; Francis, F. Oviposition deterrent activity of basil plants and their essentials oils against Tuta absoluta (Lepidoptera: Gelechiidae). Environ. Sci. Pollut. Res. 2018, 25, 29880–29888. [Google Scholar] [CrossRef] [PubMed]
- Lo Pinto, M.; Vella, L.; Agrò, A. Oviposition deterrence and repellent activities of selected essential oils against Tuta absoluta Meyrick (Lepidoptera: Gelechiidae): Laboratory and greenhouse investigations. Int. J. Trop. Insect Sci. 2022, 42, 3455–3464. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Vizueta, J.; Sánchez-Gracia, A.; Rozas, J. Bitacora: A comprehensive tool for the identification and annotation of gene families in genome assemblies. Mol. Ecol. Resour. 2020, 20, 1445–1452. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Rondon, J.J.; Moreyra, N.N.; Pisarenco, V.A.; Rozas, J.; Hurtado, J.; Hasson, E. Evolution of the odorant-binding protein gene family in Drosophila. Front. Ecol. Evol. 2022, 10, 957247. [Google Scholar] [CrossRef]
- Pandey, M.; Bhattarai, N.; Pandey, P.; Chaudhary, P.; Katuwal, D.R.; Khanal, D. A review on biology and possible management strategies of tomato leaf miner, Tuta absoluta (Meyrick), Lepidoptera: Gelechiidae in Nepal. Heliyon 2023, 9, e16474. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, Y.X.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Alonso-Valiente, M.; Vacas, S.; Gallego, C.; Rambla, J.L.; Navarro-Llopis, V.; Granell, A.; Urbaneja, A. Eliciting tomato plant defenses by exposure to herbivore induced plant volatiles. Entomol. Gen. 2021, 41, 209–218. [Google Scholar] [CrossRef]
- Sun, Z.; Lin, Y.; Wang, R.; Li, Q.; Shi, Q.; Baerson, S.R.; Chen, L.; Zeng, R.; Song, Y. Olfactory perception of herbivore-induced plant volatiles elicits counter-defences in larvae of the tobacco cutworm. Funct. Ecol. 2021, 35, 384–397. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Midega, C.A.; Hooper, A.; Pickett, J. Push-pull: Chemical ecology-based integrated pest management technology. J. Chem. Ecol. 2016, 42, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z. Study on the characters and microscopic identification of Plectranthus tomentosa. Lishizhen Med. Mater. Med. Res. 2016, 27, 1907–1908. [Google Scholar]
- Sun, Y.; Liu, T.; Sun, C.; Luo, Q. Chemical constituents of Plectranthus tomentosa extract and its control effect on Tetranychus kanzawai. J. Chem. 2022, 2022, 5609391. [Google Scholar] [CrossRef]
- Rakha, M.; Zekeya, N.; Sevgan, S.; Musembi, M.; Ramasamy, S.; Hanson, P. Screening recently identified whitefly/spider mite-resistant wild tomato accessions for resistance to Tuta absoluta. Plant Breed. 2017, 136, 562–568. [Google Scholar] [CrossRef]
- Han, P.; Desneux, N.; Becker, C.; Larbat, R.; Le Bot, J.; Adamowicz, S.; Zhang, J.; Lavoir, A.-V. Bottom-up effects of irrigation, fertilization and plant resistance on Tuta absoluta: Implications for Integrated Pest Management. J. Pest Sci. 2019, 92, 1359–1370. [Google Scholar] [CrossRef]
- Bacci, L.; Silva, É.M.; Silva, G.A.; Silva, L.J.; Rosado, J.F.; Samuels, R.I.; Picanço, M.C. Natural mortality factors of tomato leafminer Tuta absoluta in open-field tomato crops in South America. Pest Manag. Sci. 2019, 75, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Desneux, N.; Han, P.; Mansour, R.; Arnó, J.; Brévault, T.; Campos, M.R.; Chailleux, A.; Guedes, R.N.; Karimi, J.; Konan, K.A.J. Integrated pest management of Tuta absoluta: Practical implementations across different world regions. J. Pest Sci. 2022, 95, 17–39. [Google Scholar] [CrossRef]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagne, N.; Darboux, I.; et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef]
- Zhu, J.; Ban, L.; Song, L.-M.; Liu, Y.; Pelosi, P.; Wang, G. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem. Mol. Biol. 2016, 72, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Poivet, E.; Rharrabe, K.; Monsempes, C.; Glaser, N.; Rochat, D.; Renou, M.; Marion-Poll, F.; Jacquin-Joly, E. The use of the sex pheromone as an evolutionary solution to food source selection in caterpillars. Nat. Commun. 2012, 3, 1047. [Google Scholar] [CrossRef]






| Gene Name | Genome Identifier | Locus | CDS (bp) | No of Exons | No of TMSs | ||
|---|---|---|---|---|---|---|---|
| Chr. | Starting | Ending | |||||
| TabsOBP01 | Tabs020023.1 | chr1 | 20,474,931 | 20,476,482 | 483 | 4 | 0 |
| TabsOBP02 | Tabs003327.1 | chr1 | 22,042,224 | 22,045,788 | 405 | 4 | 0 |
| TabsOBP03 | Tabs020567.1 | chr1 | 22,052,312 | 22,054,427 | 402 | 4 | 0 |
| TabsOBP04 | Tabs001544.1 | chr1 | 24,733,631 | 24,739,562 | 768 | 5 | 0 |
| TabsOBP05 | Tabs020984.1 | chr2 | 6,902,003 | 6,930,209 | 774 | 5 | 0 |
| TabsOBP06 | Tabs001496.1 | chr2 | 6,933,200 | 6,936,252 | 750 | 6 | 0 |
| TabsOBP07 | Tabs011682.1 | chr2 | 6,937,697 | 6,941,224 | 789 | 5 | 0 |
| TabsOBP08 | Tabs011532.1 | chr2 | 6,944,108 | 6,949,332 | 735 | 5 | 0 |
| TabsOBP09 | Tabs001649.1 | chr2 | 6,966,595 | 6,982,926 | 750 | 5 | 0 |
| TabsOBP10 | Tabs003077.1 | chr3 | 10,972,251 | 10,992,148 | 753 | 5 | 0 |
| TabsOBP11 | Tabs004058.1 | chr4 | 7,306,416 | 7,306,929 | 402 | 2 | 0 |
| TabsOBP12 | Tabs012945.1 | chr4 | 7,307,857 | 7,309,328 | 426 | 2 | 0 |
| TabsOBP13 | Tabs014664.1 | chr4 | 8,793,786 | 8,795,957 | 588 | 5 | 0 |
| TabsOBP14 | Tabs017562.1 | chr4 | 10,087,502 | 10,091,190 | 651 | 4 | 0 |
| TabsOBP15 | Tabs018202.1 | chr4 | 10,265,535 | 10,282,898 | 771 | 6 | 0 |
| TabsOBP16 | Tabs012221.1 | chr4 | 10,349,993 | 10,352,269 | 702 | 4 | 1 |
| TabsOBP17 | Tabs000652.1 | chr4 | 10,360,251 | 10,364,950 | 477 | 6 | 0 |
| TabsOBP18 | Tabs000441.1 | chr9 | 5,168,187 | 5,198,937 | 555 | 6 | 0 |
| TabsOBP19 | Tabs006266.1 | chr10 | 15,096,649 | 15,106,364 | 798 | 6 | 0 |
| TabsOBP20 | Tabs007711.1 | chr12 | 15,334,109 | 15,335,900 | 1158 | 5 | 0 |
| TabsOBP21 | Tabs006148.1 | chr14 | 8,101,391 | 8,108,160 | 312 | 3 | 0 |
| TabsOBP22 | Tabs006809.1 | chr14 | 8,109,187 | 8,117,805 | 327 | 3 | 0 |
| TabsOBP23 | Tabs014933.1 | chr15 | 1,281,831 | 1,284,781 | 561 | 5 | 0 |
| TabsOBP24 | Tabs011481.1 | chr15 | 5,249,011 | 5,251,111 | 444 | 5 | 0 |
| TabsOBP25 | Tabs001448.1 | chr15 | 5,256,732 | 5,261,756 | 552 | 5 | 0 |
| TabsOBP26 | Tabs015427.1 | chr15 | 7,017,169 | 7,026,005 | 738 | 5 | 1 |
| TabsOBP27 | Tabs004992.1 | chr15 | 7,048,260 | 7,058,579 | 681 | 5 | 0 |
| TabsOBP28 | Tabs021922.1 | chr15 | 7,069,040 | 7,080,090 | 732 | 5 | 1 |
| TabsOBP29 | Tabs016278.1 | chr15 | 7,313,748 | 7,321,387 | 768 | 5 | 0 |
| TabsOBP30 | Tabs003939.1 | chr15 | 7,330,368 | 7,333,804 | 852 | 5 | 0 |
| TabsOBP31 | Tabs012068.1 | chr15 | 7,334,821 | 7,338,351 | 771 | 5 | 0 |
| TabsOBP32 | Tabs003951.1 | chr15 | 7,339,501 | 7,342,294 | 780 | 5 | 0 |
| TabsOBP33 | Tabs019705.1 | chr15 | 7,348,269 | 7,352,529 | 777 | 5 | 0 |
| TabsOBP34 | Tabs010627.1 | chr15 | 7,363,776 | 7,375,218 | 753 | 5 | 0 |
| TabsOBP35 | Tabs017381.1 | chr15 | 7,391,770 | 7,405,770 | 690 | 5 | 0 |
| TabsOBP36 | Tabs002475.1 | chr15 | 13,600,553 | 13,615,475 | 726 | 5 | 0 |
| TabsOBP37 | Tabs010825.1 | chr15 | 13,619,535 | 13,625,215 | 729 | 5 | 0 |
| TabsOBP38 | Tabs019998.1 | chr15 | 13,634,512 | 13,639,054 | 717 | 5 | 1 |
| TabsOBP39 | Tabs014722.1 | chr15 | 13,749,598 | 13,752,428 | 726 | 5 | 0 |
| TabsOBP40 | Tabs021282.1 | chr15 | 19,271,022 | 19,286,596 | 729 | 5 | 0 |
| TabsOBP41 | Tabs017578.1 | chr15 | 19,308,964 | 19,326,115 | 711 | 6 | 0 |
| TabsOBP42 | Tabs008070.1 | chr15 | 19,340,229 | 19,355,389 | 942 | 6 | 0 |
| TabsOBP43 | Tabs021817.1 | chr16 | 2,311,543 | 2,313,131 | 504 | 3 | 0 |
| TabsOBP44 | Tabs020287.1 | chr16 | 2,314,295 | 2,317,205 | 621 | 3 | 0 |
| TabsOBP45 | Tabs021019.1 | chr16 | 2,318,859 | 2,320,133 | 489 | 3 | 0 |
| TabsOBP46 | Tabs016106.1 | chr16 | 2,322,267 | 2,324,749 | 492 | 3 | 0 |
| TabsOBP47 | Tabs020787.1 | chr16 | 2,327,768 | 2,328,714 | 480 | 3 | 0 |
| TabsOBP48 | Tabs019694.1 | chr16 | 2,329,157 | 2,330,090 | 492 | 3 | 0 |
| TabsOBP49 | Tabs008083.1 | chr16 | 2,685,303 | 2,710,260 | 501 | 3 | 0 |
| TabsOBP50 | Tabs015113.1 | chr16 | 2,998,204 | 2,999,676 | 372 | 3 | 0 |
| TabsOBP51 | Tabs018547.1 | chr16 | 5,453,277 | 5,462,809 | 1464 | 4 | 0 |
| TabsOBP52 | Tabs002572.1 | chr16 | 10,456,814 | 10,458,608 | 477 | 3 | 0 |
| TabsOBP53 | Tabs012705.1 | chr16 | 10,473,039 | 10,473,978 | 375 | 2 | 0 |
| TabsOBP54 | Tabs011469.1 | chr16 | 10,482,872 | 10,483,856 | 381 | 2 | 0 |
| TabsOBP55 | Tabs006705.1 | chr16 | 10,487,715 | 10,489,717 | 387 | 3 | 0 |
| TabsOBP56 | Tabs011723.1 | chr16 | 10,494,905 | 10,496,370 | 393 | 3 | 0 |
| TabsOBP57 | Tabs017738.1 | chr16 | 10,504,939 | 10,506,577 | 465 | 4 | 1 |
| TabsOBP58 | Tabs004569.1 | chr16 | 10,517,393 | 10,525,941 | 390 | 2 | 0 |
| TabsOBP59 | Tabs016148.1 | chr16 | 10,526,599 | 10,527,497 | 369 | 2 | 0 |
| TabsOBP60 | Tabs002679.1 | chr16 | 10,530,590 | 10,531,705 | 360 | 2 | 0 |
| TabsOBP61 | Tabs016850.1 | chr16 | 10,533,210 | 10,534,500 | 384 | 2 | 0 |
| TabsOBP62 | Tabs006415.1 | chr16 | 10,547,657 | 10,549,207 | 396 | 4 | 0 |
| TabsOBP63 | Tabs000522.1 | chr16 | 10,554,299 | 10,556,471 | 261 | 3 | 0 |
| TabsOBP64 | Tabs011176.1 | chr16 | 10,560,330 | 10,561,033 | 366 | 2 | 0 |
| TabsOBP65 | Tabs021726.1 | chr16 | 10,562,757 | 10,563,449 | 363 | 2 | 0 |
| TabsOBP66 | Tabs016239.1 | chr16 | 10,569,165 | 10,570,123 | 363 | 2 | 0 |
| TabsOBP67 | Tabs008941.1 | chr16 | 10,571,905 | 10,572,952 | 378 | 2 | 0 |
| TabsOBP68 | Tabs020095.1 | chr16 | 10,654,204 | 10,658,840 | 753 | 6 | 0 |
| TabsOBP69 | Tabs012728.1 | chr16 | 11,298,069 | 11,299,744 | 378 | 2 | 0 |
| TabsOBP70 | Tabs003652.1 | chr16 | 11,407,380 | 11,408,982 | 378 | 2 | 0 |
| TabsOBP71 | Tabs009636.1 | chr17 | 4,447,737 | 4,453,664 | 441 | 5 | 0 |
| TabsOBP72 | Tabs013199.1 | chr17 | 4,461,601 | 4,476,794 | 906 | 9 | 0 |
| TabsOBP73 | Tabs009535.1 | chr17 | 4,479,265 | 4,484,324 | 273 | 4 | 0 |
| TabsOBP74 | Tabs001054.1 | chr17 | 4,487,417 | 4,489,144 | 330 | 4 | 0 |
| TabsOBP75 | Tabs013797.1 | chr17 | 4,498,725 | 4,501,477 | 366 | 4 | 0 |
| TabsOBP76 | Tabs006184.1 | chr17 | 4,504,823 | 4,507,643 | 366 | 4 | 0 |
| TabsOBP77 | Tabs003612.1 | chr17 | 4,522,428 | 4,526,310 | 453 | 5 | 0 |
| TabsOBP78 | Tabs003709.1 | chr17 | 4,527,188 | 4,531,697 | 435 | 6 | 0 |
| TabsOBP79 | Tabs021931.1 | chr17 | 4,535,639 | 4,539,656 | 480 | 5 | 0 |
| TabsOBP80 | Tabs014350.1 | chr17 | 4,540,637 | 4,544,986 | 462 | 5 | 1 |
| TabsOBP81 | Tabs018171.1 | chr17 | 4,548,567 | 4,556,919 | 450 | 5 | 0 |
| TabsOBP82 | Tabs016909.1 | chr17 | 4,557,328 | 4,563,313 | 435 | 5 | 0 |
| TabsOBP83 | Tabs011627.1 | chr17 | 4,566,965 | 4,576,767 | 432 | 5 | 0 |
| TabsOBP84 | Tabs005154.1 | chr17 | 4,930,978 | 4,938,967 | 699 | 5 | 1 |
| TabsOBP85 | Tabs000196.1 | chr18 | 10,446,352 | 10,450,296 | 420 | 6 | 0 |
| TabsOBP86 | Tabs016953.1 | chr20 | 2,752,027 | 2,757,025 | 453 | 4 | 0 |
| TabsOBP87 | Tabs019360.1 | chr20 | 3,696,624 | 3,697,265 | 642 | 1 | 1 |
| TabsOBP88 | Tabs003259.1 | chr20 | 5,312,448 | 5,312,897 | 450 | 1 | 0 |
| TabsOBP89 | Tabs015434.1 | chr20 | 5,314,670 | 5,315,573 | 342 | 2 | 0 |
| TabsOBP90 | Tabs007749.1 | chr20 | 5,328,286 | 5,336,568 | 582 | 5 | 0 |
| TabsOBP91 | Tabs014543.1 | chr20 | 8,925,425 | 8,931,904 | 660 | 3 | 0 |
| TabsOBP92 | Tabs014416.1 | chr21 | 1,643,734 | 1,656,139 | 747 | 6 | 0 |
| TabsOBP93 | Tabs006243.1 | chr21 | 12,411,678 | 12,418,079 | 657 | 5 | 0 |
| TabsOBP94 | Tabs003544.1 | chr21 | 12,420,711 | 12,425,861 | 588 | 4 | 0 |
| TabsOBP95 | Tabs015492.1 | chr21 | 12,628,571 | 12,631,367 | 459 | 4 | 0 |
| TabsOBP96 | Tabs017882.1 | chr21 | 14,305,708 | 14,309,043 | 1083 | 3 | 0 |
| TabsOBP97 | Tabs017358.1 | chr29 | 7,179,442 | 7,199,087 | 633 | 6 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, R.; Li, D.; Peng, C.; Wang, S.; Chen, Y.; Gui, F.; Sun, Z. Genome-Wide Identification of the Genes of the Odorant-Binding Protein Family Reveal Their Role in the Olfactory Response of the Tomato Leaf Miner (Tuta absoluta) to a Repellent Plant. Agronomy 2024, 14, 231. https://doi.org/10.3390/agronomy14010231
Ma R, Li D, Peng C, Wang S, Chen Y, Gui F, Sun Z. Genome-Wide Identification of the Genes of the Odorant-Binding Protein Family Reveal Their Role in the Olfactory Response of the Tomato Leaf Miner (Tuta absoluta) to a Repellent Plant. Agronomy. 2024; 14(1):231. https://doi.org/10.3390/agronomy14010231
Chicago/Turabian StyleMa, Ruixin, Donggui Li, Chen Peng, Shuangyan Wang, Yaping Chen, Furong Gui, and Zhongxiang Sun. 2024. "Genome-Wide Identification of the Genes of the Odorant-Binding Protein Family Reveal Their Role in the Olfactory Response of the Tomato Leaf Miner (Tuta absoluta) to a Repellent Plant" Agronomy 14, no. 1: 231. https://doi.org/10.3390/agronomy14010231
APA StyleMa, R., Li, D., Peng, C., Wang, S., Chen, Y., Gui, F., & Sun, Z. (2024). Genome-Wide Identification of the Genes of the Odorant-Binding Protein Family Reveal Their Role in the Olfactory Response of the Tomato Leaf Miner (Tuta absoluta) to a Repellent Plant. Agronomy, 14(1), 231. https://doi.org/10.3390/agronomy14010231






